Abstract
Cell signaling proteins are often modular, containing distinct catalytic and regulatory domains. Recombination of such biological modules has been proposed to be a major source of evolutionary innovation. We systematically analyzed the phenotypic diversity of a signaling response that results from domain recombination by using eleven proteins in the yeast mating pathway to construct a library of 66 chimeric domain recombinants. Domain recombination resulted in greater diversity in pathway response dynamics than did duplication of genes, of single domains, or of two unlinked domains. Domain recombination led also to changes in mating phenotype – including recombinants with increased mating efficiency over wild-type. Thus, novel linkages between pre-existing domains may have a major role in the evolution of protein networks and novel phenotypic behaviors.
Domains are the basic functional and structural modules in proteins (1). In signaling networks, domains generally encode one of two major functions: (i) regulation or localization and (ii) catalysis. Catalytic domains directly transmit signaling information (e.g. through phosphorylation), whereas regulatory domains mediate interactions that either target or regulate this catalytic activity. The vast number of domain combinations found in the proteome suggests that domain shuffling could be a major source of evolutionary innovation in signaling behaviors (2–4). Three principal lines of evidence support this view. First, specific changes in protein functions have been associated with domain recombination (5). Second, mutations leading to the fusion of protein-coding genes may lead to the improper activation of signaling networks that result in oncogenic transformations (6, 7). Third, fusions of diverse regulatory and catalytic domains can yield synthetic proteins with non-natural input/output relationships, both in vitro (8) and in vivo (9–11).
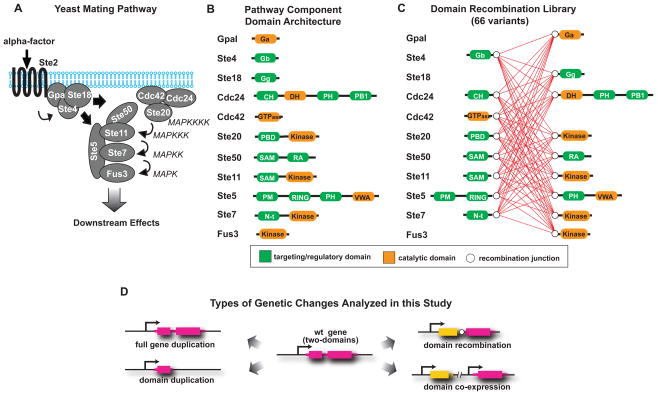
To test whether recombination of signaling protein domains provides a route for evolutionary innovation, analogous to the swapping of cis-regulatory elements and coding sequences in transcriptional circuits (12–14), we have systematically determined the effects of domain recombination on the behavior of a well-understood signaling network, the yeast mating pathway (Fig. 1A), and compared it to the effects brought about by gene or domain duplication. We used the domains of eleven proteins belonging to the mating pathway to construct a library of 66 recombinant proteins (Fig. 1B). Specifically, all native proteins composed of at least two domains were split in a manner that separated regulatory and catalytic domains. The split points were chosen to ensure that domains were left intact and therefore are located within interdomain connecting regions. We then created a library of chimeric proteins that includes all possible recombinations of “N-terminal” and “C-terminal” blocks to systematically map the resulting phenotypic effects (Fig. 1C and Fig. S1). Each protein was transformed into a yeast strain that retained the endogenous copies of the eleven mating pathway genes, such that an additional protein (with altered domain combination) was added to the existing network. To distinguish the effects of domain recombination from those of gene or domain duplication, we created three additional sets of strains (Fig. 1D): in the first one, each of the eleven genes analyzed was duplicated; in the second one, each of the “N- “ or “C-terminal” blocks was duplicated; and in the third one, each possible pair of “N-“ and “C-terminal” blocks were duplicated and co-expressed (all 66 combinations lacking domain recombination). To prevent any bias that might be related to differential transcriptional control, we expressed all constructs at low abundance using a 250 bp segment of the constitutive cycI promoter.
Fig. 1. Design of the recombination library of protein domains belonging to the yeast mating pathway.
