Abstract
Living cells have evolved a broad array of complex signaling responses, which allows them to survive diverse environmental challenges and to execute specific physiological functions. Our increasingly sophisticated understanding of the molecular mechanisms of cell signaling networks in eukaryotes has revealed a remarkably modular organization, and synthetic biologists are exploring how this can be exploited to engineer cells with novel signaling behaviors. This approach is beginning to reveal the logic of how cells might evolve innovative new functions, and moves us towards the exciting possibility of engineering custom cells with precise sensing–response functions that could be useful in medicine and biotechnology.
Keywords: cell signaling, engineering, synthetic biology, therapeutic, MAP kinase, scaffold, modules, protein interactions, N-WASP, receptors, GPCR, Notch, RTK, Cancer, adoptive immunotherapy, Chimeric Antigen Receptors, T-cells, optical control, bioproduction
Living cells are highly dynamic systems that use complex molecular signaling circuits to monitor external and internal states, and to execute the appropriate physiological responses. Like any sensory machine, evolved or man-made, these cellular signaling circuits contain decision making subsystems that act as sensors and processors (such as receptors and their downstream effectors) that ultimately control various response subsystems (such as gene transcription and cytoskeletal dynamics) (Figure 1A). A major goal of modern cell biology is to understand how these molecular signaling systems achieve their complex responses, which are optimally tuned for their physiological role. While the vast majority of research is aimed at dissecting, mapping and analyzing cell signaling networks, our increasing understanding of how these systems work has led to the emergence of a radical new approach – efforts to design and build custom synthetic signaling circuits [1,2].
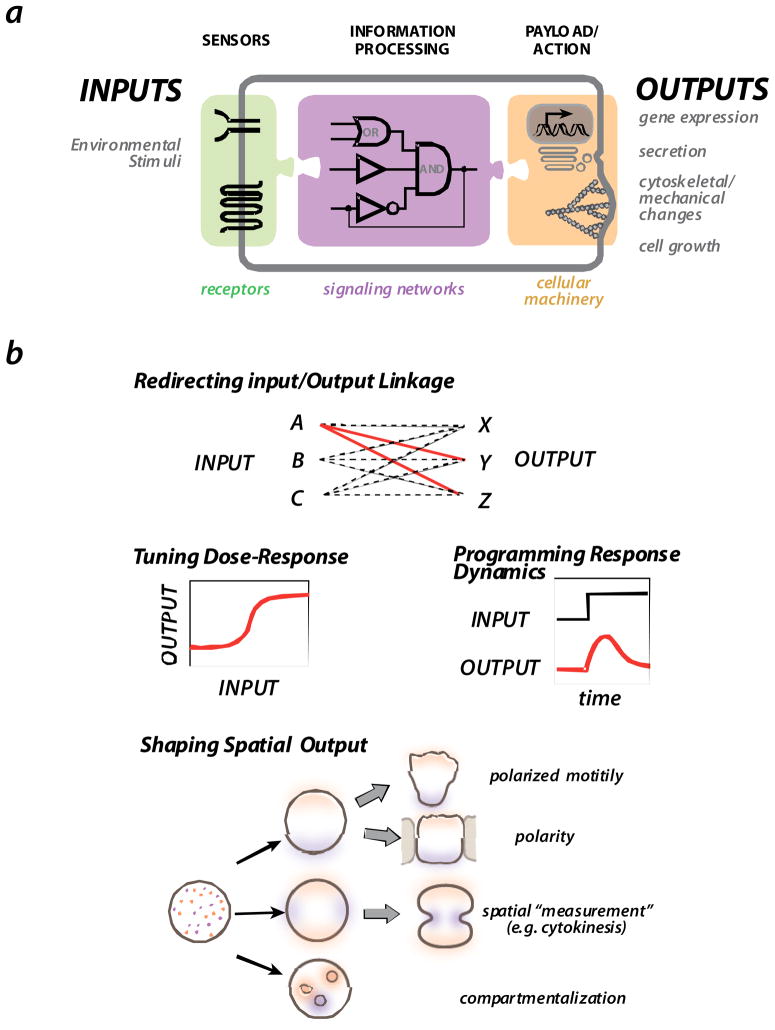
FIGURE 1. The general organization and behaviors of cell signaling circuits.
a| Cells generally sense environmental stimuli via receptors and other sensors. This information is then processed by intracellular signaling networks, which in turn engage various cellular outputs, including gene expression, secretion, cytoskeletal changes, and cell growth.
b| Some of the major challenges in the evolution or engineering of novel signaling circuits are: achieving the correct linkage of specific inputs and specific outputs; tuning quantitative behaviors of the signaling response - dose-response and dynamics -- so that they are optimal for the physiological function; and generating robust spatially self-organizing processes, such as those associated with cell polarization, directed motility, cell division, and cell compartmentalization.
Here we focus on the synthetic biology of signaling and look at how the signaling circuitry of eukaryotic cells can be engineered to construct cells with designed signaling behaviors. Eukaryotic cells use signaling protein networks to sense their environment and mediate rapid responses. As signal processing networks in cells function in a three dimensional setting, they also control complex spatial or morphological cellular responses. We will look at how signaling circuits with precise response behaviors can be generated by considering how the specificity of a response is determined (that is, what sets of outputs are linked to a specific input), how the precisely tuned dose-response or temporal dynamic profiles of responses are optimized for particular physiological functions, and how complex spatial and morphological control can be achieved (Fig. 1B). We will also consider why efforts to design and build custom synthetic signaling circuits have emerged, how they might provide a deeper perspective on the design principles and mechanisms of molecular signaling systems and how customized response behaviors could be applied in medicine and biotechnology. Finally we consider how future tools and methods could be developed to make the engineering of cellular behaviors easier.
Why Engineer Cell Signaling?
Before considering specific examples of engineered signaling pathways, it is useful to discuss the motivations for engineering cell signaling. Attempting to create new signaling behaviors in cells can seem like an audacious and foolish goal, given that we do not yet have a complete or reliably predictive understanding of the cells natural signaling circuits. However, the engineering of cell signaling is not simply a process for applying an already well-developed understanding, but it offers an approach for ‘understanding by building’. While biology has traditionally been a science of analysis and deconstruction to identify genes and molecules that are important for a particular process, synthetic biology offers an inverse approach, focusing on how individual molecular parts can be assembled into systems that carry out complex behaviors. As we currently have fully sequenced genomes and a vast amount of proteomic data we do not lack a complete list of molecular parts, but rather an understanding of how these parts fit together in a functionally coherent way. Engineering new cell signaling networks offers an approach for us to test and expand our understanding of the organizational principles of complex molecular systems.
In this sense, the synthetic biology of signaling is not simply oriented towards achieving an application goal, such as building a cell with a target function, but it is also an exploratory science in which it is important to understand what designs ‘work’, and how they relate to designs that ‘don’t work’. If, for example, one has a natural signaling network that carries out a complex behavior of interest, traditional genetic deconstruction can be used to identify molecules and linkages that are necessary and important for function (Fig. 2A). However, synthetic approaches can then be used to systematically explore many types of changes - alternative network linkages, the tuning of linkage strength, the addition of new linkages - to test which networks are compatible with this behavior of interest. By dissecting the natural network, or engineering a single successful circuit, one is unlikely to gain the deeper understanding of the functional landscape that a more complete and systematic synthetic circuit exploration can yield (Fig. 2B) [3–5]. In this sense, attempting to engineer cellular behaviors is akin to the early history of synthetic organic chemistry, where synthesis of new or modified molecules provided a complimentary approach to chemical analysis in the development of the fundamental theories of chemical bonding, structure, and reactivity [6]
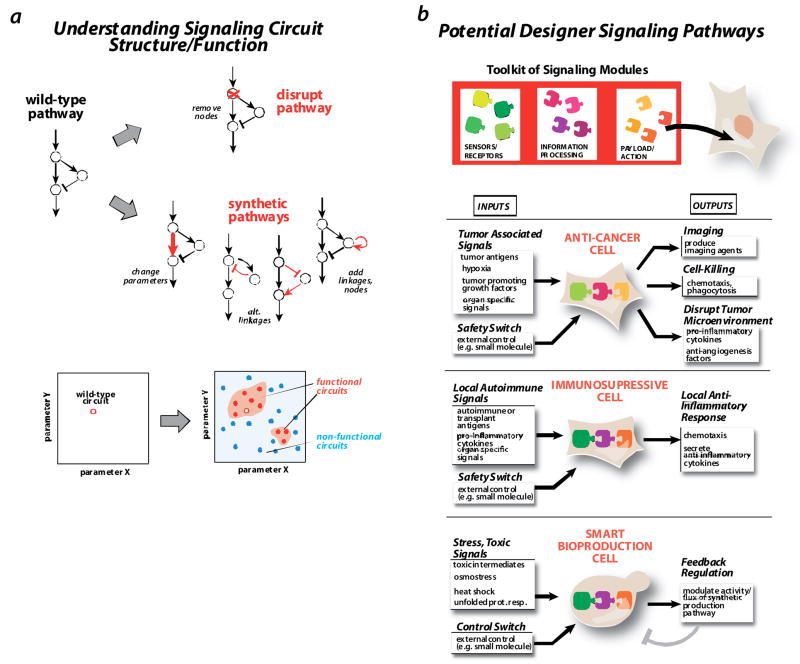
FIGURE 2. Why rewire cell signaling circuits?
