Abstract
Over a decade has passed since publication of the last review on “Cytometry in cell necrobiology.” During these years we have witnessed many substantial developments in the field of cell necrobiology such as remarkable advancements in cytometric technologies and improvements in analytical biochemistry. The latest innovative platforms such as laser scanning cytometry, multispectral imaging cytometry, spectroscopic cytometry, and microfluidic Lab-on-a-Chip solutions rapidly emerge as highly advantageous tools in cell necrobiology studies. Furthermore, we have recently gained substantial knowledge on alternative cell demise modes such as caspase-independent apoptosis-like programmed cell death (PCD), autophagy, necrosis-like PCD, or mitotic catastrophe, all with profound connotations to pathogenesis and treatment. Although detection of classical, caspase-dependent apoptosis is still the major ground for the advancement of cytometric techniques, there is an increasing demand for novel analytical tools to rapidly quantify noncanonical modes of cell death. This review highlights the key developments warranting a renaissance and evolution of cytometric techniques in the field of cell necrobiology.
Keywords: necrobiology, necrosis, apoptosis, autophagy, cytomics, cytometry, imaging, spectroscopy, microfluidics, Lab-on-a-Chip
The term cell necrobiology has been introduced in late 1990s to collectively define: “various modes of cell death; the biological changes which predispose, precede, and accompany cell death; as well as the consequences and tissue response to cell death” (1,2). Our prior reviews on the subject of cytometry in cell necrobiology received wide recognition and the term cell necrobiology became even included in a free encyclopedia Wikipedia http://en.wikipedia.org/wiki/Necrobiology. It also inspired the renowned artist Julie Newdoll, the author of numerous cover page illustrations to Nature and Cell journals, to create a series of artworks devoted to necrobiology (available at: www.brushwithscience.com).
Undoubtedly, the speed and wealth of multiparameter data collection contributed to the dramatic expansion and popularity of cytometric techniques in studies of cell death (1–4). Cytometry, however, still remains a viable and dynamic field of engineering where advancements prove beneficial for both the basic biology and clinical diagnostics (5–7). Expectedly, the current pace in development of novel cytometric systems will open new horizons for cell biology, clinical diagnostics, and drug discovery (8,9). We expect that this comprehensive update of our earlier reviews reporting advances in the field will be of interest to many researchers in diverse areas of biology, biophysics, biotechnology, and medicine.
THE BIOLOGY OF CELL DEATH
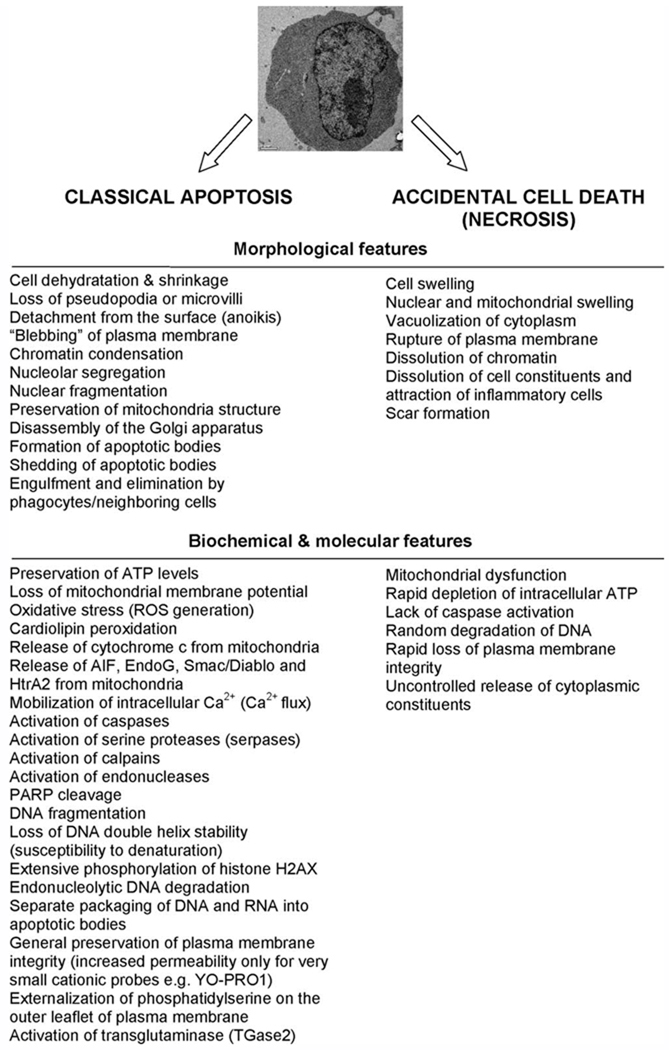
Cells are archetypically known to disassemble in two morphologically and biochemically distinct processes: apoptosis and necrosis (1,10,11). Both were initially identified based on characteristic changes in cell morphology (1,10). Figure 1 outlines major morphological and molecular changes occurring during apoptosis versus accidental cell death (herein termed necrosis). Alterations in cellular parameters, as presented in Figure 1, also become a basis for development of a variety of markers for fluorescence microscopy and flow cytometry (1–4). Despite subsequent introduction of numerous molecular assays, the morphological changes, detected by light and electron microscopy, still remain the “gold standard” to differentiate these two distinct modes of cell death (1,12). Noteworthy, recent reports led to the characterization of alternative cell demise modes such as caspase-independent apoptosis-like programmed cell death (PCD), autophagy, necrosis-like PCD, and mitotic catastrophe (Table 1) (13–16). Although still a matter of debate, these noncanonical pathways appear to have wide reaching connotations in pathogenesis and treatment of human diseases (11,17,18). Moreover, they present an increasingly complex network of molecular cross-talks reflecting in a diversity of phenotypes. These discoveries raised also an ongoing debate aiming at the classification of cell death programs (19,20). It must be acknowledged that the general term apoptosis, commonly exploited in many research articles, tends sometimes to misinterpret the actual mechanisms underlying cellular suicide (13,19,20). It is, thus, postulated to restrict the term apoptosis to only the traditional cascade featuring all canonical “hallmarks of apoptotic cell death,” such as (i) activation of caspases as an absolute marker of cell death; (ii) high degree of compaction of chromatin; (iii) activation of endonucleases(s) causing internucleosomal DNA cleavage and extensive DNA fragmentation; (iv) appearance of distinctive cellular morphology with preservation of organelles, (v) cell shrinkage, (vi) plasma membrane blebbing, and (vii) nuclear fragmentation followed by formation of apoptotic bodies (Table 1) (13,19–21).
Figure 1.
Morphological and biochemical hallmarks of apoptosis and accidental cell death (necrosis). Note that some features characterizing apoptosis may not be present and depend heavily on a particular cell type, stimuli, and cellular microenvironment.
Table 1.
Current concepts on the complexity of cell demise modes
| CELL DEMISE MODE | DISTINCTIVE MORPHOLOGICAL FEATURES | DISTINCTIVE BIOCHEMICAL FEATURES |
|---|---|---|
| Classical apoptosis | Strong condensation of chromatin, cell shrinkage; preservation of cellular organelle; cell membrane blebbing; formation of apoptotic bodies |
Absolute requirement of caspase cascade activation; internucleosomal DNA fragmentation; phosphatidylserine exposure |
| Caspase-independent apoptosis-like programmed cell death |
Chromatin condensation less pronounced than in classical apoptosis; varying gradation and combination of apoptotic features possible |
Activation of caspases not necessary to execute the program although possible; common activation of other proteases: cathepsins, calpains, serine proteases; DNA fragmentation less pronounced; phosphatidylserine exposure often observed |
| Autophagy | Partial chromatin condensation; formation of double/multilayered autophagosome vacuoles dependent on ATG genes; cell membrane blebbing possible |
Initially perceived as caspase independent although recent reports indicate possible cross-talk with classical apoptosis; lack of DNA fragmentation; increased lysosomal activity |
| Mitotic catastrophe | Formation of giant polykaryons; lack of chromatin condensation; lack of cell membrane blebbing |
At initial stages caspase independent although final rerouting to caspase dependent execution is possible; initiated by a violation of G2 checkpoint of the cell cycle and premature entry to mitosis |
| Necrosis-like programmed cell death (classified also as aborted apoptosis) |
Lack of geometric chromatin condensation or condensation forming loose speckles; varying scale and combination of apoptotic features possible |
Initial caspase cascade activation possible with common subsequent inhibition and rerouting to alternative pathways; predominantly random degradation of DNA; phosphatidylserine exposure possible |
| Necrosis (classified also as accidental cell death or cell lysis) |
Lack of geometric chromatin condensation, dissolution of chromatin; organelle and cell swelling; lack of cell membrane blebbing; rapid rupture of plasma membrane |
Lack of protease cascade activation; random degradation of DNA (no DNA laddering); rapid and uncontrolled release of cell constituents |
| Senescence (“cell zombie”) | Appearance of characteristic heterochromatic foci; characteristic flattened cytoplasm; increased cellular granularity; lack of cell membrane blebbing |
Caspase independent; initiated by a shortening of telomeres and cell entry into irreversible cell cycle arrest (replicative senescence); profound changes in metabolism and activation of senescence- associated β-galactosidase (SA-β-gal) |
Nevertheless, single cytometric assays such as the estimation of sub-G1 fraction or Annexin V binding are still being exploited in many research articles to solely characterize and quantify cell death (1,2,4). Moreover, data obtained from such single assays are persistently referred to as “apoptotic cells.” In view of the new discoveries dependence on single cytometric readouts can lead to substantial analytical artifacts (2,4). As discussed earlier, it was only recently proposed to define apoptosis as a “caspase mediated cell death” (19). Logically, caspase activation would be the most specific marker of apoptosis (22). There are, however, many examples of cell death that resembles classical apoptosis, yet there is no evidence of caspase activation (11,13,23). Similarly, externalization of posphatidylserine residues as detected by Annexin V binding is not an absolute marker of apoptosis. Extensive DNA fragmentation is also often considered to be a specific marker of apoptosis where labeling in the TUNEL reaction ensures discrimination from cells that underwent primary necrosis (24). There are, however, mushrooming examples where apoptotic or apoptotic-like cell death proceeds without internucleosomal DNA degradation (25–29). In these instances, the intensity of cell labeling in TUNEL assay will be inadequate to positively identify apoptotic cells (1,2).