A, the yeast mating pathway is activated by binding of the mating pheromone (α-factor) to the membrane receptor Ste2 (in “a” cells, or Ste3 in “α” cells), which causes the dissociation of the G protein alpha subunit (GpaI) from the G beta (Ste4) and gamma (Ste18) complex (20, 25). The scaffold protein Ste5 is then recruited to the membrane-localized Ste4, bringing along the MAPKKK Ste11, MAPKK Ste7 and MAPK Fus3. In addition, Ste11 interacts with the bridging protein Ste50, which by binding to the small GTPase Cdc42, positions Ste11 near its upstream activator, the PAK kinase Ste20 (26). Activated Ste11 phosphorylates Ste7, which in turn phosphorylates Fus3. The activated MAPK translocates to the nucleus where it phosphorylates a number of transcription factors, leading to changes in gene transcription, cell cycle progression and cell morphology and culminating in the fusion between “a” and “α” cells. B, domain architecture of the yeast mating signaling pathway components. Regulatory domains are shown in green, catalytic domains are shown in orange. Fully annotated domain maps are given in Fig. S8. C, domain recombination library: recombination junctions are depicted as white circles, all possible recombinations are shown as red connecting lines. D, possible evolutionary events analyzed in this work. Gene duplication, domain duplication, domain recombination and co-expression of two duplicated domains.
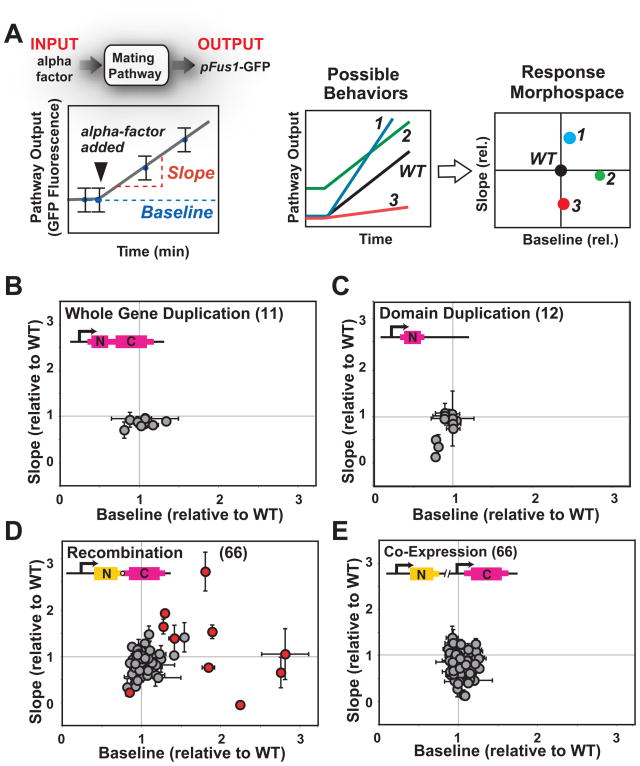
As a metric for how each additional protein altered signaling behavior, we measured the dynamics of mating pathway activation by flow cytometry. A green fluorescent protein (GFP) reporter was controlled by a mating-responsive promoter from the fus1 gene (15) in an “a-type”, Δfar1 strain (to prevent cell cycle arrest and shmooing that could affect flow cytometry measurements (16)). We measured the intensity of GFP fluorescence before and after activation of the mating pathway with α-factor, and used those values to calculate the baseline and slope of activation (Fig. 2A). The normalized baseline and slope values for each variant in our libraries (relative to wild type) were plotted on a “morphospace” diagram. Gene and domain duplications had little effect on the dynamics of pathway activation (Fig. 2B and C). Only three domain duplication variants showed changes, slightly inhibiting pathway activation (variants with lower slopes in Fig. 2C), perhaps by acting as dominant negative fragments. In contrast, recombination of domains resulted in a wide range of altered dynamic behaviors, with variants that, either prevented pathway activation, or led to stronger activation of the mating pathway (Fig. 2D). These altered signaling behaviors appear to depend on domain recombination, because co-expression of all analogous pairs of unlinked N- and C-terminal domain blocks had limited effects on pathway activation (Fig. 2E). At least for the genes and signaling pathway analyzed here, gene or domain duplication alone may contribute little to the immediate diversification of signaling phenotypes -- changes in pathway behaviors probably require sequence divergence of the duplicates (e.g. by neo-functionalization or by differential transcriptional regulation of sub-functionalized duplicates (17, 18) see Fig. S2). In contrast, shuffling of domains provides a more direct path to functional divergence (19), resulting in readily available alterations in signaling behaviors.