a| understanding design principles. Traditionally, methods like gene disruption are used to dissect a signaling network. Synthetic approaches offer complementary information by creating alternative versions of a network that differ both in the network connectivity or in the strength of links. By mapping the space of functional (red circles) vs. nonfunctional (blue circles) variants, one gains a deeper understanding of functional requirements.
b | constructing designer signaling pathways for therapeutic or biotechnology applications. We hope to assemble a toolkit of signaling modules that can be used to create cells with designed signaling responses. An anti-cancer cell might detect a combination of tumor signals, and yield responses such as production of imaging reagents, cell killing, or secretion of factors that disrupt the tumor microenvironment. Such a cell might also have safety switches that could disable the cell if needed. An immunosuppressive cell might detect a combination of auto-immune response or transplant rejection signals, and trigger localized countermeasures, such as secretion of anti-inflammatory cytokines. A smart bioproduction (fermentation) cell would be engineered to precisely modulate the flux in growth versus production pathways in response to the stress state of the cell, thus optimizing overall yield.
Exploring the plasticity of signaling pathways, and how their functions can be tuned, is also relevant to the pathology and treatment of disease. Many cancers harbor oncogenic mutations that effectively ‘rewire’ the cell signaling networks that control the balance between cell growth, differentiation and death [7]Similarly, many intracellular pathogens, including bacteria and viruses, produce specific proteins that ‘rewire’ endogenous signaling pathways [8–10]. Many bacterial pathogen proteins that interface with host cellular signaling kinase and actin regulatory pathways, often to suppress the host immune response or to enhance infection (see Supplemental Box1). Thus, by using synthetic biology to understand the plasticity of pathways, and how their behavior is changed by network perturbations, we can gain a better framework for understanding of the strategies that pathogen’s adopt to exploit the inherent fragilities of signaling networks. Moreover, we can develop strategies for shifting a diseased network back to a stable, non-pathological behavior. The most stable network-based therapies may not involve simply blocking the primary oncogenic protein with a drug, but reshaping the network so that it lies in a new and stable region of behavior space.
Applications of engineered signaling in therapy and biotechnology
Another motivation for engineering cell signaling behaviors is the potential to construct cells programmed to execute precisely designed applications (Fig. 2B). Imagine if we could mimic and exceed evolution by using a toolkit of molecular parts to genetically engineer cells that carry out custom designed responses. As stem cell biology matures [11–12], and techniques such as adoptive immunotherapy develop [13–14],, the possibility of using cell-based therapeutics gets closer, but this will require sophisticated cellular engineering to precisely control cell behavior. For example, without novel control, how could proper stem cell migration and differentiation be directed for regenerative medicine, given the absence normal developmental signals? Moreover, as more industrial production processes engage biological organisms (such as biofuel or materials production) [15], it might be possibility to engineer smarter production strains that, like macroscopic production facilities, have cellular control systems that monitor external and internal states to optimize production. This may be particularly critical as we ask fermentation organisms, such as yeast, to produce a wide range of materials that may have toxic effects.
Designed anti-cancer cells
If we focus on designing custom therapeutic cells, which can sense disease signals and execute highly targeted and precisely calibrated therapeutic programs, what behaviors would we want? Immune cells such as T-lymphocytes or Natural Killer cells could be modified to identify and kill tumor cells. Such cells can already be removed from patients, genetically modified, expanded ex vivo, and adoptively transferred back to the patient [16–17]. An anti-cancer cell could be designed to detect a combination of tumor associated signals, including specific tumor antigens, hypoxia, organ specific antigens, as well as specific growth factors and cytokines that are secreted by tumors to evade normal immune responses and to create a tumor promoting microenviroment [18],. Engineering cells that recognize these factors, but are linked to an anti-tumor response, would be ideal. It is also critical to engineer external control (for example, small molecule) or safety switches into these therapeutic cells, so that their behavior can be shut off or attenuated in response to undesirable side effects, or to titrate the magnitude of their response.
Designed cells that detect these tumor specific inputs could be engineered to yield a number of different responses, such as the production of imaging agents that aid in identifying tumors and metastases, and the control of endogneous immune cell responses, such as chemotaxis, phagocytosis and cell killing. Perhaps most importantly, these therapeutic cells might be programmed to secrete factors that disrupt the local tumor microenvironment, such as pro-inflammatory cytokines and anti-angiogenesis factors, making it untenable for sustained tumor growth. This would be equivalent to creating a custom immune cell that disables the tumor cells and the microenviroment at multiple levels.
Targeted immunosupression
An immune cell could also be designed to block autoimmune disease or the rejection of transplanted organs. Normal immunosupressive drug therapy has broad and serious systemic effects. An engineered cell could be programmed to react in a local immunosupressive manner, perhaps in response to specific autoimmune or transplant antigens in combination with the cytokine signatures of a strong autoimmune response. Such cells might be programmed to chemotax to the sites of these signals, and respond by secreting anti-inflammatory cytokines that would disable the inflammatory positive feedback loops that would normally lead to a full-blown autoimmune or rejection response.
Although custom designed therapeutic cells lie in the future, it is useful to think about which detection and response behaviors would be valuable, as they provide useful target milestones in the development of tools and strategies for cellular rewiring.
Are Cell Signaling Networks Engineerable?
There is broad disagreement as to whether cells are actually engineerable. Are cell signaling systems so finely optimized that our intervention will lead to catastrophic malfunctions, or so robustly designed by evolution that the addition of new genes and network links will not be able to significantly alter function? Clearly, evolution has been able to rewire cell signaling pathways to yield diverse responses -- at some level they are relatively plastic and evolvable. Thus prior to trying to engineer new cellular behaviors, it may be instructive to consider how evolution can achieve innovative new functions.
A hallmark of signaling proteins, which is thought to play a major role in evolution, is their modular structure. They are almost always composed of multiple modular domains, some of which have catalytic function and many of which have specific regulatory or interaction functions [19, 20]. Throughout different signaling proteins, these modular domains are found in highly varied combinations. This has led to the model that diversity in signaling function could evolve via recombination of this toolkit of domains. Thus in principle, if we could understand how evolution works with these modules, we might be able to exploit the same toolkit to find regions of behavior space that evolution has, to our knowledge, not yet explored.
Why are signaling proteins and systems so modular? Most agree that, on an evolutionary timescale, organisms are under fitness pressure to develop innovative cellular signaling responses that might lead to advantages in changing environments and against competing organisms. Under this kind of changing fitness pressure, modular systems might spontaneously evolve as a way to facilitate the more rapid diversification of function [21]. Alon and co-workers have simulated biological network evolution using evolutionary algorithms to search for simple computational networks that solve a target goal [22]. When they repeatedly switch the target goal, the resultant networks spontaneously develop more modular solutions -- networks that have within them funtional subnetworks. These pre-formed subnetworks -- the modules -- can be rapidly reconnected in novel ways to shift from one target function to another. In essence, modules appear to provide a way to rapidly move from one function space to another, while jumping over vast regions of non-functional network space. Thus, the modular organization of signaling proteins and networks may reflect the pressure on these systems to generate behaviors that fit the needs of a constantly changing environment.