As evidenced by the examples the cellular disintegration represents a diverse and interconnected pathway with many unexpected fails-safe mechanisms (13,30). Proper experimental approaches and understanding of assays’ principles should aid to avoid perplexing artifacts during development and exploitation of novel assays (1–4).
INNOVATIONS IN CYTOMETRIC TECHNOLOGIES
Last decade, in particular, has brought many innovations to the field of cytometry (5–9). Rapid development of multispectral imaging cytometry and the slide-based cytometers offer great hopes for improved data quality and throughput (31,32). Probably the most fascinating, however, are prospects of spectroscopic cytometry and innovative micro- and nanofluidic technologies (33,34). In the following section, we summarize the most important advancements with major implications to cell necrobiology.
Flow Cytometry
Modern high-end flow cytometers are often capable of analyzing up to 16 optical parameters from a single cell, with maximal acquisition rates reaching approximately 100,000 events per second (7,8). Yet, probably the most striking advancement in the field of flow cytometry is the current miniaturization and simplification of analytical components (5). Such technological milestones bring flow cytometry out of centralized core facilities while making it more affordable and user friendly. Importantly, near-UV, violet, green, yellow, orange, and red lasers are becoming an increasingly affordable solution with many modular and portable designs with two or even three excitation lines being brought to the markets (5,35–39). These trends will surely drive the renaissance of flow cytometry and the development of new multiplexed assays.
In-Flow Imaging—Multispectral Imaging Flow Cytometry
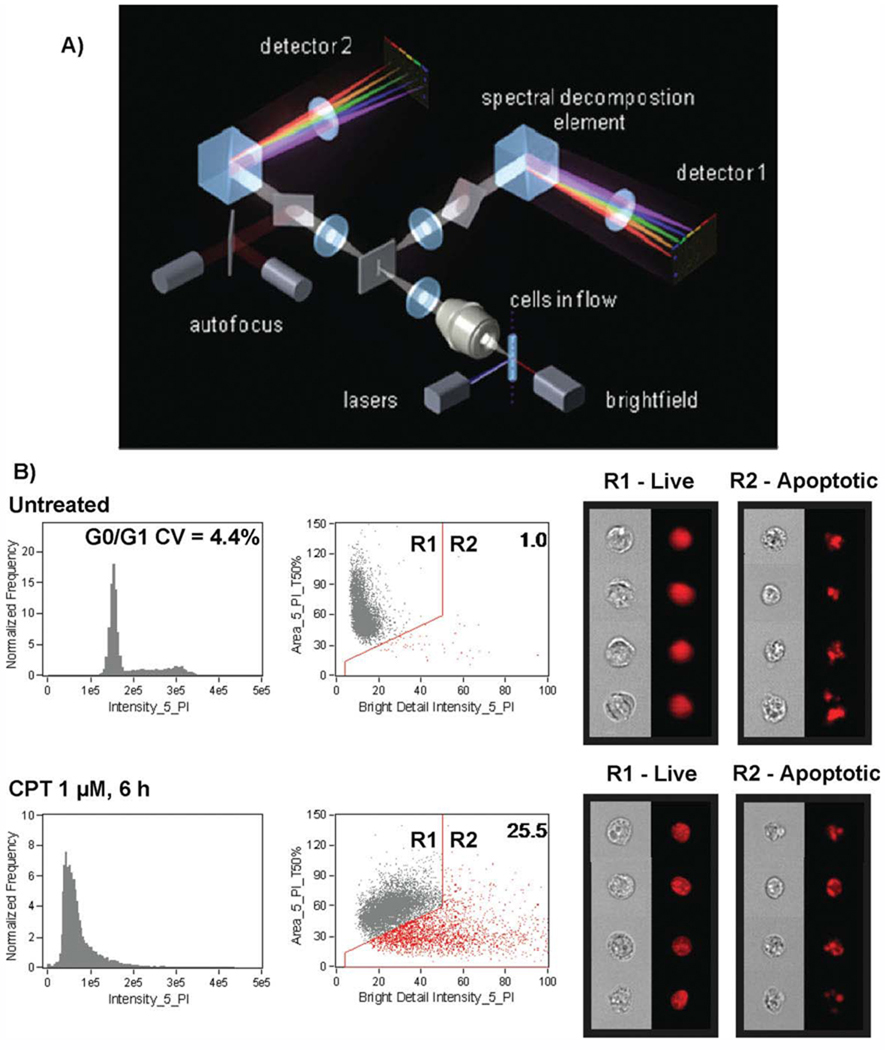
A common drawback of flow cytometry is that the cells flow suspended in a laminar stream of fluid and only integrated fluorescence is collected by photomultiplier tubes (PMTs) without collecting the valuable information on subcellular and morphological features. A noteworthy improvement, that can in many aspects revolutionize conventional flow cytometry, has recently been proposed by Morrissey’s group at Amnis (Amnis Corp, Seattle, WA) (Fig. 2). The Image Stream System integrates a fast in-flow imaging with the conventional layout of the flow cytometer (31,40). At its core the time-delay-integration (TDI) technology preserves sensitivity and image quality during imaging of fast moving cells (31,40,41) (Fig. 2). In combination with a fast charged couple device (CCD), instead of PMTs, it is capable of simultaneously acquiring six spectrally decomposed images from each cell (42) (Fig. 2). The proprietary “virtual sorting mode” provides a valuable visual verification of each cell while the number of morphometric parameters greatly surpasses that of conventional analyzers. By combining attributes of both flow cytometry and image analysis multispectral imaging cytometry appears to be a very attractive instrumentation for multiparameter studies on cell demise (31,40 — 43) (Fig. 2).
Figure 2.
Multispectral imaging flow cytometer. (A) Layout and key components of the ImageStream high speed imaging system. Cells are hydrodynamically focused into a core stream and orthogonally illuminated with lasers for side scatter (SSC) and fluorescence imaging, and transilluminated for brightfield imaging. Light is collected from the cells with an imaging objective lens (20×, 40×, or 60×) and projected onto a charge-coupled detector (CCD). Before projection on the CCD, the light is passed through a spectral decomposition optical system that directs light of different wavelengths to different lateral positions across the detector, enabling simultaneous capture of up to six spectrally distinct images per detector. In the example shown, cells are illuminated by spatially separated lasers resulting in the generation of two composite images per cell. Each image is spectrally decomposed and projected onto separate detectors, enabling collection of up to 12 images per cell. (B) Morphology-based identification of apoptotic cells using Image Stream. Jurkat cells in midexponential growth were left untreated (top) or were incubated with 1 µM CPT for 6 hours, fixed and stained with PI, then collected on the ImageStream. DNA content histograms of single cells are shown in the left panels. Cells exhibiting nuclear fragmentation (low nuclear area and bright detail intensity) are gated in the histograms at right, with the percentage of apoptotic cells indicated in the upper right corner of the plot. Representative brightfield and PI image of nonapoptotic cells from the untreated sample and apoptotic cells from the treated sample are shown at right (data courtesy of Dr. Tad George and Dr. Brian Hall, Amnis Corporation, Seattle, WA) (31,40). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Raman Flow Cytometry
Nondestructive spectroscopies such as infrared microspectroscopy and Raman microspectroscopy attract an increasing interest for a multispectral analysis of cells and tissues (44,45). Nonresonant Raman spectroscopy is one of the most dynamically developing technologies based on inelastic scattering of incident photons and resulting vibrational excitation of the biomolecules (46). Energy from the excited molecule is subsequently reemitted as a wavelength of higher wavelength. Complex biological molecules exhibit many vibrational modes that result in characteristic Raman spectra. These provide precise chemical fingerprints without the use of fluorescent labels (44,47). The advent of surface-enhanced Raman spectroscopy (SERS) introduced expended opportunities for signal enhancement and multiplexing over nonenhanced Raman scattering (47 — 49). Interestingly only a handful of reports have employed Raman spectroscopy to investigate cells undergoing apoptosis (50). Furthermore, a noninvasive method of detecting necrotic cell death has recently been proposed by Kunapareddy et al (51). The ability of Raman microspectroscopy to noninvasively track cell cycle dynamics in living cells has also been recently investigated (52). Only recently, Watson et al. were the first to show application of surface-enhanced Raman spectroscopy to flow cytometry (47,53). Although still largely in its infancy, Raman Spectral Flow Cytometry is an innovative approach to multiplexed chemical cytometry on living cells (47,53). It remains to be seen whether Raman Spectral Flow Cytometry can find wider applications in studies of programmed cell death.