Fig. 2. Recombination of protein domains results in diversification in signaling behaviors.
A, mating pathway activation was measured by flow cytometry, using a GFP reporter under the control of a promoter (from the Fus1 gene) that responds to pathway activation, in an “a-type” ΔFar1 strain. Time course measurements of GFP fluorescence were done to calculate the baseline and slope of pathway activation, under conditions of linear pathway response. Baseline and slopes were then normalized relative to wild type values and the resulting values were plotted in pathway morphospace. B, gene duplications had minor effects on mating pathway response dynamics, with most values clustered around wild type. C, domain duplications had also minor effects on mating pathway response dynamics; duplication of Ste50 [N] (Ste50’s SAM domain), Ste5 [N] (which includes Ste5’s RING domain) and Ste11 [N] (Ste11’s SAM domain) are exceptions with low slopes and may act as dominant negative. D, domain recombination led to a diverse set of novel signaling behaviors. Recombination variants with dynamic behaviors most different from wild type and from the corresponding co-expressed N- and C-domain pair (Fig. S3) are shown in red. E, these behaviors could not be recapitulated by co-expression of the unlinked corresponding pairs of domains. Fluorescent values were measured in at least two independent experiments, each time in triplicates. Mean values +/− standard deviation (error bars) are depicted.
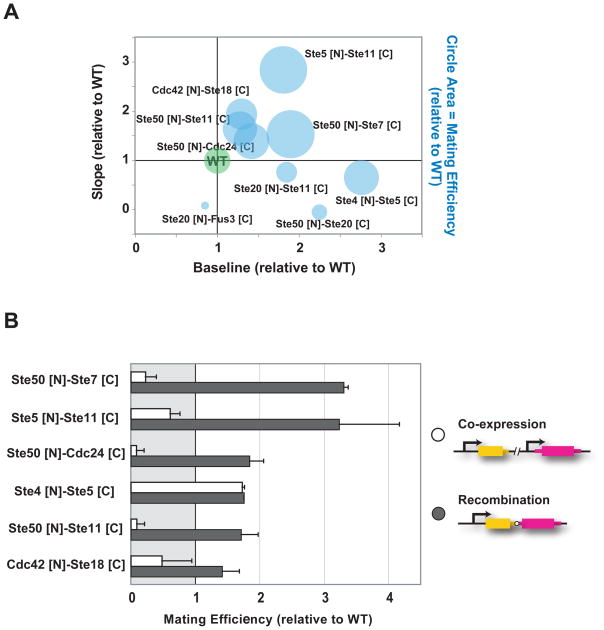
Beyond changes in gene expression, activation of the mating pathway leads to a coordinated response that arrests cell cycle, alters cell morphology, and ultimately results in the fusion of mating partners (20). To determine if changes in reporter gene expression dynamics caused by domain recombination were mirrored by changes in overall pathway outcome, we measured the efficiency with which “a” strains, expressing domain recombination variants, mated with wild type “α” cells. We focused on the ten recombination variants with dynamic behaviors most different from wild type and from the corresponding co-expressed N- and C-domain pair (Fig. S3 and Fig. S4) and measured the percentage of “a” cells that successfully mated when co-incubated with “α” cells (21). Yeast strains expressing domain recombination variants with slopes of pathway activation greater than that of wild type mated more efficiently than did wild type yeast (Fig. 3A and Table S1). The same was true for one variant with high baseline of pathway activation but slightly lower slope (Ste4 [N]-Ste5 [C]). In contrast, yeast strains expressing variants with activation slopes lower than that of wild type mated more poorly. The observed changes in mating efficiency also appeared to depend on domain recombination, because there were marked differences between the mating efficiencies of corresponding recombination and co-expression variants (Fig. 3B). Thus, domain recombination can alter complex pathway outputs, such as the biochemical and morphological changes needed for mating. At least under laboratory conditions, recombination of protein domains can lead to strains that mate more efficiently than wild type, though further work is needed to determine whether the changes in mating efficiency we observed could confer a selective advantage.