The importance of modularity in facilitating the evolution of new functions fits with concepts in evolution and development in which it is argued that much of the diversification of function and morphology of organisms evolves via the alternative regulation of existing components, rather than on the invention of radically new components [23]. While many of these ideas have developed focusing primarily on the regulation of genes by diverse cis-acting modules, they could also apply to the regulation of key catalytic signaling modules by diverse localization and regulatory modules [24,25]. Not surprisingly, many of the efforts to engineer new signaling behaviors, outlined below, exploit strategies of recombining modular functional units in novel ways, thus, in effect, harnessing an evolutionary strategy to engineer new function.
Engineering New Sensor Systems
One of the most critical tools for rewiring cellular behavior will be the ability to engineer novel sensors and receptors for targeted inputs. However, this is perhaps the least characterized element in engineering cell signaling, because the universe of possible inputs is so vast and it often involves the challenge of working with relatively complex membrane-associated membrane proteins. We describe, below, recent progress in modifying or constructing diverse receptor molecules.
Redirecting the output of natural receptors
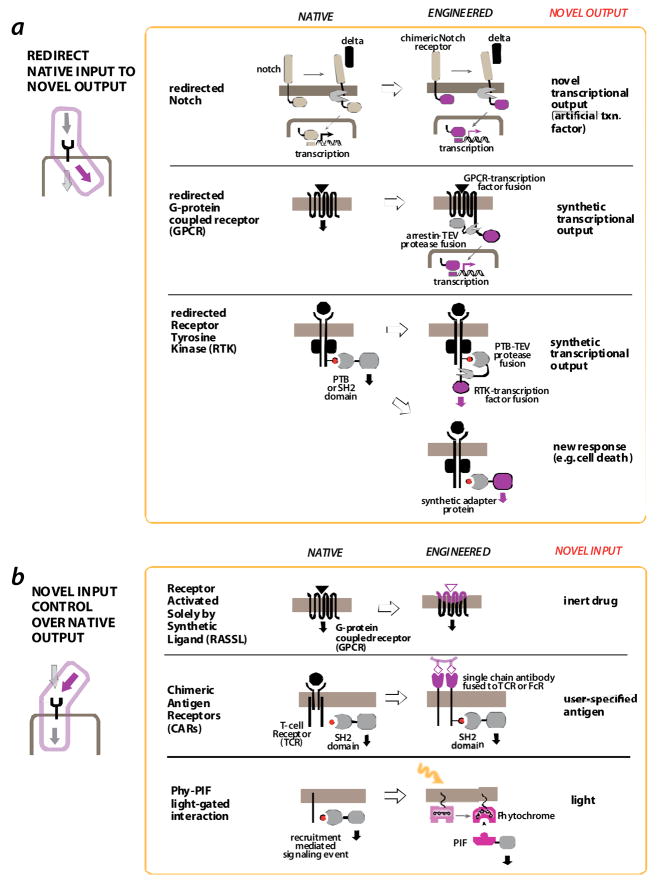
Natural receptors, which detect specific endogenous inputs, can be engineered to generate a non-native output response. There are several examples of a native receptor being redirected to elicit a novel transcriptional response. One such approach exploits the modular structure of the receptor protein Notch. Notch is a transmembrane receptor that detects the Delta protein presented on neighboring cells -- a critical cell-cell communication channel in development and differentiation. When Delta binds Notch the Notch transmembrane region is cleaved by a membrane protease, releasing the Notch C-terminal domain into the cytoplasm. This domain can enter the nucleus and activate gene transcription. Struhl et al, showed that this notch receptor transcription factor module can be replaced by a synthetic transcription factor (Gal4-AD) so that, when activated in vivo, this chimeric notch receptor can activate genes targeted by the new transcription factor [26,27]. While this construct was used as a reporter for Notch activation, it could easily be used to link detection of the native delta ligand to a completely novel set of non-native target genes.
Barnea et al, have expanded on this Notch-inspired modular strategy, by engineering novel transcriptional outputs for other receptors that normally do not use this type of protease activation mechanism [28]. When G-protein coupled receptors (GPCRs) are activated by their specific ligands they often recruit β-arrestin which is involved in downregulating GPCR signalling. Barnea et al, fused arrestin to a highly specific protease from the tobacco etch virus (TEV), so that it was co-recruited to activated GPCRs. A synthetic transcription factor was also fused to the GPCR cytoplasmic tail, linked by a TEV cleavage site. Thus, when the engineered GPCR fusion protein is activated by its endogenous ligand, it recruits the arrestin-TEV protease partner, which cleaves and releases the transcription factor domain from the GPCR, whereby it can enter the nuclease and activate target genes. This system has been used successfully to link new transcriptional reporters to the activation of a wide range of specific GPCRs. The response is highly specific, owing to the specificity of TEV cleavage. In principle, this strategy could be used to link any endogenous GPCR mediated signal to the expression of desired target genes.
Barnea et al also used this strategy to link endogenous receptor tyrosine kinase (RTK) signaling to novel transcriptional outputs [28]. Most RTKs, when stimulated, activate their kinase domains, which mediate autophosphorylation on cytoplasmic tyrosines to recruit SH2 domain containing proteins. Here, the TEV protease was fused to recruited SH2 domains, and a synthetic transcription factor was fused to the cytoplasmic tail of the RTK through the TEV protease cleavage site. So, RTK activation leads to the recruitment of the SH2-domain–TEV fusion, the release the receptor-associated transcription factor and engineered gene transcription. It is remarkable that this simple modular strategy can be applied to several receptor classes, as long as they recruit a specific partner protein upon activation.
Howard et al, harnessed the modularity of RTK signaling to redirect an oncogenic growth signal to an apoptotic response [29]. They engineered a novel SH2 adapter protein in which an SH2 domain that recognized an activated RTK was fused to a death effector domain from Fadd. Thus, activation of the RTK led to membrane recruitment of the death domain, which induced a cell death response. The possibility of linking other novel outputs to these key recruitment events has not been well explored.
Receptors that detect novel small molecule inputs
The above strategies focus on ways to take receptors that detect endogenous signaling molecules and engineer them to elicit novel responses. However, in many cases, cellular engineering may require receptors that detect novel signals for which there are no endogenous receptors. These novel signals include small molecules that we may want to ensure external control of an engineered system.
Relatively good success has been achieved in using GPCR’s as a platform for engineering small molecular controlled receptors. Certain GPCR’s, such as opioid receptors, can be activated by their endogenous ligands and specific small molecular agonists. Conklin, Roth and co-workers have engineered molecules known as receptors activated solely by synthetic ligands (RASSLs) [30,32]. These receptors are mutated so that they cannot bind their endogenous ligand, but are activated by, and elicit their endogenous downstream effect in response to, a small, pharmacologically inert, molecule agonist.
GPCR’s differ in their outputs, in part because individual receptors communicate with specific heterotrimeric G-proteins. Further engineering has yielded versions of RASSLs that are specifically coupled to each of these distinct downstream pathways, thus allowing small molecule control of a highly diverse set of outputs. These RASSLs have been successfully deployed in transgenic mice - essentially rewiring signaling in a full living organism -- mostly as a diagnostic and analytical tool. The applications have been diverse, given the broad usage of GPCRs throughout different tissues. For example, mice bearing taste neurons expressing RASSLs showed specific sweet (attractive) or bitter (aversive) responses to water mixed with the agonist (spiradoline), depending on which type of neuron they were expressed in [33]. In addition, expression of RASSLs in heart cells allowed for control of heart rate by administration of spiradoline [34]. That these receptors work so robustly in vivo, hints at their potential utility in more complex cellular engineering.
Chemical dimerizers form another strategy for achieving small molecule control over signaling. Such strategies have been reviewed elsewhere [35,36], and will not be discussed here.