Laser Scanning and Imaging Cytometry
Inherent limitations of traditional flow cytometry have recently fuelled the rapid development of static, slide-based cytometers that combine advantages of flow cytometry (FCM) and fluorescence image analysis (FIA) (32). Laser scanning cytometer (LSC) was a brainchild of Louis Kamentsky who in early 1990s constructed the first model of this cytometer (32). Its construction led to technological breakthrough that paved the way to more advanced LSC instruments such as iCys™ Research Imaging Cytometer or iCyte™ Automated Imaging Cytometer. LSC is a microscope, slide-based cytometer featuring the ability to spatially analyze fluorescence of large numbers of adherent cells (32,54). By having many attributes of both flow cytometry and fluorescent imaging, LSC appears to be an optimal instrumentation for multiparameter cell necrobiology studies as described by us earlier (54). LSC has recently found interesting applications in clinical pathology, tissue microarrays, and in the analysis of living-cell microar-rays (55,56). Combination with the latter provides a budding outlook for a very high throughput and multidimensional analysis on rare populations of cells (57—59).
Since the pioneering introduction of LSC other innovative scanning cytometers have been developed including microplate laser scanning cytometer Accumen eX3 (TTP Lab-Tech, Melbourn, UK). This innovative technology employs cytometric principles rather than conventional image analysis to collect high-content data. Multiple laser configuration and up to 12 detection channels offer a noteworthy advancement toward a fast and user-friendly approach to high content scanning cytometry. There are also several other innovative scanning, event-based systems that include i.e. the LEAP™ and Celigo™ systems (Cyntellect Inc., San Diego, CA), the Oper-a™ High Content Screening system (Perkin-Elmer, Waltham, MA), InCell Analyzer 1000™ (GE Healthcare Biosciences, Pittsburgh, PA), and the BD Pathway™ 435 and BD Pathway™ 855 analyzers (BD Biosciences, San Jose, CA). These are all single-cell and moderate-throughput instruments that utilize comparable cytometric technologies. New scanning and imaging cytometers reportedly feature increasing throughput and number of acquisition channels that makes them especially suitable for high-content screening of cell death features. Some of the most advanced platforms have data processing and presentation capabilities similar to flow cytometers. When combined with slide based and/or microplate scanning and increasing integration with Nipkow spinning disk confocal modules, these innovative systems provide reasonable throughput and high-resolution screening capabilities at a subcellular level.
Capacitance and Impedance Cytometry
Recent reports suggest that early apoptotic changes in composition of phospholipids in plasma membrane can be detected with much greater sensitivity using dielectric spectroscopy than using fluorescently conjugated Annexin V (60,61). Measuring electrical properties of cells emerges, thus, as a pioneering way to develop label-free assays (62).
Capacitance cytometry is one of such innovative approaches. It is based on a linear relationship between the nuclear DNA content and the change in capacitance evoked by the cell passage across a 1-kHz electric field (62). DNA is a highly charged molecule and in low-frequency AC electric field, its polarization response can be measured with a great precision. Comparisons made with flow cytometry have already demonstrated sensitivity of this technique in measurements of DNA content (62). Capacitance measurements require no cell processing and can be readily performed on live cells making it applicable for the real-time monitoring of the effects of pharmacological agents on cell cycle distribution (62).
Another emerging electric spectroscopy relies on local changes of the ionic environment at the electrode-solution interface that affect the impedance value of the electrode (63). The xCELLigence system (ACEA Biosciences, San Diego, CA and Roche Applied Science, Penzberg, Germany) based on these principles allows a label-free and nondestructive analysis of adherent cells on microelectrode arrays (63,64). It offers several advantages such as (i) avoidance of fluorescent labels and subsequent imaging, (ii) real-time kinetic monitoring of cytotoxicity, (iii) potential for sequential drug challenge, and (iv) straightforward automation (65). A disadvantage of this innovative technology is, however, the restriction to only adherent cells. Moreover, impedance cytometry, being in principle a bulk technique, measures the average impedance value and cannot be used to characterize or quantify e.g. apoptotic cell death. This technology can nevertheless prove very valuable in kinetic screening of anticancer drugs and readily detect drugs that induce irreversible or reversible cell cycle arrest such as senescence (66).
Application of dielectric spectroscopy to flow cytometry has recently been implemented by Cheung et al. at the Institute of Microelectronics and Microsystems (Lausanne, Switzerland) (61). Polarization of flowing cells in an alternatecurrent (AC) electric field, results in the accumulation of charges at the boundaries between the plasma membrane and the aqueous medium. Analysis of the amplitude, opacity, and phase information is subsequently used in Impedance Spectroscopy Flow Cytometer (Axetris, Kaegiswil, Switzerland) for discrimination between different cell subpopulations without the use of fluorescent markers (61).
Microfluidic (Lab-on-a-Chip) Cytometry
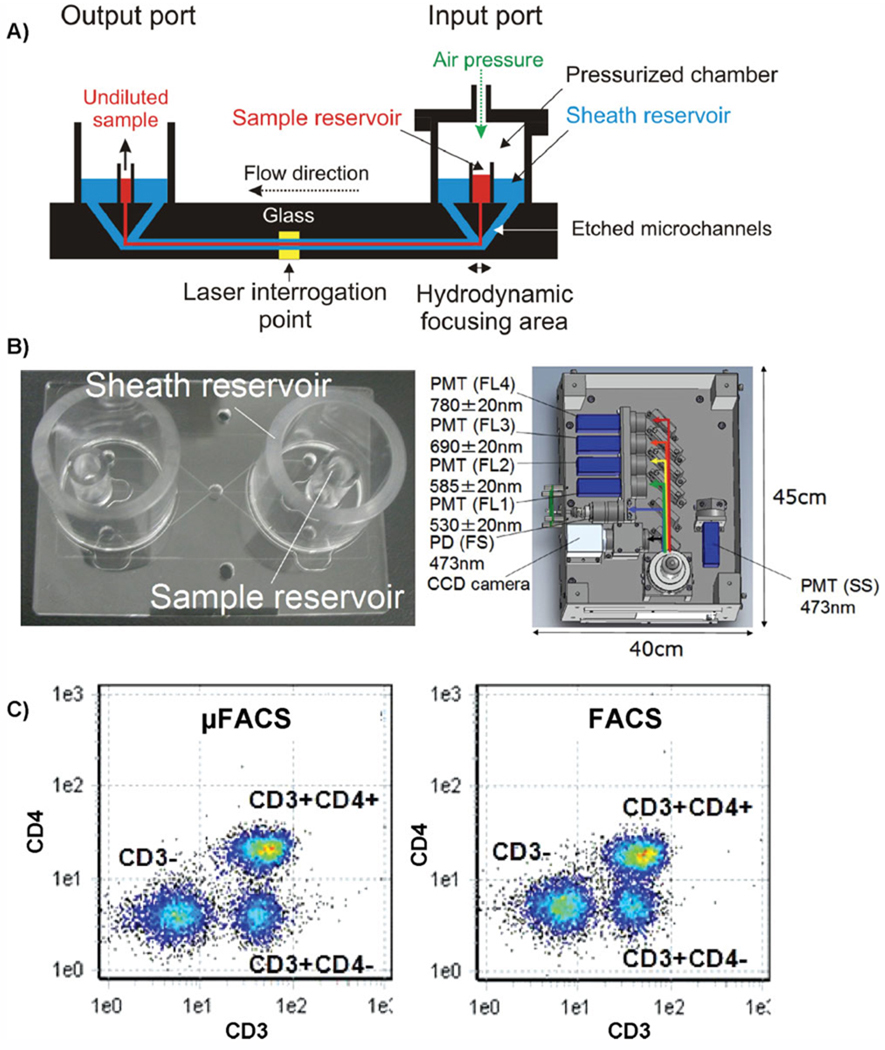
The advent of microfluidic and nanofluidic technologies (Lab-on-a-Chip, LOC) is one of the most innovative cytometric approaches to the analysis of rare cells and organelles. LOC devices promise greatly reduced equipment costs, increased sensitivity, and throughput by implementing parallel processing (67—69). Most importantly, as only low cell numbers and operational reagent volumes are required, dynamic cytomics on a single-cell level finally appears within investigational reach (67—69). Microfluidics has already been named the emerging technology with multitude applications in high-throughput drug screening routines; content-rich personalized clinical diagnostics, and improved analytical capabilities for resource-poor areas (70). It also provides exceptional evolutionary avenues for microfluidic flow cytometry (µFACS), cell sorting, and revolutionary cell microarrays (33,34). Mushrooming reports have demonstrated chip-based flow cytometers and cell sorters with both electro-osmotic and hydrodynamic-driven flows (33,34,71). Microcytometers are, however, often criticized by much lower detection sensitivities than conventional analyzers, particularly for FITC fluorescence (>2,000,000 MESF of FITC, as opposed to <500 MESF, respectively) (72). This can lead to unambiguous discrimination of dim fluorescent events. In our recent work, we have proven that sensitivity of µFACS is adequate to detect subtle changes in fluorescence intensity during microcytometric analysis of apoptosis (73). Noteworthy, a dramatic leap forward in µFACS has recently been made by the group of Takeda at On-chip Biotechnologies (Tokyo, Japan) by introducing a user-friendly chip-based system with (i) hydrodynamic sheath focusing, (ii) multilaser excitation, (ii) up to four color detection, and (iv) detection with fluorescence area, height, and width parameters for all fluorescent channels (Fig. 3) (74,75). Sensitivity of this portable microfluidic cytometer falls within specifications of conventional flow cytometers (FITC < 600 MESF, PE< 200 MESF). Interestingly, this is also the very first µFACS that includes true Forward Site Scatter and Side Site Scatter detection (74,75). Moreover, an innovative chip design facilitates the collection of cells after flow cytometric analysis, feature not attainable in any conventional system (Fig. 3) (74,75).