Fig. 3. Domain recombination can lead to strains that mate more efficiently than wild type.
A, mating efficiencies were measured for recombination variants with slope and baseline values that were substantially different from wild type (more than one standard deviation) and also different from the slope and baseline values of the corresponding co-expressed N- and C- pair (Fig. S3 and Fig. S4). Mating efficiencies of wild type and recombination variants are depicted as circles, with areas representing relative mating efficiencies. B, Comparison of domain recombination to co-expression of the corresponding domain pairs (wild type values are set to one). Ste50 SAM domain interacts with the Ste11 (MAPKKK) N-terminal SAM domain, facilitating the interaction of Ste11 with Ste20, its upstream activator. Thus, it is possible that, as an isolated domain, Ste50 [N] (as well as Ste11 [N]) act as dominant-negatives, competing for the interaction between the wild type proteins.
Activation of the mating pathway response alters the regulation of the cell cycle (16). In addition, the mating pathway shares several proteins with other signaling pathways, such as the high osmolarity pathway. Thus, domain recombination variants that alter the mating pathway response could also have pleiotropic effects on other cellular processes. To test this possibility, we measured growth rate, as well as the response to high osmolarity stimulus, for the recombination variants that most substantially affected mating response. We found that variants with growth rate deficiencies of only 2–3% compared to WT (Fig. S5A), mate up to ~3 times better than WT. This suggests that, in some cases and under laboratory conditions, the cost in asexual growth likely imposed by recombination-induced network remodeling could be compensated in part by the benefit in mating efficiency it confers (Fig. S5B). In addition, we observed that the response to high osmolarity is only marginally affected (Fig. S5C). We cannot rule out the possibility that some of the variants analyzed might have detrimental effects on other signaling pathways or cellular processes.
Signaling responses are often characterized by their dynamics of temporal activation, as well as by the specific dose response profile: whereas some pathways follow a graded dose response, others have switch like activation profiles (22). To explore whether domain recombination could also alter the dose response profile, we measured pathway response at different concentrations of pheromone for two of the domain recombination variants that most markedly affected the mating pathway temporal response. We found that cells expressing the domain recombination variants Ste50 [N]-Ste7 [C] and Ste5 [N]-Ste11 [C] have dose response profiles similar to that of WT cells, with only a small shift towards lower concentrations of pheromone for cells expressing Ste5 [N]-Ste11 [N] (Fig. S6A). In addition, we observed a wider cell-to-cell variation in pathway response for the domain recombination variant Ste5 [N]-Ste11 [C] (Fig. S6B). These results suggest that domain recombination might slightly alter the sensitivity of the mating pathway to pheromone levels.
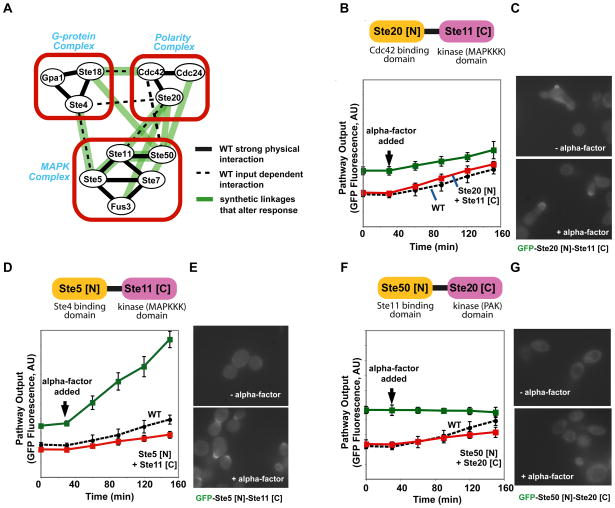
We investigated the mechanisms by which recombination variants might alter the dynamics of the response. We first measured protein abundance for some duplication or recombination variants and found that there is no clear correlation between changes in mating response and protein abundance (Fig. S7). Mating pathway signaling requires interactions between three major functional complexes: the membrane-bound “G-protein complex”, the “Mitogen-activated protein kinase (MAPK) complex”, and the membrane-bound “polarity complex” (Fig. 1). Recruitment of the MAPK complex to the membrane, by its interaction with the G-protein complex, positions the MAPKKK, Ste11, close to its PAK kinase activator, Ste20 (also referred to as the MAPKKKK), a member of the polarity complex (20, 23).