Receptors that detect user specified antigens
It would be ideal to engineer receptors that can sense disease-associated antigens, such as a protein expressed strongly in a tumor or infectious agent. If receptors could be engineered to achieve the same diversity and selectivity of recognition as antibodies, a wide range of inputs could be detected and linked to specific responses. Chimeric antigen receptors (CARs) -- receptors designed with single-chain antibodies (scFv’s) as part of their detection mechanism – have been developed as just this type of multi-purpose framework. This strategy stems from the modularity of immune cell receptors, such as the T-cell receptor. Although the T-cell receptor is a complex multiprotein complex, crosslinking of the cytoplasmic region of the CD3 zeta chain subunit is sufficient to induce T-cell signaling [37]. The CD3 zeta chain contains motifs that are phosphorylated upon activation, by tyrosine kinases such as Lck, to induce recruitment of SH2 domain containing proteins such as the ZAP-70 kinase. Fusion of the cytoplasmic region of the CD3 zeta chain to an extracellular single chain antibody (scFv) yields a receptor often referred to as a “T-body”, which, when expressed in T-cells, leads to the targeted killing of cells expressing the recognized antigen (presumably the surface antigens crosslink and activate the chimeric receptors) [38,39]. Fusion of scFv’s to the intracellular region of the Fc receptor (gamma chain) can yield a similar type of chimeric antigen responsive receptor. These studies highlight the modularity of these receptors: linkage of a novel extracellular recognition element to downstream intracellular signaling elements leads to a novel input/output sensor.
These first generation CARs are relatively primitive and have met with mixed results. T-cells expressing CARs directed towards tumor antigens have moderate signaling capability compared to endogenous TCR responses, proliferate moderately ex vivo and in vivo and have poor survival upon repeated antigen exposure [16,17]]. Improvements in these behaviors have been made by incorporating additional modular domains in the intracellular regions of the CARs, including domains from co-receptor molecules that are part of normal TCR activation, thus perhaps mimicking a more complete activated intracellular assembly [40,41]. Cells g these next-generation CARs more effectively control xenograft tumors in mice, and are now being ported to clinical trials [16]. More sophisticated engineering of CARs may lead to even further improvement in therapeutic function.
Sensors that detect physical signals such as light
Another fascinating area of exploration is the development of genetically encoded sensors that can detect light and transduce this to a specific biological response, an area referred to as optogenetics. Naturally occurring photosensitive proteins from plants, algae and bacteria can be modified for use in higher organisms, including mammals. These tools are extremely useful as spatiotemporal dials to control and analyze complex cellular and organismal behavior, especially when they are expressed from cell-type specific promoters. In the long-term, optogenetic tools could be used to remotely control cells used for therapeutic applications, although there are major technical challenges, such as how light can be delivered within an organism, that would have to be surmounted. The most commonly utilized optogenetic tools today are the microbial channelrhodopsin and halorhodopsin proteins, which have been used extensively to control neuronal function. These are reviewed elsewhere [42] and will not be discussed in detail here.
More recently, additional optogenetic tools have emerged that can be applied to a broader range of cell signaling systems. Airan et al constructed a set of light activated GPCRs that can communicate with both downstream Gs and Gq heterotrimeric G-proteins [43]. Chimeras of the light sensitive visual system GPCR, rhodopsin (bovine), were made that contain intracellular loops from both Gq and Gs-coupled adrenergic receptors. The endogenous retinal molecule is the light sensitive chromophore. These new tools significantly expand the signaling “vocabulary” that can be controlled by light, given the importance of Gq and Gs signalling pathways in diverse cell types.
An even more generalized strategy for light control involves the use of light-controlled protein interactions. The transient interaction of specific partner proteins is the basis of many intracellular signaling events (see below), and receptors can be bypassed so that light directly controls such intracellular interactions. Levskaya et al used the plant derived Phytochrome interaction system — the binding of this photoreceptor to its partner PIF domain can be toggled on and off by specific wavelengths of light — to recruit specific proteins to the membrane in a precise spatialtemporal manner [44]. In the case of guanine nucleotide exchange factors (GEFs), which control Rho-family GTPases, this can be used to trigger GTPase activation and downstream cytoskeletal changes, leading to light-guided cell protrusion. Although this technique is powerful and potentially applicable to many signaling interactions, the Phy-PIF system requires addition of a cell permeable chromophore that is not endogenous to mammalian cells. Wu et al used a photosensitive LOV (light-oxygen-voltage) domain (found in plants, algae and bacteria) to conformationally occlude the Rac GTPase in a light controlled manner [45]. This flavin binding domain provides another potentially generic conformational light control element, which could be coupled to control diverse signaling proteins.
Engineering Signal Processing Systems
Ultimately cells decide what response programs to execute based on intracellular signaling networks that receive and process signals from sensor molecules (see above). Recent work in cellular engineering has focused on understanding how these networks function to make decisions, and how they can be rewired.
Modular logic of signal processing
Intracellular signaling proteins are highly modular (see above). Most modules fall into two classes (Fig. 4a). The first class are enzymatic domains, such as kinases and phosphatases, which catalyze the post-translational modifications or conformational changes by which information is stored. In most cases these catalytic domains come in pairs: “writer” enzymes (like kinases) make a modification and “eraser” enzymes (like phosphatases) remove the modification. The second class are regulatory or interaction domains that modulate the activity of catalytic domains, or target them to specific partners or sites in the cell. These modules can mediate specific protein-protein interactions (either constitutive interactions or those dependent on post-translational modifications like phosphorylation) or protein-membrane interactions. Thus, it is predominantly the regulatory and interaction domains that determine when and where the catalytic domains are activated, and to what partners they transmit information [5].
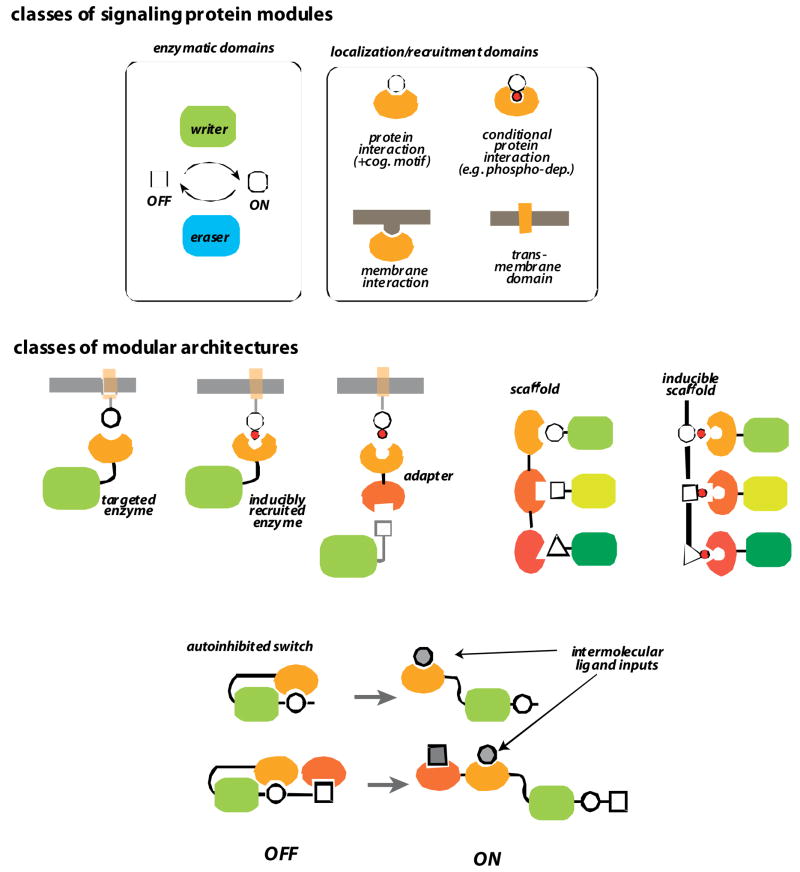
FIGURE 4. The modular logic of intracellular signaling components.
a | enzymatic and regulatory domains. Modular eukaryotic signaling proteins are generally composed of enzymatic domains and localization domains. Enzymatic domains, like kinases and phosphatases, and GEFs and GAPs, catalyze regulatory modifications such as phosphorylation and GTPase activation, respectively (enzymatic domains often come in “writer” and “eraser” pairs that have opposing activities). These enzymatic domains are regulated and targeted by interaction domains, including protein-protein interaction domains, membrane interaction domains, or transmembrane domains.
b | different classes of multidomain architectures. Enzymatic domains can be directly targeted to specific substrates, partners or subcellular locations by interaction domains. Alternatively they can be indirectly targeted via adapters or scaffold proteins, which contain multiple interaction domains. Interaction domains can also allosterically regulate catalytic domains by engaging in intramolecular autoinhibitory interactions. Such switch proteins can be activated by competing ligands that relieve autoinhibition.