Figure 3.
Advanced microfluidic flow cytometer (Fishman-R). (A) Cross-sectional view of the microfluidic chip. (B) Disposable microfluidic cartridge (left panel) and an optical layout of the microcytometer (right panel). Note FSC, SSC, and four fluorescence detectors used in combination with spatially separated solid state 473 nm and 640 nm lasers. Side scatter detection is performed using innovative Side scattered Light detection using Edge Reflection (SLER) technology. (C) Immunophenotyping performed on the microfluidic Fishman-R flow cytometer as compared with conventional flow cytometer. Note that µFACS analysis requires merely 20 µl of blood and yields comparative multiparameter data to FACS (data courtesy of Dr. Kazuo Takeda, On-Chip Biotechnologies Co, Tokyo, Japan) (74,75). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Such technological innovations dramatically improve throughput and open new opportunities for cell necrobiology field. We have recently reported application of µFACS technology (Caliper LabChip®, Caliper Life Sciences, Hopkinton, MA) in rapid, two-color quantification of apoptosis on very small cell samples (73). This technology allows for a simultaneous analysis of up to six low volume and independent samples on disposable cartridge chips without laser aligning or operator engagement (33,73).
An increasing interest also attracts live-cell microarray technology that rapidly change modern cytomics and cytometry. In our most recent study, we have reported the development of microfluidic cell arrays (microfluidic array cytometers) for the kinetic analysis of apoptosis (76,77). These microfabricated chips were tailored for dynamic observations of individually positioned cells during sequential pharmacological stimuli, while greatly reducing both number of cells and reagents required to conduct dynamic studies on a single-cell level (76,77). We anticipate that a new generation of cytometers will prospectively employ microfluidic and nanofluidic systems with integrated on-chip cell culture, drug delivery, cytometric analysis, and sorting modules (71,76).
ADVANCEMENT IN ANALYTICAL BIOCHEMISTRY
Recently, many innovative fluorescent assays have been introduced to characterize and quantify programmed cell death. Below we outline only a handful of the noteworthy developments that probe characteristic features of programmed cell demise.
Cell Membrane, Annexin V, and Beyond
The overlapping spectra of organic fluorochromes profoundly hamper the proliferation of multiplexed apoptotic assays. Development of semiconductor nanocrystals commonly called Quantum Dots (QD) has recently superseded the common problem of spectral mismatch (78,79). Successful attempts have already been made to implement semiconductor nanocrystals in multiparameter flow cytometry (80). Annexin V conjugates are so far sole examples of QD application in cell necrobiology and they attract considerable attention in both cytometric and time-lapse imaging studies (78—80).
A recent development from Upstate (Waltham, MA) introduced fluorescently labeled monoclonal antibodies against phosphatidylserine residues that alleviates dependence on calcium-supplemented buffers without compromising sensitivity of detection. Progress is also being made in the field of inorganic zinc coordination complexes (fluorescent Zn2+-dipicolyamines, DPA) that under Ca2+-free conditions selectively bind to membranes enriched in anionic phospholipids (81,82). Finally, a small cationic molecule merocyanine 540 (MC540) emerges as a new probe to detect membrane phospholipids rearrangements in apoptotic cells (83).
A new assay based on selective membrane permeability to substituted cyanine probes YO-PRO 1 and PO-PRO 1 has recently been developed (Fig. 4) (84). Entry of YO-PRO 1 cation (629 Da) into early apoptotic cell probably depends on the activation of P2X7 ion-gated channel, an event concurrent with scramblase activation and phosphatidylserine externalization (85). Early changes in lipid composition, structural relaxation, or even impaired active dye efflux cannot be, however, excluded (84,86,87). Interestingly, our recent studies revealed that the time-window of apoptosis detection by YO-PRO 1 is substantially wider than assessed by Annexin V conjugates and coincides with the loss of ΔΨm as detected by tetramethylrhodamine (TMRM) probe (Fig. 4). Caution should be, however, exercised as dye uptake and/or efflux may vary between different cell types and some cells may not exhibit differential staining with YO-PRO 1 during apoptosis.
Figure 4.
Apoptotic changes in plasma membrane. (A) Detection of apoptosis by concurrent staining with annexin V-APC and PI. Human B-cell lymphoma cells were untreated (left panel) or treated with dexamethasone (right panel), as described previously (73). Cells were subsequently stained with Annexin V—APC conjugate and PI and their far-red and red fluorescence was measured by flow cytometry. Live cells (V) are both Annexin V and PI negative. At early stage of apoptosis (A) the cells bind Annexin V while still excluding PI. At late stage of apoptosis (N) they bind Annexin V-FITC and stain brightly with PI. (B) Detection of apoptosis by concurrent staining with YO-PRO 1 and PI. Cells were treated as in A) and supravitaly stained with YO-PRO 1 and PI probes. Their green and red fluorescence was measured by flow cytometry. Live cells (V) are both YO-PRO 1 and PI negative. Early apoptotic cells (A) are permeant to YO-PRO 1, but still exclude PI. Late apoptotic/ secondary necrotic cells (N) are permeant to both probes (double positive events). (C) Comparison between Annexin V and YO-PRO 1 based assays (for details refer to text). Cells were treated as in (A) and supravitally stained with Annexin V-APC, YO-PRO 1, and PI probes. PI positive cells were electronically excluded from subsequent analysis. Note that essentially all cells that responded to dexamethasone by increase in Annexin V binding were YO-PRO 1 positive. Nevertheless, large fraction of YO-PRO 1 positive cells did not bind Annexin V (arrow). These data indicate that time-window for YO-PRO 1 assay is wider than for Annexin V binding, and precedes the latter. For details refer to text.
Detection of Activated Caspases
One of the hallmarks of classical apoptosis is the activation of unique cysteine aspartyl-specific proteases having a conserved QACXG consensus site containing active cysteine, called caspases (from cysteinyl aspartate-specific proteases; Fig. 1).
Under normal physiological conditions, caspases are constitutively expressed in the cytoplasm as zymogens with very low intrinsic activity. They become activated upon transcatalytic cleavage followed by dimerization. Several methods have recently been developed to detect activation of caspases by flow and laser scanning cytometry (3,4).
Fluorogenic caspase substrates
Recently some innovative fluorogenic substrates, designed to measure caspases activation in living cells, have become commercially available. While certain substrates can be used to detect activation of multiple caspases (pan-caspase probes), other have the peptide moieties that make them specific for particular caspases (88–94). There is currently a wide choice of the substrates with different excitation and emission spectra suitable to be used in combination with other fluorochromes for multiparametric analyses. In principle, these peptide substrates are not fluorescent but upon the cleavage by caspase they generate fluorescing products (88–94). Two currently available on the market reagents include the Phi-PhiLux system (OncoImmunin Inc, Gaithersburg, MD) and the NucView 488 system (Biotium Inc, Hayward, CA).