Close examination of the ten recombination variants that most markedly changed signaling behavior, revealed that seven of the ten created novel links between the different signaling complexes, whereas only three created linkages within an individual functional complex (Fig. 4A). Thus, new behaviors may arise when key components change in their localization or complex formation. To explore this hypothesis we examined three recombinant variants in greater detail. The Ste20[N]-Ste11[C] fusion (which tethers the Cdc42 binding domain of Ste20 to the kinase domain of Ste11, Fig. S8) resultsed in higher baseline output (Fig. 4B). This fusion protein may result in the recruitment of Ste11 kinase domain to the polarity complex, even in the absence of α-factor stimulation, where it can be constitutively activated by Ste20 (the MAPKKKK, Fig. S9A). This relocalization to sites of polarity was confirmed by microscopy experiments with the GFP-labeled fusion protein (Fig. 4C). The Ste5[N]-Ste11[C] fusion (which tethers the Ste4 binding domain of Ste5 to the kinase domain of Ste11, Fig. S8) resulted in a large increase in the slope of output (Fig. 4D). This fusion may result in an additional population of Ste11 kinase domain that, because it is covalently fused to Ste5 [N], is more efficiently recruited to the membrane upon alpha-factor stimulation, which may increase signaling (Fig. S9B). Microscopy studies confirmed that this fusion protein is inducibly localized to membrane sites of polarization (Fig. 4E). The Ste50[N]-Ste20[C] fusion (which tethers Ste11 binding SAM domain of Ste50 to the kinase domain of Ste20, Fig. S8) resulted in high constitutive activation (Fig. 4F). This fusion protein may bring the Ste20 kinase domain to the MAPK complex, where it will constitutively activate Ste11 and trigger the MAPK cascade, without the need for membrane recruitment of the MAPK complex (Fig. S9C). Microscopy studies confirmed that this fusion localizes to the cytoplasm both with and without α-factor stimulation (Fig. 4G). Overall, these more detailed observations are consistent with a model in which shuffling of a catalytic domain with different regulatory domains results in novel regulation or localization of the catalytic domain, leading to distinct changes in signaling behavior and cellular phenotype.
Fig. 4. Mechanisms of recombination-derived changes in signaling behavior.
A, activation of the mating pathway requires interactions between three multi-protein complexes: the membrane-bound G-protein complex, the membrane bound polarity complex and the MAPK complex. Seven novel connections between the three multiprotein complexes and three novel connections within an individual complex formed by the recombination variants analyzed. B, D and F, flow cytometry time course of pFus1-GFP for strains expressing Ste20 [N]-Ste11 [C], Ste5 [N]-Ste11 [C], and Ste50 [N]-Ste20 [C], respectively. C, E and G, fluorescence microscopy of strains expressing GFP-labeled Ste20 [N]-Ste11 [C], Ste5 [N]-Ste11 [C], and Ste50 [N]-Ste20 [C], respectively.
The high frequency with which the limited diversity encoded in our recombination library led to novel signaling behaviors suggests that domain recombination might have an important role in the generation of phenotypic novelty from simple genotypic changes, and could likely complement the role of cis-regulatory elements in the evolution of global cellular regulatory networks composed of both transcriptional and signaling elements (24). Further work will be needed to compare in quantitative terms the contributions of gene duplication and recombination to the evolutionary process.