These different classes of modules are found in diverse combinations and arrangements in signaling proteins (Fig. 4b). Catalytic domains fused to targeting domains can be recruited to specific complexes or membrane locations, where they will modify specific targets; often these catalytic domains have a high intrinsic Michaelis constant (Km’sand thus require targeting by accessory interaction domains for efficient catalysis. Sometimes these targeting interactions are regulated, if, for example, the interaction is dependent on a post-translational modification, such as the targeting of SH2 domain proteins to autophosphorylated pTyr sites on activated RTK’s.. Proteins with two interaction domains can act as adapters that translate one interaction into a second one, leading to increased response flexibility depending on the adapter proteins that are expressed in a particular cell type. Multiple interaction domain proteins can also function as scaffold proteins, which organize multiple proteins in a pathway into a complex. These interactions might be constitutive or preformed, or induced by factors such as phosphorylation, or conformational changes that expose interaction sites. Thus scaffold proteins can in principle determine the wiring linkages of signaling proteins, as well as control when or where signaling happens [24, 20].
A second major role for interaction and regulatory domains is to directly control the activity of catalytic domains. In many cases, the interaction domains participate in intramolecular autoinhibitory interactions that sterically occlude the catalytic domain or conformationally perturb it - a type of regulation referred to as modular allostery [46]. Binding of competing intermolecular ligands to the interaction domains induces the proteins catalytic activity. Often multiple interaction domains participate in the autoinhibition of a catalytic domain in a cooperative or hierarchical manner [47,48]. These proteins can function as complex multi-input switches that require a specific combination of inputs for proper activation. In addition, since external ligands activate these proteins, localization (driven by these interactions) can be directly coupled to activation.
Engineering new protein switches
Lim and co-workers have explored whether the modular allosteric logic of many natural eukaryotic signaling proteins can be exploited to design new signaling switches by domain recombination (Fig. 5). Indeed, the catalytic domains of the actin regulatory protein N-WASP and of Rho-family GEFs can be linked to novel autoinhibitory domains to yield proteins whose activity is gated by novel ligands [49,50]. The intramolecular linkage of either of these catalytic domains to a PDZ domain and a PDZ ligand peptide can yield a switch that is activated by competing PDZ peptide. Similarly, multiple interaction domains can be appended to yield a combinatorial switch that displays AND-gate control. Depending on the exact configuration of the domains and intramolecular interactions, the types of regulation can be different in response to different competing external ligands -- one ligand could activate the protein, while another represses it. These types of diverse relationships between regulatory domains is reminiscent of the diverse behaviors observed in natural signaling proteins, supporting the notion that this kind of switch architecture facilitates the evolution of diverse combinatorial regulatory switches [48]. Dueber et al, have also shown that synthetic autoinhibitory switches, utilizing multivalent interactions of the same type, leads to switches, the activation behavior of which can be tuned cooperativity from a linear to a digital-like response [51].
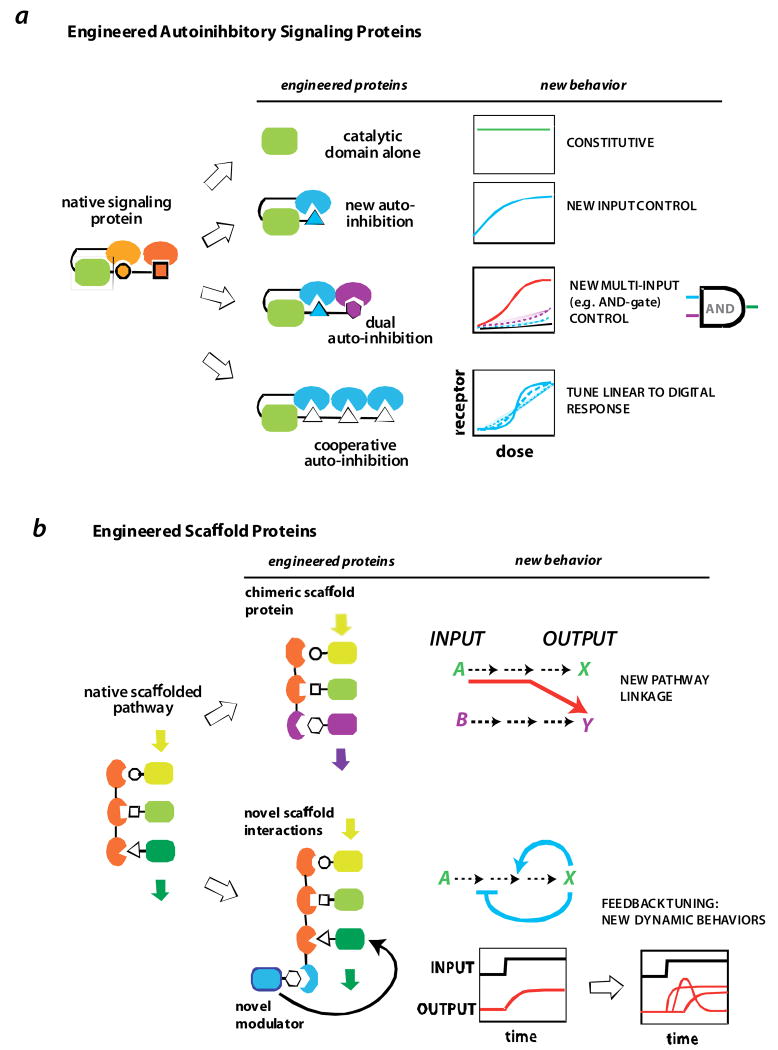
FIGURE 5. Engineering signal processing circuits.
a | engineered allosteric protein switches. Dueber et al [49,51] showed that the allosterically regulation of the signaling protein N-WASP could be reprogrammed by recombining the catalytic domain from N-WASP with different combinations of interaction domains. Novel behaviors included multi-input (AND-gate) control and highly cooperative switch-like activation.
b | using scaffold proteins as a molecular circuitboard for reshaping signaling output. The input/output linkage of a MAP kinase pathway in yeast could be redirected via an engineered chimeric scaffold that assembled a novel combination of kinases [58]. Novel interaction sites can also be appended on to scaffolds to recruit additional modulatory factors. These additional factors can be build synthetic feedback loops that can be used to generate pathways that display diverse signaling dynamics [62].
Scaffold proteins as molecular circuitboards
Intracellular signaling circuits can also be directly controlled by harnessing regulatory interactions to rewire pathway connections. For example, the catalytic domain of the Src family kinase, Hck, which is normally regulated by SH2 and SH3 domains, can be fused to a PDZ domain and directed in vivo to specifically phosphorylate substrates with a PDZ ligand motif [52].
Scaffold proteins can also be used to generate new pathway input/output relationships. In yeast, there are multiple functionally distinct mitogen activated protein (MAP) kinase pathways that regulate responses to mating pheromone and osmotic stress [53,54]. These pathways share common kinase components, but remain specific because each pathway is organized by a distinct scaffold protein [55–57]. A chimeric scaffold protein that organizes select members of the mating and osmotic stress pathways yields a non-natural pathway in which mating pheromone specifically induces the osmostress response program in vivo [58]. Similary covalent fusions that, like a scaffold, force the interaction between two signaling proteins, can be used to force signal transmission down a single pathway [59].