The PhiPhiLux system utilizes xanthenes dyes forming H-type dimmers that exhibit fluorescence quenching associated with their formation (89,93). Two xanthenes dyes such as rhodamine fluorophores are separated by caspase recognition and cleavage sequence. Proximity enforced formation of H-type rhodamine dimers results in the formation of a quenched and cell permeant caspase substrate (89,93). Upon cleavage of the liker by the activated caspases both rhodamine molecules are released and become highly fluorescent (89,93). Importantly, cleaved fluorescent fragments are well retained on the side of the membrane where the cleavage took place, which provides an opportunity to detect low-level, compartmentalized caspase activity in living cells (93).
The NucView488 system is based on a new DNA-binding dye (NucView 488) directly linked to the caspase-3 recognition peptide DEVD (90). NucView488 probe is a derivative of thiazole orange and when attached to negatively charged group of Ac-DEVD, its DNA binding ability is inactivated. Substrate cleavage by active caspase-3 initiates dye release, restoration of its high positive charge and translocation to the nucleus. Upon binding to DNA, NucView488 becomes highly fluorescent (90).
Immunocytochemical detection of the caspase cleavage products (“death substrates”)
Another approach to detect caspase activation relies on detection of the specific cleavage products, e.g. such as of poly(ADP-ribose)polymerase (PARP-1). PARP-1 is a nuclear enzyme involved in DNA repair that is activated in response to DNA damage. During apoptosis, PARP-1 is cleaved, primarily by caspase-3, to the specific 89-kDa and 24-kDa products (95,96). For many years, the electrophoretic detection of these PARP-1 products served as hallmark of apoptotic mode of cell death. The Ab recognizing the 89-kDa product became has been later adapted as a probe identifying apoptotic cells by cytometry (97). The multiparameter analysis of the cells differentially stained for PARP-1 p89 and DNA makes it possible not only to identify and quantify apoptotic cells, but also to correlate apoptosis with the cell cycle phase.
Immunocytochemical detection of activated caspases
Still another approach is based on immunocytochemical detection of the epitope of activated caspases. Their activation involves cleavage followed by assembly of the cleaved parts into the enzymatically active heterotetramer (98). Abs specific to several activated caspases have been developed and are commercially available. Similar to immunocytochemical detection of the PARP-1 cleavage (p89) (97), the multiparameter analysis of caspases activation and DNA content by cytometry allows one to correlate the activation with the cell cycle phase (99).
Fluorochrome-labeled inhibitors of caspases
Activation of caspases can also be detected with the use of fluorochrome-labeled inhibitors of caspases (FLICA) (99,100). Together with PhiPhiLux system and the NucView 488 system they constitute most live cell caspase detection assays available to date. In contrast to PhiPhiLux and NucView 488, however, FLICAs were designed as affinity ligands to the active enzyme centers of individual caspases. Each FLICA has three functionally distinct moieties: (a) the fluorochrome (carboxyfluorescein; FAM or sulforhodamine; SR), (b) the caspase recognition domain comprising a four amino-acid peptide, and (c) the covalent binding moiety which consists of chloro- or fluoromethyl ketone (CMK or FMK) that covalently binds to cysteine of the respective caspase forming thiomethyl ketone (101). FLICAs are plasma membrane permeant and relatively nontoxic. Several FLICAs kits are commercially available, including FAM (or SR) VAD-FMK which contains the valyl-alanyl-aspartic acid residue sequence. This three amino acid target sequence allows the inhibitor to bind to activated caspase-1, -3 -4, -5, -7, -8, and -9 making it a pan-caspase marker.
Exposure of live cells to FLICAs results in the uptake of these reagents followed by their covalent binding to cells having activated caspases, while unbound FLICAs are removed from cells that lack activated caspases by rinsing with buffer. Concurrent probing of the plasma membrane integrity with propidium iodide (PI) allows one to distinguish at least two sequential stages of apoptosis, FLICA+/PI− and FLICA+/PI+. FLICA may also be used concurrently with a probe of mitochondrial potential, such as Mitotracker Red CMXRos (99).
Because FLICAs bind covalently, they remain attached after cell fixation (with formaldehyde) and permeabilization. The FLICA assay therefore can be combined with the analysis of the attributes that can be measured after cell fixation e.g. such as DNA content (cell cycle phase) or DNA fragmentation (TUNEL assay).
A caution should be exercised, however, in interpreting the specificity of FLICA binding (102). It was recently shown that in addition to caspases, FLICAs can bind to other targets which become accessible in the course of caspases activation (99,102). The reactivity of fluoromethyl ketone moiety with intracellular thiols may provide some explanation of these interactions. In this context, opening of the disulfide cysteine bridges (inter- and/or intramolecular) may provide as yet unidentified affinity sites (102). Noncovalent hydrophobic interactions between fluorochrome domain and intracellular targets have also been postulated to play a role in FLICA retention (102). In any case FLICA reagents have proven to be reliable and sensitive markers of apoptotic cell death. Necrotic cells do not exhibit FLICA staining and caspase-3 activation assay correlates well with results obtained by fluorochrome-labeled inhibitors of caspases. Recently, data suggest also superior applicability of FLICAs in a plethora of multiparametric applications. Moreover, the covalent labeling of apoptotic cells with FLICA make these probes, called FLIVO, the markers of choice for detection of apoptosis in vivo, both in real-time and after fixation of the tissue. Nevertheless, in light of recent reports one should be aware that staining with FLICA apparently does not represent affinity labeling of individual caspase active centers (102).
GFP-FRET caspase activation assay
Activation of caspases can also be assayed using the tandem molecules of green fluorescent protein (GFP) and blue fluorescent protein (BFP), or cyan-(CFP) and yellow-fluorescent protein (YFP), linked by a short peptide, the target of a particular caspase (103–106). The fluorescence resonance energy transfer (FRET) between the pairs of these fluorescent proteins when they are linked by the peptide is terminated upon cleavage of the linker. Caspase activation, thus, is revealed by loss of FRET (103–106). The advantage of this approach is that the marker of caspase activation is intrinsic, operating in the live cell. The method, thus, is simple and rapid, as there is no need for cell fixation, application of any external fluorochromes, etc. The panel of different cells lines transfected to constitutively express the tandem FRET fluorescing proteins would represent the optimal setting for drug screening, by monitoring caspase activation/apoptosis in the treated cells.
Phosphorylation of Histone H2AX on Ser-139 as a Marker of Apoptosis
Extensive DNA fragmentation is a hallmark of apoptosis and there are several ways to assess it. The gold standard assay is based on labeling 3′OH termini of DNA strand breaks with fluorochrome-tagged deoxynucleotides in the reaction utilizing terminal deoxynucleotidyl transferase (“TUNEL” assay) (24,107). Because DNA fragmentation may lead to extraction of small molecular weight DNA fragments from the cell or their extrusion via apoptotic bodies, apoptotic cells are often characterized by fractional DNA content and their population on the DNA frequency histograms is then defined as “subG1 cells.” Extensive fragmentation of DNA during apoptosis also triggers DNA damage response (DDR). One of the characteristic features of DDR is phosphorylation of histone H2AX on Ser-139. The extent of DNA fragmentation and therefore H2AX phosphorylation during apoptosis is at least an order of magnitude greater than that induced by pharmacological doses of ionizing radiation or DNA damaging drugs (108,109). As shown in Figure 5, the immunocytochemical detection of Ser-135 phosphorylated H2AX (defined as γH2AX) using phospho-specific Ab, provides the means to identify apoptotic cells based on their extremely high fluorescence intensity. It should be noted, however, that during late stages of apoptosis the level of γH2AX immunofluorescence may decline as a result of progressive proteolysis that leads to histones degradation (109).
Figure 5.
Identification of apoptotic cells based on high level of expression of γH2AX. Leukemic HL-60 cells were untreated (Ctrl) or treated with 150 nM topotecan (Tpt) for 1 or 3 hours. Expression of γH2AX and of activated (cleaved) caspase-3 (caspase-3*) was detected immunocytochemically using fluorochromes of different emission wavelength, DNA was counterstained with DAPI, fluorescence measured by laser scanning cytometry. Induction of primary DNA double-strand breaks (Pr-DSB) by Tpt triggers H2AX phosphorylation (induction of γH2AX) which is evident after 1 hour. DNA fragmentation during apoptosis which occurs after 3 hours generates high number of DSB which leads to an additional, very intense response by expression of γH2AX. Apoptotic cells, Ap, are characterized by both, activation of caspase-3 and very high level of γH2AX. The dashed straight lines show the maximal threshold of γH2AX and caspase-3* expression (mean + 3 SD) of the untreated cells. Compared with early apoptotic cells expression of γH2AX is reduced during late phase of apoptosis (108—110).