The strategy used here of targeted domain recombination between proteins that belong to a specific signaling network could facilitate the engineering of other protein networks of interest, a fundamental goal of synthetic biology. Genes known to belong to a target pathway could be deconstructed into domains and used to build small libraries of domain recombinations that are subsequently screened for the desired function
Methods
For detailed information on all methods see Supplementary Online Material.
Supplementary Material
Acknowledgments
We thank Angi Chau for her help with Matlab, Noah Helman and Jessica Walter for their help with fluorescence microscopy and members of the Lim laboratory for critical reading of this manuscript and assistance. This work was supported by grants from the Howard Hughes Medical Institute, the Packard Foundation, the NIH, and the NIH Nanomedicine Development Centers (W. A. L.). S. G. P. was supported by a Long-Term Postdoctoral Fellowship from the Human Frontier Science Program. P. W. was supported by the Li Foundation.
Footnotes
Authors Contributions
S. G. P. and W. A. L. conceived the project. S. G. P and J. G. performed the experiments. P. W. assisted with initial experiments. S. G. P., J. G. and W. A. L. analyzed the results. S. G. P. and W. A. L. wrote the manuscript.
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Chothia C, Gough J, Vogel C, Teichmann SA. Science. 2003 Jun 13;300:1701. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 2.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Curr Opin Struct Biol. 2004 Apr;14:208. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T, Nash P. Science. 2003 Apr 18;300:445. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 4.Voigt CA, Martinez C, Wang ZG, Mayo SL, Arnold FH. Nat Struct Biol. 2002 Jul;9:553. doi: 10.1038/nsb805. [DOI] [PubMed] [Google Scholar]
- 5.Bashton M, Chothia C. Structure. 2007 Jan;15:85. doi: 10.1016/j.str.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Pawson T, Warner N. Oncogene. 2007 Feb 26;26:1268. doi: 10.1038/sj.onc.1210255. [DOI] [PubMed] [Google Scholar]
- 7.Maher CA, et al. Nature. 2009 Mar 5;458:97. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dueber JE, Yeh BJ, Chak K, Lim WA. Science. 2003 Sep 26;301:1904. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 9.Harris K, et al. Curr Biol. 2001 Nov 27;11:1815. [PubMed] [Google Scholar]
- 10.Howard PL, Chia MC, Del Rizzo S, Liu FF, Pawson T. Proc Natl Acad Sci U S A. 2003 Sep 30;100:11267. doi: 10.1073/pnas.1934711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh BJ, Rutigliano RJ, Deb A, Bar-Sagi D, Lim WA. Nature. 2007 May 31;447:596. doi: 10.1038/nature05851. [DOI] [PubMed] [Google Scholar]
- 12.King MC, Wilson AC. Science. 1975 Apr 11;188:107. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 13.Carroll SB. PLoS Biol. 2005 Jul;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prud’homme B, Gompel N, Carroll SB. Proc Natl Acad Sci U S A. 2007 May 15;104(Suppl 1):8605. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashor CJ, Helman NC, Yan S, Lim WA. Science. 2008 Mar 14;319:1539. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 16.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. Cell. 1993 May 21;73:747. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 17.Lynch M, Force A. Genetics. 2000 Jan;154:459. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Genome Biol. 2002;3:RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers RL, Bedford T, Hartl DL. Genetics. 2009 Jan;181:313. doi: 10.1534/genetics.108.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dohlman HG, Thorner JW. Annu Rev Biochem. 2001;70:703. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 21.Sprague GF., Jr Methods Enzymol. 1991;194:77. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi S, Pryciak PM. Curr Biol. 2008 Aug 26;18:1184. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pryciak PM, Huntress FA. Genes Dev. 1998 Sep 1;12:2684. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerhart J, Kirschner M. Proc Natl Acad Sci U S A. 2007 May 15;104(Suppl 1):8582. doi: 10.1073/pnas.0701035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh L, Neiman AM, Herskowitz I. Annu Rev Cell Biol. 1991;7:699. doi: 10.1146/annurev.cb.07.110191.003411. [DOI] [PubMed] [Google Scholar]
- 26.Bardwell L. Peptides. 2005 Feb;26:339. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.