More recently scaffold proteins have been shown to not only mediate the linear input/output relationship of pathways, but also to coordinate the recruitment of modulatory factors that shape the dose dependence and dynamics of pathway response [60,61]. Inspired by these natural examples, Bashor et al showed that the yeast mating MAP kinase scaffold, the Ste5 protein, can be used as a molecular circuitboard to flexibly reshape the quantiative behavior of the mating response [62]. Fusing an additional synthetic interaction site to the Ste5 scaffold (using a leucine zipper heterodimer pair) facilitates the recruitment of novel modulatory factors such as a MAPK phosphatase, which suppresses the pathway response. However, if expression and recruitment of the phosphatase is linked to pathway output, a negative feedback loop is generated that leads to adaptation – a transient response followed by the automatic return to lower output levels, which is a key behavior in many biological sensory systems. By linking positive and negative pathway modulators in different ways, this small toolkit of scaffold control elements could be used to generate highly diverse dose response and dynamic behaviors, including highly cooperative switching, delayed responses, accelerated responses and pulse generation. These studies show how organizing centers such as scaffold are a rich platform for processing and shaping intracellular signaling, either through evolution or engineering.
Engineering spatial self-organization
One of the most poorly understood aspects of cell signaling, is how circuits made of diffusible molecules can lead to highly precise spatial organization in the cell, such as directed polarization and migration. This type of self-organization is an aspect of control circuitry where there are no good electronic or engineered counterparts, and where biology can instruct engineering.
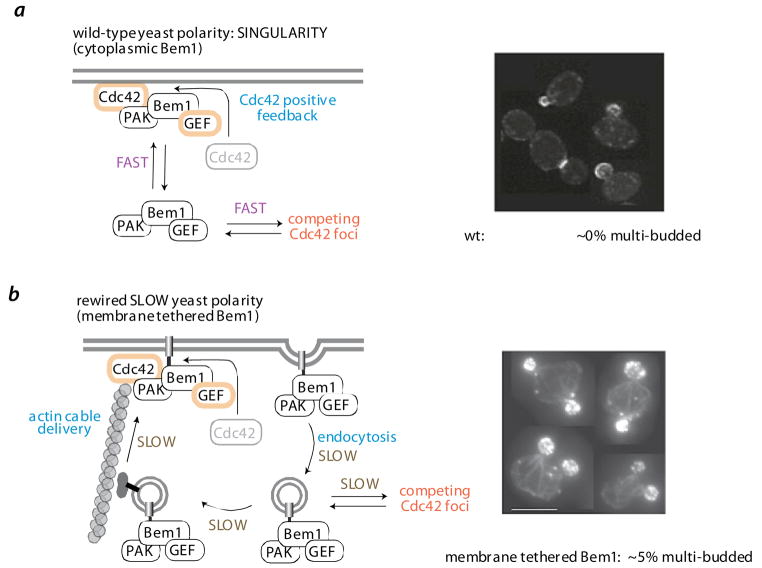
Engineering principles are being applied to understand the mechanism of polarization in the budding yeast, S. cerevisae. Polarization is controlled by the GTPase Cdc42, which ultimately localizes to one site on the mother cell, leading to the formation of a single bud that grows into the daughter cell [63]. Remarkably this process leads to the formation of a singular bud with nearly 100% reliability. A positive feedback circuit involving the Cdc42 GTPase is key in polarization: active Cdc42 at the membrane recruits the cytoplasmic GEF protein Bem1, which activates and localizes additional Cdc42 [64]. While this kind of feedback loop leads to the formation of foci of Cdc42, the rapid diffusion and redistribution of Bem1 between competing foci might be important to allow one foci to become dominant, leading to singularity of budding. The effect of slowing down Bem1’s diffusion and redistribution, by linking it to a transmembrane motif, has been analyzed [65]. Membrane tethered Bem1 could rescue the lethality of a Bem1 knockout, but could not undergo diffusion in the cytoplasm. Instead, it was delivered to the plasma membrane in vesicles via actin cables (also coordinated by Cdc42 foci), and away from membrane foci by endocytosis, and thus redistributed much more slowly. Severe defects in singularity, such as many persistent competing Cdc42 foci, were observed, and the frequency of multi-budded cells increased to ~5%. Studies such as these help reveal the requirements for precisely controlled spatial self-organization, and suggest that we can learn how to engineer signaling circuits that produce customized spatial outcomes with important therapeutic behaviors (such as regenerative medicine that requires specific cellular morphology and orientation).
Making engineering of signaling predictable
The studies above show that signaling systems are highly modular and plastic and recombining modules, particularly catalytic domains with novel regulatory domains, can lead to distinct response behaviors. Thus, the question is no longer whether signaling systems can be engineered to yield new behaviors, but whether they can be engineered in a way that allows us to predict what behaviors will emerge, and how successful each designed circuit will be.
Challenge of unanticipated crosstalk
One of the major issues with engineering cell signaling is that natural components -- the toolkit of available domains – are being reused, which can result in unanticipated crosstalk. Will the interactions that you engineer lead to specific phosphorylation of the desired protein, or will the domain used also cross-interact with other targets, competitively titrating out important physiological interactions and leading to unanticipated effects or failure of the designed circuit? Often natural parts do not have absolute specificity, and evolution most likely uses complex networks of cross-reactivity to yield important coordinated regulation. While this kind of complex neural net-like system may provide advantages for a cell, it is an anathema to predictive engineering.
Envisioning the signaling toolkit of the future
One solution to this problem is to assemble a toolkit of parts that are specifically optimized for engineering. This issue is important for any type of signaling part, but we will focus on how to assemble a useful toolkit of protein-interaction parts (Fig. 7a).
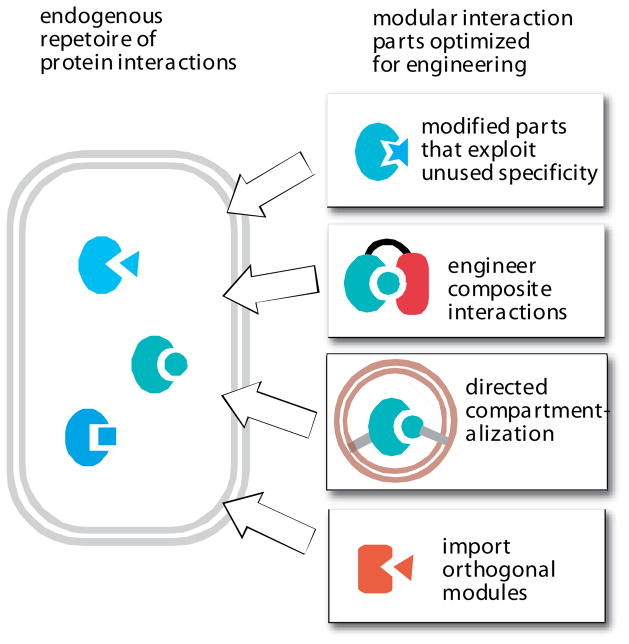
FIGURE 7. Improving the toolkit for predictable engineering of cell signaling: orthogonal interaction parts.
A native cell has its own native repetoire of protein interaction modules, and thus it is challenging to engineer new functions using related interaction modules that might show inadvertent and unintended crosstalk in the cell. An optimized toolkit of interaction parts could significantly increase the predictability of cellular engineering, by eliminating the chance for unintended crosstalk. Several strategies for optimization include engineering of interaction modules that exploit untapped specificity; engineering of composite, multi-domain interactions; combining interaction modules with novel subcellular targeting; and importing orthogonal interaction modules (either synthetically constructed or from other organisms) that are not found in the host cell.
Although nature has repeatedly used families of parts, such as interaction domains of a particular type, recent studies indicate that in some cases family members contain unused recognition sites within these domains. These could be exploited to engineer domain-peptide pairs that are simultaneously optimized to interact with their correct partner, while avoiding cross-interaction with other members of the family [66,67]. In fact PDZ domain-ligand pairs and heterodimerizing leucine zipper pairs have been constructed that are optimized to avoid cross reaction with natural domains of the same type [68,69]. The selectivity and predictability of existing interaction domains can also be improved by engineering composite interactions. Certainly, multi-domain cooperation is a natural mechanism for increased specificity. But a new twist on this is the engineering of composite two domain clamshell interactions. Koide et al have taken a PDZ domain and fused it to a fibronectin domain [70]. Using phage display they selected for variants of this tandem domain that bind a specific peptide so that it is sandwiched between the two domains. The dramatically enlarged recognition surface area leads to interactions with much higher specificity and affinity. Another solution for specificity, which is observed in nature, is differential compartmentalization. If targeting motifs could be used to localize partner proteins to specific organelles or cellular locations, then interaction motifs are likely to function in a more specific manner, especially if few or no competing interactions of this type take place at this location or organelle.