Supravital Assessment of DNA Content
Measurement of many apoptotic features can be then directly correlated with the cell cycle phase or DNA ploidy of the tumor cell population. The most common problem associated with analysis of DNA content is the need for cell permeabilization to stain nuclei with fluorescent probes such as PI, 7-AAD, or TO-PRO 3. This often precludes concurrent analysis with supravital markers and/or kinetic analysis. Until very recently the only probe that permitted for supravital estimation of DNA content on intact cells was Hoechst 33342 (110). This fluorochrome is lipophilic, and unlike most other DNA probes, crosses the intact cell membrane. Hoechst 33342 requires, however, costly true UV excitation line and dedicated optics still uncommon on many flow cytometers. Moreover, while limited binding of Hoechst to DNA of stem cells appears to be nontoxic, the interactions with nonstem cells was shown to induce single-strand DNA breaks, particularly in combination with UV irradiation (111—113).
The DRAQ5 is an example of new fluorochromes, developed to overcome disadvantages of Hoechst 33342 (114) (Fig. 6). Unlike Hoechst 33342, DRAQ5 can be excited at longer a wavelength (488 and 633 nm) which markedly extends its applications (114). It belongs, however, to a class of the DNA intercalating agents of the anthraquinone family, very closely related to the anticancer drug mitoxantrone (2). This raises the concerns about its applicability in the long-term cell culture and viable cell sorting (118). Indeed, it was recently observed that changes in chromatin after exposure to DRAQ5 are largely incompatible with cell survival (115,116,119). This does not, however, diminishes the value of DRAQ5 as an useful plasma membrane permeant DNA dye for short-term experiments, especially when multiplexed with other spectrally favorable fluorochromes (115,116,119) (Fig. 6).
Figure 6.
Supravital assessment of DNA content. (A) Comparison between propidium iodide (PI), DRAQ5, and DyeCycle Orange (DCO) probes. Human histiocytic leukemia U937 cells were challenged with topoisomerase II inhibitor (Etoposide); topoisomerase I inhibitor (Camptothecin), or left untreated (Control). Cells were live stained with DRAQ5 and DCO for 20 minutes at RT. For PI staining cells were harvested, permeabilized in 70% EtOH, and digested with RNase A. Note that resolution of DNA profile when using DRAQ5 and DCO is satisfactory to assess cell cycle block in response to Etoposide and Camptothecin. Interestingly both DRAQ5 and DCO permit also for supravital assessment of sub-G1 fraction. (B) Comparison between DRAQ5 and ethidium bromide in a HTS screen of selected anticancer drugs. Note that permeabilisation and fixation result in loss of informative cells in the sub-G1 fraction (thick arrow). The same population is retained when DRAQ5 is applied supravitaly for DNA content analysis (data courtesy of Roy Edward, Biostatus Limited Shepshed, UK) (114—117). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Vybrant Dye Cycle probes are yet another example of cell permeable fluorochromes excited with inexpensive violet and blue diode lasers (120—122). Vybrant DyeCycle Violet (DCV) together with two other related dyes Vybrant DyeCycle Green (DCG) and Vybrant DyeCycle Orange (DCO) are widely advertised for assessment of the cell cycle distribution in live cells (Fig. 6). In our recent work we showed, however, that DCO can cause profound cytotoxic effects in U937 and HL-60 cells that render long-term experiments impossible (117). Moreover, the most prominent effect of treatment of carcinoma A549 cells with DCV was suppression of cell cycle progression through G2M and S (116). Exposure to DCV can also activate DNA damage response kinases ATM and Chk2 and phosphorylation of p53 protein (116). In summary, we speculate that probably all supravital stochiometric DNA probes induce varying degrees of DNA damage response and caution should be taken when designing dynamic experiments on live cells with these probes (116).
Fixable Viability Stains
The need for sample transportation and storage often requires cell fixation. Cell fixation can be also used to obtain information on the cell cycle phase specificity of apoptosis by a concurrent analysis with the cellular DNA content. This makes, however, most of the supravital methods such as i.e. the assays of plasma membrane integrity incompatible with subsequent processing. Recently, a considerable progress has been made by the introduction of amine reactive, fixable viability dyes (ViD) (123). Prior staining with ViD facilitates lucid discrimination of cells with intact and damaged plasma membrane even in fixed specimens. Reportedly, ViD probes span a broad range of visible excitation and emission spectra. Their uptake by cells with permeabilized membranes is followed by covalent binding to cytoplasmic amine residues that withstand formaldehyde fixation and alcohol permeabilisation procedures (123).
Dynamic Tracking of Apoptosis
Cell death is a stochastic process initiated and executed through multiple and interconnected signaling pathways. Majority of techniques to quantify cell death exploit the endpoint principle where experiment is interrupted before staining is performed. This approach reveals only a frequency of live versus apoptotic or necrotic cells at the time of cell harvesting (73,117). The possibility to continuously track individual cells up to the point of their demise would offer a superior sensitivity and accuracy allowing one to assess dynamics of the apoptotic process (76,77). Reduction of sample processing steps achieved with real-time assays can be also beneficial for preservation of fragile apoptotic population (73,76,77).
As discussed earlier in this review, several approaches to dynamically track caspase activation in living cells have been developed. Among them are cell permeable fluorogenic caspase substrates such as PhiPhiLux, NucView 488, and fluorochrome-labeled inhibitors of caspases. They are all relatively nontoxic to live cells but identify cells with activated caspases. By virtue of slowing down the process of disintegration of apoptotic cells (“stathmo-apoptosis”) FLICA can be used to estimate kinetics apoptosis in cell populations (124). Because of paucity of fluorescent probes that can be applied to measure extent of apoptosis in living organisms the use in vivo FLICA reagents (defined “FLIVO”) is of particular interest (125,126). Another group of the probes applicable to live cells are also already mentioned tandem molecules exploiting FRET of green fluorescent proteins. These probes can be reportedly used in real-time or near real-time assays featuring superior sensitivity and wealth of information as compared to standard end-point assays.
We have recently reported that also substituted cyanine SYTO probes represent a promising class of markers that do not adversely affect normal cellular physiology. When preloaded into cells or continuously present in medium, SYTOs do not interfere with cell viability while their intracellular retention can be easily used for kinetic analysis of caspase-dependent apoptosis (73,117). SYTO 16-based sorting of intact apoptotic cells provides also an innovative approach to supravitally track progression of apoptotic cascade (73).
Autophagy and Cytometry
Autophagy is an intracellular bulk degradation system for long-lived proteins and whole organelles (127). Emerging evidence suggests that autophagy may enhance survival of cancer cells exposed to nutrient deprivation, hypoxia, or certain chemotherapeutics, but contribute to cell death when induced above an acceptable for cellular homeostasis threshold (128). Accurate estimation of autophagosome formation and/or functional catabolic autophagy emerges, thus, as an important measurement in preclinical drug screening (129,130). To date, only a handful of methods have been introduced to quantify autophagy, including electron and fluorescent microscopy to follow steady-state accumulation of autophagosomes, and long-lived protein degradation assay to access the catabolic autophagic activity (131,132). Fluorescent microscopy is widely used to follow autophagosome accumulation using markers such as LC3 protein tagged with fluorescent proteins. Upon induction of autophagy cytoplasmatically localized LC3-I is cleaved and lipidated to form LC3-II that associates with the isolation membrane (131). By using, for e.g., adeno-viral delivery of LC3-GFP, it is possible to follow the changes in LC3-GFP distribution from diffuse cytoplasmic into punctuate, the latter indicative of autophagosome accumulation. Current methods designed to quantify autophagic activity using LC3 are, however, time consuming, labor intensive, and require substantial expertise in accurate data interpretation (133,134).
Recently, several attempts have been made to quantify autophagy in cells stably expressing GFP-LC3 reporters using flow cytometry (133,134). As discussed however, in flow cytometry, only integrated fluorescence over each cell is collected. This in turn may not be sensitive enough to detect subtle redistribution at subcellular level. More recently, a successful attempt has been made to employ the multispectral imaging flow cytometry to quantify autophagosome formation (135). Authors utilized the “virtual sort” capability to enumerate cells exhibiting the bright, punctuate spots of GFP-LC3. The in-flow imaging is the first example of an automated and unbiased detection of autophagy in rare subpopulations of cells (135).
SUMMARY
In closing, key technological advances allowed unparallel proliferation of cytometry in studies of cell death. Although the present array of techniques has delivered many options, there are still numerous areas for technological improvements. Discovery of alternative cell death modes have brought potential for development of innovative bioassays. Regrettably only the classical caspase-dependent apoptosis has so far been a major area that drives assay development for the cytometry. As exploration of novel therapeutic targets based on noncanonical pathways of cell death is ongoing, there is a dire need for novel bioassays. Development of virtually inert fluorescent probes with enhanced photostability would also open up new horizons for many functional and real-time cytometric assays.
There is a reason to believe, that progress in novel technologies like laser scanning cytometry (LSC), multispectral imaging cytometry, and spectroscopic cytometry is just a prelude to a major transformation of the field in the years to come. Expectedly, we are soon to witness the rise of micro-and nanotechnologies with vast potential in many analytical platforms. Although still in their infancy, these warrant a major “quantum leap” for cell necrobiology.
ACKNOWLEDGMENTS
The authors thank Dr. Tad George and Dr. Brian Hall (Amnis Corporation, Seattle, WA) for generously providing Image StreamX data; Dr. Kazuo Takeda (On-Chip Biotechnologies Co., Tokyo, Japan) for providing data on Fish-manR microfluidic cytometer; and Roy Edward (Biostatus Limited Shepshed, UK) for sharing DRAQ5 and ethidium bromide data.
LITERATURE CITED
- 1.Darzynkiewicz Z, Juan G, Li X, Gorczyca W, Murakami T, Traganos F. Cytometry in cell necrobiology: Analysis of apoptosis and accidental cell death (necrosis) Cytometry. 1997;27:1–20. [PubMed] [Google Scholar]
- 2.Darzynkiewicz Z, Bedner E, Traganos F. Difficulties and pitfalls in analysis of apoptosis. Methods Cell Biol. 2001;63:527–546. doi: 10.1016/s0091-679x(01)63028-0. [DOI] [PubMed] [Google Scholar]
- 3.Telford WG, Komoriya A, Packard BZ. Multiparametric analysis of apoptosis by flow and image cytometry. Methods Mol Biol. 2004;263:141–160. doi: 10.1385/1-59259-773-4:141. [DOI] [PubMed] [Google Scholar]
- 4.Darzynkiewicz Z, Huang X, Okafuji M, King MA. Cytometric methods to detect apoptosis. Methods Cell Biol. 2004;75:307–341. doi: 10.1016/s0091-679x(04)75012-8. [DOI] [PubMed] [Google Scholar]
- 5.Bonetta L. Flow cytometry smaller and better. Nat Methods. 2005;2:785–795. [Google Scholar]
- 6.Eisenstein M. Divide and conquer. Nature. 2006;441:1179–1185. doi: 10.1038/4411179a. [DOI] [PubMed] [Google Scholar]
- 7.Melamed MR. A brief history of flow cytometry and sorting. Methods Cell Biol. 2001;63:3–17. doi: 10.1016/s0091-679x(01)63005-x. [DOI] [PubMed] [Google Scholar]
- 8.Robinson JP. Multispectral cytometry: The next generation. Biophotonics Int. 2006;10:36–40. [Google Scholar]
- 9.Shapiro H. The evolution of cytometers. Cytometry Part A: J Intl Soc Anal Cytol. 2004;58A:13–17. doi: 10.1002/cyto.a.10111. [DOI] [PubMed] [Google Scholar]
- 10.Kerr JF, Wyllie AH, Currie AR. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockshin RA, Zakeri Z. Programmed cell death and apoptosis: Origins of the theory. Nat Rev Mol Cell Biol. 2001;2:545–550. doi: 10.1038/35080097. [DOI] [PubMed] [Google Scholar]
- 12.Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am J Pathol. 1995;146:3–16. [PMC free article] [PubMed] [Google Scholar]
- 13.Leist M, Jaattela M. Four deaths and a funeral: From caspases to alternative mechanisms. Nat Rev Mol Cell Biol. 2001;2:589–598. doi: 10.1038/35085008. [DOI] [PubMed] [Google Scholar]
- 14.Kroemer G, Martin SJ. Caspase-independent cell death. Nat Med. 2005;11:725–730. doi: 10.1038/nm1263. [DOI] [PubMed] [Google Scholar]
- 15.Levine B, Yuan J. Autophagy in cell death: An innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broker LE, Kruyt FA, Giaccone G. Cell death independent of caspases: A review. Clin Cancer Res. 2005;11:3155–3162. doi: 10.1158/1078-0432.CCR-04-2223. [DOI] [PubMed] [Google Scholar]
- 17.Okada H, Mak TW. Pathways of apoptotic and non-apoptotic death in tumour cells. Nat Rev Cancer. 2004;4:592–603. doi: 10.1038/nrc1412. [DOI] [PubMed] [Google Scholar]
- 18.Edinger AL, Thompson CB. Death by design: Apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Blagosklonny MV. Cell death beyond apoptosis. Leukemia. 2000;14:1502–1508. doi: 10.1038/sj.leu.2401864. [DOI] [PubMed] [Google Scholar]
- 20.Zhivotovsky B. Apoptosis, necrosis and between. Cell Cycle. 2004;3:64–66. [PubMed] [Google Scholar]
- 21.Ziegler U, Groscurth P. Morphological features of cell death. News Physiol Sci. 2004;19:124–128. doi: 10.1152/nips.01519.2004. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y. Mechanisms of caspases activation and inhibition during apoptosis. Mol Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 23.Joza N, Susin SA, Gaugas E, Stanford WL, Cho SK, Li CYI, Sasaki T, Elia AJ, Cheng HYM, Ravagnan L, Ferri KF, Zamzani N, Wakeham A, Hakem R, Yoshida H, Kong YY, Mak TW, Zuniga-Pflucker JC, Kroemer G, Penninger JM. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410:549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 24.Gorczyca W, Bruno S, Darzynkiewicz R, Gong J, Darzynkiewicz Z. DNA strand breaks occurring during apoptosis: Their early in situ detection by the terminal deoxynucleotydil transferase and nick translation assays and prevention by serine protease inhibitors. Int J Oncol. 1992;1:639–648. doi: 10.3892/ijo.1.6.639. [DOI] [PubMed] [Google Scholar]
- 25.Catchpoole DR, Stewart BW. Etoposide-induced cytotoxicity in two human T-cell leukemic lines. Delayed loss of membrane permeability rather than DNA fragmentation as an indicator of programmed cell death. Cancer Res. 1993;53:4287–4296. [PubMed] [Google Scholar]
- 26.Cohen GM, Su XM, Snowden RT, Dinsdale D, Skilleter DN. Key morphological features of apoptosis may occur in the absence of internucleosomal DNA fragmentation. Biochem J. 1992;286:331–334. doi: 10.1042/bj2860331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins RJ, Harmon BV, Gobe GC, Kerr JFR. Internucleosomal DNA cleavage should not be the sole criterion for identifying apoptosis. Int J Radiat Biol. 1992;61:451–453. doi: 10.1080/09553009214551201. [DOI] [PubMed] [Google Scholar]
- 28.Knapp PE, Bartlett WP, Williams LA, Yamada M, Ikenaka K, Skoff RP. Programmed cell death without DNA fragmentation in the jimpy mouse: Secreted factors can enhance survival. Cell Death Differ. 1999;6:136–145. doi: 10.1038/sj.cdd.4400457. [DOI] [PubMed] [Google Scholar]
- 29.Ormerod MG, O’Neill CF, Robertson D, Harrap KR. Cisplatin induced apoptosis in a human ovarian carcinoma cell line without a concomitant internucleosomal degradation of DNA. Exp Cell Res. 1994;211:231–237. doi: 10.1006/excr.1994.1082. [DOI] [PubMed] [Google Scholar]
- 30.Ferri KF, Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol. 2001;3:E255–E263. doi: 10.1038/ncb1101-e255. [DOI] [PubMed] [Google Scholar]
- 31.George TC, Basiji DA, Hall BE, Lynch DH, Ortyn WE, Perry DJ, Seo MJ, Zimmerman CA, Morrissey PJ. Distinguishing modes of cell death using the ImageStream multispectral imaging flow cytometer. Cytometry Part A: J Intl Soc Anal Cytol. 2004;59A:237–245. doi: 10.1002/cyto.a.20048. [DOI] [PubMed] [Google Scholar]
- 32.Kamentsky LA, Kamentsky LD. Microscope-based multiparameter laser scanning cytometer yielding data comparable to flow cytometry data. Cytometry. 1991;12:381–387. doi: 10.1002/cyto.990120502. [DOI] [PubMed] [Google Scholar]
- 33.Chan SD, Luedke G, Valer M, Buhlmann C, Preckel T. Cytometric analysis of protein expression and apoptosis in human primary cells with a novel microfluidic chip-based system. Cytometry Part A: J Intl Soc Anal Cytol. 2003;55A:119–125. doi: 10.1002/cyto.a.10070. [DOI] [PubMed] [Google Scholar]
- 34.Huh D, Gu W, Kamotani Y, Grotberg JB, Takayama S. Microfluidics for flow cytometric analysis of cells and particles. Physiol Meas. 2005;26:R73–R98. doi: 10.1088/0967-3334/26/3/R02. [DOI] [PubMed] [Google Scholar]
- 35.Kapoor V, Subach FV, Kozlov VG, Grudinin A, Verkhusha VV, Telford WG. New lasers for flow cytometry: Filling the gaps. Nat Methods. 2007;4:678–679. doi: 10.1038/nmeth0907-678. [DOI] [PubMed] [Google Scholar]