An alternative approach to achieving reliable specificity is to import domains from other organisms that do not exist in the host being engineered. For example, PDZ domains can be imported into yeast (which lack most such domains), although the possibility of fortuitous cross reacting partners cannot be ruled out [58]. An example of an orthogonal molecular system that has been successfully ported to a novel host is the bacterial Cre-Lox recombinase system, which is reliably used to engineer complex chromosomal rearrangements in complex organisms, including mice [71].
Thus, imagining the toolkit of the future, one might want a set of ten or so protein interaction pairs that are optimized for a particular organism of choice (for example, E.coli, S.cerevisae, Mammals) in that they are orthogonal, that is, known to not cross-react with the host proteome or proteins within the toolkit, except for their cognate ligand. It is also important for these interactions to be tunable, so a series of ligands for each interaction domain that vary in affinity, over several orders of magnitude, would be ideal. This would allow the systematic exploration of how recruitment affinity alters system behavior.
Combinatorial design vs. prediction
Another different, but still complementary, approach to predictably engineer cell signaling is to utilize combinatorial variability. In natural evolution, the recombination of signaling modules to generate new function was presumably not designed or guided, but rather was relatively random, and it was natural selection that identified rewiring events that led to fitness advantages. Thus, a very fruitful approach, given the lack of predictability in cellular engineering, might be to construct combinatorial libraries of synthetic circuits, and to select for the desired function [25,72].. Moreover, this approach could be combined with semi-predictive design, where the overall architectures of engineered circuits could be design, but combinatorial methods used to search a broader range of parameter space (using variants of each module in the library). Focusing on combinatorial selections may also provide a very useful strategy in the early days of the field of synthetic biology, as it may help us learn more rapidly about core design principles.
Outlook
The goal of understanding how cells communicate and make decisions remains very attractive, especially because understanding the molecular language within the cell may allow us to communicate with cells and instruct them to carry out new programmed functions. Our ability to rewire cell signaling could provide many powerful applications, such as therapeutic cells programmed to detect a selective set of disease related signaling and to locally respond in a precisely tailored way.
Although evolution has achieved this kind of innovation and precise engineering of cellular function, we are only beginning to understand how to execute this kind of goal. We have a good foundational understanding of the logic of cell signaling machinery and the sources of functional plasticity. In addition, major first steps have been made in engineering new receptor/sensor systems, as well as new or modified intracellular signal processing circuits. Despite these tools, very few efforts have been made, to date, to link these types of components in new ways to yield larger integrated circuits capable of highly refined, precision responses. Such efforts are underway. For example, the Cell Propulsion Lab is an NIH nanomedicine center http://nihroadmap.nih.gov/nanomedicine/devcenters/cellularcontrol.asp) that is attempting to take the relatively simple anti-tumor immune cells engineered with synthetic chimeric antigen receptors (CARs), and improve their signal processing and the suite of responses that are elicited by them to optimize the cells for ex vivo expansion, in vivo survival, anti-tumor cytotoxicity and the disruption of a hospitable tumor microenvironment. It will be exciting to see how these types of efforts unfold, and how the challenges will improve the sophistication and reliability of cellular engineering.
Supplementary Material
FIGURE 3. Engineering novel signaling sensors.
a | redirecting native inputs to novel outputs. The C-terminal domain of the notch receptor is transcription factor that is released by transmembrane proteolysis upon activation by the ligand, delta. Replacement with an alternative transcription factor domain yields a new gene expression response [26]. G-protein coupled receptor (GPCR) output can be redirected in a similar way by fusing a transcription factor domain via a tether with a TEV protease site. Activation of the GPCR results in recruitment of the protein arrestin. If an arrestin-TEV protease fusion is expressed in the cell, GPCR activation results in release of the transcription factor, and a novel gene expression output [28]. Thus GPCR activation can be arbitrarily linked to a novel transcriptional output. Receptor tyrosine kinase (RTK) output can be redirected by harnessing the recruitment of synthetic SH2 or PTB domain adapters to the activated and tyrosine phosphorylated receptor. The SH2 domain could be used to recruit a TEV protease (to again release an artificially tethered transcriptional domain) [28] or to recruit novel effector domains, such as those involved in cell death [29].
b | engineering novel input control over native responses. GPCRs have been engineered to be controlled by small molecule agonists by mutating their extracellular surface such that they no longer bind their endogenous ligands (Receptors activated solely by sythetic ligand – RASSL [32]). Receptors that activate T-cells in response to arbitrary inputs can be generated by fusing engineered single chain antibodies (scFv’s) to the intracellular region of the T-cell receptor (CD3 zeta chain) – referred to as chimeric antigen receptors (CARs [16, 17]). A recruitment mediated signaling event can be placed under light control by replacing the endogenous interaction with the light-gated interaction Phytochrome-PIF interaction pair from plants [44].
FIGURE 6. Engineering spatial regulation.
a | wild-type polarization circuit controls single bud formation. In budding yeast, localized activation of the polarity GTPase Cdc42 is amplified by a positive feedback loop - active Cdc42 recruits the scaffold protein Bem1, co-assembles the p21 activated kinase (PAK – Ste20) and the Cdc42 GEF (Cdc24). Although a cell may have multiple Cdc42 foci, these are quickly resolved into one dominant foci, which develops into the cells only bud. A fast rate of interchange of the diffusible Bem1/PAK/GEF complex between competing Cdc42 foci, is hypothesized to be critical for resolution into a single dominant foci.
b | a synthetic slow polarization circuit leads to multiple bud formation. To test this hypothesis, Bem1 was artificially tethered to the membrane via a fused membrane targeting motif [65]. Although this membrane tethered Bem1 can properly assemble the Bem1/PAK/GEF complex at sites of Cdc42 activity (i.e. the positive feedback loop), the exchange of the complex between competing Cdc42 foci is slow (dependent on vesicular transport via actin cables and endocytosis). This synthetic polarization circuit therefore leads to poor resolution of competing Cdc42 foci and a much higher frequency (5% vs ~ 0%) of multibudded cells (micrographs from [65]).
References
- 1.Pryciak PM. Designing new cellular signaling pathways. Chem Biol. 2009;16:249–54. doi: 10.1016/j.chembiol.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiel C, Yus E, Serrano L. Engineering signal transduction pathways. Cell. 2010;140(1):33–47. doi: 10.1016/j.cell.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 3.Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–8. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- 4.Mukherji S, van Oudenaarden A. Synthetic biology: understanding biological design from synthetic circuits. Nat Rev Genet. 2009;10:859–71. doi: 10.1038/nrg2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. Rewiring Cells: Synthetic Biology as a Tool to Interrogate the Organizational Principles of Living Systems. Annu Rev Biophys. 2010 Feb 16; doi: 10.1146/annurev.biophys.050708.133652. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh BJ, Lim WA. Synthetic biology: lessons from the history of synthetic organic chemistry. Nat Chem Biol. 2007;3:521–5. doi: 10.1038/nchembio0907-521. [DOI] [PubMed] [Google Scholar]
- 7.Weinberg R. The Biology of Cancer. Garland Science; 2006. [Google Scholar]
- 8.Galán JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–9. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Münter S, Way M, Frischknecht F. Signaling during pathogen infection. Sci STKE. 2006;2006:re5. doi: 10.1126/stke.3352006re5. [DOI] [PubMed] [Google Scholar]
- 10.Shan L, He P, Sheen J. Intercepting host MAPK signaling cascades by bacterial type III effectors. Cell Host Microbe. 2007;1:167–74. doi: 10.1016/j.chom.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Kiskinis E, Eggan K. Progress toward the clinical application of patient-specific pluripotent stem cells. J Clin Invest. 2010;120:51–9. doi: 10.1172/JCI40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindvall O, Kokaia Z. Stem cells in human neurodegenerative disorders — time for clinical translation? J Clin Invest. 2010;120:29–40. doi: 10.1172/JCI40543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.June CH. Adoptive T cell therapy for cancer in the clinic. J Clin Invest. 2007;117:1466–76. doi: 10.1172/JCI32446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varela-Rohena A, Carpenito C, Perez EE, Richardson M, Parry RV, Milone M, Scholler J, Hao X, Mexas A, Carroll RG, June CH, Riley JL. Genetic engineering of T cells for adoptive immunotherapy. Immunol Res. 2008;42:166–81. doi: 10.1007/s12026-008-8057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol. 2008;19:556–63. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Sadelain M, Brentjens R, Rivière I. The promise and potential pitfalls of chimeric antigen receptors. Curr Opin Immunol. 2009;21:215–23. doi: 10.1016/j.coi.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.June CH, Blazar BR, Riley JL. Engineering lymphocyte subsets: tools, trials and tribulations. Nat Rev Immunol. 2009;9:704–16. doi: 10.1038/nri2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–50. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 19.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–80. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 21.Gerhart J, Kirschner M. The theory of facilitated variation. Proc Natl Acad Sci U S A. 2007;104 (Suppl 1):8582–9. doi: 10.1073/pnas.0701035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashtan N, Alon U. Spontaneous evolution of modularity and network motifs. Proc Natl Acad Sci U S A. 2005;102:13773–8. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 24.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;326:1220–4. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peisajovich S, Garbarino J, Wei P, Lim WA. Rapid Diversification of Cell Signaling Phenotypes by Modular Domain Recombination. Science. doi: 10.1126/science.1182376. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–60. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 27.Sprinzak D, Lakhanpal A, LeBon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis Interactions between Notch and Delta Generate Mutually Exclusive Signaling States. Nature. doi: 10.1038/nature08959. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, Axel R, Lee KJ. The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci USA. 2008;105:64–9. doi: 10.1073/pnas.0710487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howard PL, Chia MC, Del Rizzo S, Liu FF, Pawson T. Redirecting tyrosine kinase signaling to an apoptotic caspase pathway through chimeric adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:11267–72. doi: 10.1073/pnas.1934711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conklin BR, Hsiao EC, Claeysen S, Dumuis A, Srinivasan S, Forsayeth JR, Guettier JM, Chang WC, Pei Y, McCarthy KD, Nissenson RA, Wess J, Bockaert J, Roth BL. Engineering GPCR signaling pathways with RASSLs. Nat Methods. 2008;5:673–8. doi: 10.1038/nmeth.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104:5163–8. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coward P, Wada HG, Falk MS, Chan SD, Meng F, Akil H, Conklin BR. Controlling signaling with a specifically designed Gi-coupled receptor. Proc Natl Acad Sci U S A. 1998;95:352–7. doi: 10.1073/pnas.95.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–66. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 34.Redfern CH, Coward P, Degtyarev MY, Lee EK, Kwa AT, Hennighausen L, Bujard H, Fishman GI, Conklin BR. Conditional expression and signaling of a specifically designed Gi-coupled receptor in transgenic mice. Nat Biotechnol. 1999;17:165–9. doi: 10.1038/6165. [DOI] [PubMed] [Google Scholar]
- 35.Corson TW, Aberle N, Crews CM. Design and Applications of Bifunctional Small Molecules: Why Two Heads Are Better Than One. ACS Chem Biol. 2008;3:677–692. doi: 10.1021/cb8001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollock R, Clackson T. Dimerizer-regulated gene expression. Curr Opin Biotechnol. 2002;13:459–67. doi: 10.1016/s0958-1669(02)00373-7. [DOI] [PubMed] [Google Scholar]
- 37.Irving BA, Weiss A. The cytoplasmic domain of the T cell receptor zeta chain is sufficient to couple to receptor-associated signal transduction pathways. Cell. 1991;64:891–901. doi: 10.1016/0092-8674(91)90314-o. [DOI] [PubMed] [Google Scholar]
- 38.Gross G, Gorochov G, Waks T, Eshhar Z. Generation of effector T cells expressing chimeric T cell receptor with antibody type-specificity. Transplant Proc. 1989;21:127–30. [PubMed] [Google Scholar]
- 39.Eshhar Z, Waks T, Bendavid A, Schindler DG. Functional expression of chimeric receptor genes in human T cells. J Immunol Methods. 2001;248:67–76. doi: 10.1016/s0022-1759(00)00343-4. [DOI] [PubMed] [Google Scholar]
- 40.Maher J, Brentjens RJ, Gunset G, Rivière I, Sadelain M. Human T-lymphocyte cytotoxicity and proliferation directed by a single chimeric TCRzeta/CD28 receptor. Nat Biotechnol. 2002;20:70–5. doi: 10.1038/nbt0102-70. [DOI] [PubMed] [Google Scholar]
- 41.Carpenito C, Milone MC, Hassan R, Simonet JC, Lakhal M, Suhoski MM, Varela-Rohena A, Haines KM, Heitjan DF, Albelda SM, Carroll RG, Riley JL, Pastan I, June CH. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc Natl Acad Sci U S A. 2009;106:3360–5. doi: 10.1073/pnas.0813101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K. Circuit-breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007;8:577–81. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- 43.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–9. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 44.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–8. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim WA. The modular logic of signaling proteins: building allosteric switches from simple binding domains. Curr Opin Struct Biol. 2002;12:61–8. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]
- 47.Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–6. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- 48.Yu B, Martins IR, Li P, Amarasinghe GK, Umetani J, Fernandez-Zapico ME, Billadeau DD, Machius M, Tomchick DR, Rosen MK. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 2010;140:246–56. doi: 10.1016/j.cell.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301:1904–8. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 50.Yeh BJ, Rutigliano RJ, Deb A, Bar-Sagi D, Lim WA. Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature. 2007;447:596–600. doi: 10.1038/nature05851. [DOI] [PubMed] [Google Scholar]
- 51.Dueber JE, Mirsky EA, Lim WA. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat Biotechnol. 2007;25:660–2. doi: 10.1038/nbt1308. [DOI] [PubMed] [Google Scholar]
- 52.Yadav SS, Yeh BJ, Craddock BP, Lim WA, Miller WT. Reengineering the signaling properties of a Src family kinase. Biochemistry. 2009;48:10956–62. doi: 10.1021/bi900978f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–8. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz MA, Madhani HD. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet. 2004;38:725–48. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- 55.Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78(3):499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 56.Printen JA, Sprague GF., Jr Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138(3):609–19. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276(5319):1702–5. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 58.Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–4. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 59.Harris K, Lamson RE, Nelson B, Hughes TR, Marton MJ, Roberts CJ, Boone C, Pryciak PM. Role of scaffolds in MAP kinase pathway specificity revealed by custom design of pathway-dedicated signaling proteins. Curr Biol. 2001;11(23):1815–24. [PubMed] [Google Scholar]
- 60.Dodge-Kafka KL, Soughayer J, Pare GC, Carlisle Michel JJ, Langeberg LK, Kapiloff MS, Scott JD. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437(7058):574–8. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mishra P, Socolich M, Wall MA, Graves J, Wang Z, Ranganathan R. Dynamic scaffolding in a G protein-coupled signaling system. Cell. 2007;131(1):80–92. doi: 10.1016/j.cell.2007.07.037. [DOI] [PubMed] [Google Scholar]
- 62.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319(5869):1539–43. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 63.Chant J. Cell polarity in yeast. Annu Rev Cell Dev Biol. 1999;15:365–91. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- 64.Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol. 2008;18(22):1719–26. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009;139(4):731–43. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zarrinpar A, Park SH, Lim WA. Optimization of specificity in a cellular protein interaction network by negative selection. Nature. 2003;426(6967):676–80. doi: 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- 67.Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317(5836):364–9. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ernst A, Sazinsky SL, Hui S, Currell B, Dharsee M, Seshagiri S, Bader GD, Sidhu SS. Rapid evolution of functional complexity in a domain family. Sci Signal. 2009;2(87):ra50. doi: 10.1126/scisignal.2000416. [DOI] [PubMed] [Google Scholar]
- 69.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458(7240):859–64. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proc Natl Acad Sci U S A. 2008;105(18):6578–83. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14(4):381–92. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 72.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99(26):16587–91. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.