- 36.Telford WG, Babin SA, Khorev SV, Rowe SH. Green fiber lasers: An alternative to traditional DPSS green lasers for flow cytometry. Cytometry Part A: J Intl Soc Anal Cytol. 2009;75A:1031–1039. doi: 10.1002/cyto.a.20790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapoor V, Karpov V, Linton C, Subach FV, Verkhusha VV, Telford WG. Solid state yellow and orange lasers for flow cytometry. Cytometry Part A: J Intl Soc Anal Cytol. 2008;73A:570–577. doi: 10.1002/cyto.a.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Telford WG. Analysis of UV-excited fluorochromes by flow cytometry using near-ultraviolet laser diodes. Cytometry Part A: J Intl Soc Anal Cytol. 2004;61A:9–17. doi: 10.1002/cyto.a.20032. [DOI] [PubMed] [Google Scholar]
- 39.Telford W, Kapoor V, Jackson J, Burgess W, Buller G, Hawley T, Hawley R. Violet laser diodes in flow cytometry: An update. Cytometry Part A: J Intl Soc Anal Cytol. 2006;69A:1153–1160. doi: 10.1002/cyto.a.20340. [DOI] [PubMed] [Google Scholar]
- 40.George TC, Fanning SL, Fitzgerald-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y, Hall BE, Frost K, Basiji D, Ortyn WE, Morrissey PJ, Lynch DH. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods. 2006;311:117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 41.McGrath KE, Bushnell TP, Palis J. Multispectral imaging of hematopoietic cells: where flow meets morphology. J Immunol Methods. 2008;336:91–97. doi: 10.1016/j.jim.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuba-Surma EK, Kucia M, Abdel-Latif A, Lillard JW, Jr, Ratajczak MZ. The Image-Stream System: A key step to a new era in imaging. Folia Histochem Cytobiol. 2007;45:279–290. [PubMed] [Google Scholar]
- 43.Henery S, George T, Hall B, Basiji D, Ortyn W, Morrissey P. Quantitative image based apoptotic index measurement using multispectral imaging flow cytometry: A comparison with standard photometric methods. Apoptosis. 2008;13:1054–1063. doi: 10.1007/s10495-008-0227-4. [DOI] [PubMed] [Google Scholar]
- 44.Mattha¨us C, Chernenko T, Newmark JA, Warner CM, Diem M. Label-free detection of mitochondrial distribution in cells by nonresonant Raman microspectroscopy. Biophys J. 2007;93:668–673. doi: 10.1529/biophysj.106.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toms SA, Konrad PE, Lin WC, Weil RJ. Neuro-oncological applications of optical spectroscopy. Technol Cancer Res Treat. 2006;5:231–238. doi: 10.1177/153303460600500306. [DOI] [PubMed] [Google Scholar]
- 46.Notingher I, Hench LL. Raman microspectroscopy: A noninvasive tool for studies of individual living cells in vitro. Expert Rev Med Devices. 2006;3:215–234. doi: 10.1586/17434440.3.2.215. [DOI] [PubMed] [Google Scholar]
- 47.Jett JH. Raman spectroscopy comes to flow cytometry. Cytometry Part A: J Intl Soc Anal Cytol. 2008;73A:109–110. doi: 10.1002/cyto.a.20526. [DOI] [PubMed] [Google Scholar]
- 48.Kneipp J, Kneipp H, Wittig B, Kneipp K. Novel optical nanosensors for probing and imaging live cells. Nanomedicine. 2010 doi: 10.1016/j.nano.2009.07.009. in press [DOI:10.1016/j.nano.2009.07.009] [DOI] [PubMed] [Google Scholar]
- 49.Kneipp J, Kneipp H, Kneipp K. SERS-a single-molecule and nanoscale tool for bioanalytics. Chem Soc Rev. 2008;37:1052–1060. doi: 10.1039/b708459p. [DOI] [PubMed] [Google Scholar]
- 50.Uzunbajakava N, Lenferink A, Kraan Y, Volokhina E, Vrensen G, Greve J, Otto C. Nonresonant confocal Raman imaging of DNA and protein distribution in apoptotic cells. Biophys J. 2003;84:3968–3981. doi: 10.1016/S0006-3495(03)75124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kunapareddy N, Freyer JP, Mourant JR. Raman spectroscopic characterization of necrotic cell death. J Biomed Opt. 2008;13:054002. doi: 10.1117/1.2978061. [DOI] [PubMed] [Google Scholar]
- 52.Swain RJ, Jell G, Stevens MM. Non-invasive analysis of cell cycle dynamics in single living cells with Raman micro-spectroscopy. J Cell Biochem. 2008;104:1427–1438. doi: 10.1002/jcb.21720. [DOI] [PubMed] [Google Scholar]
- 53.Watson DK, Brown LO, Gaskill DF, Naivar M, Graves SW, Doorn SK, Nolan JP. A flow cytometer for the measurement of Raman spectra. Cytometry Part A: J Intl Soc Anal Cytol. 2008;73A:119–128. doi: 10.1002/cyto.a.20520. [DOI] [PubMed] [Google Scholar]
- 54.Bedner E, Li X, Gorczyca W, Melamed MR, Darzynkiewicz Z. Analysis of apoptosis by laser scanning cytometry. Cytometry. 1999;35:181–195. doi: 10.1002/(sici)1097-0320(19990301)35:3<181::aid-cyto1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 55.Oode K, Furuya T, Harada K, Kawauchi S, Yamamoto K, Hirano T, Sasaki K. The development of a cell array and its combination with laser-scanning cytometry allows a high-throughput analysis of nuclear DNA content. Am J Pathol. 2000;157:723–728. doi: 10.1016/S0002-9440(10)64585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takita M, Furuya T, Sugita T, Kawauchi S, Oga A, Hirano T, Tsunoda S, Sasaki K. An analysis of changes in the expression of cyclins A and B1 by the cell array system during the cell cycle: comparison between cell synchronization methods. Cytometry Part A: J Intl Soc Anal Cytol. 2003;55A:24–29. doi: 10.1002/cyto.a.10066. [DOI] [PubMed] [Google Scholar]
- 57.Wheeler DB, Carpenter AE, Sabatini DM. Cell microarrays and RNA interference chip away at gene function. Nat Genet. 2005;(37 Suppl):S25–S30. doi: 10.1038/ng1560. [DOI] [PubMed] [Google Scholar]
- 58.Wheeler DB, Bailey SN, Guertin DA, Carpenter AE, Higgins CO, Sabatini DM. RNAi living-cell microarrays for loss-of-function screens in Drosophila melanogaster cells. Nat Methods. 2004;1:127–132. doi: 10.1038/nmeth711. [DOI] [PubMed] [Google Scholar]
- 59.Castel D, Pitaval A, Debily MA, Gidrol X. Cell microarrays in drug discovery. Drug Discov Today. 2006;11:616–622. doi: 10.1016/j.drudis.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Becker FF, Gascoyne PRC. Membrane dielectric changes indicate induced apoptosis in HL-60 cells more sensitively than surface phosphatidylserine expression or DNA fragmentation. Biochim Biophys Acta. 2002;1564:412–420. doi: 10.1016/s0005-2736(02)00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheung K, Gawad S, Renaud P. Impedance spectroscopy flow cytometry: On-chip label-free cell differentiation. Cytometry Part A: J Intl Soc Anal Cytol. 2005;65A:124–132. doi: 10.1002/cyto.a.20141. [DOI] [PubMed] [Google Scholar]
- 62.Sohn LL, Saleh OA, Facer GR, Beavis AJ, Allan RS, Notterman DA. Capacitance cytometry: Measuring biological cells one by one. Proc Natl Acad Sci USA. 2000;97:10687–10690. doi: 10.1073/pnas.200361297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Atienza JM, Zhu J, Wang X, Xu X, Abassi Y. Dynamic monitoring of cell adhesion and spreading on microelectronic sensor arrays. J Biomol Screen. 2005;10:795–805. doi: 10.1177/1087057105279635. [DOI] [PubMed] [Google Scholar]
- 64.Zhu J, Wang X, Xu X, Abassi YA. Dynamic and label-free monitoring of natural killer cell cytotoxic activity using electronic cell sensor arrays. Immunol Methods. 2006;309:25–33. doi: 10.1016/j.jim.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 65.Atienza JM, Yu N, Kirstein SL, Xi B, Wang X, Xu X, Abassi YA. Dynamic and label-free cell-based assays using the real-time cell electronic sensing system. Assay Drug Dev Technol. 2006;4:597–607. doi: 10.1089/adt.2006.4.597. [DOI] [PubMed] [Google Scholar]
- 66.Xi B, Yu N, Wang X, Xu X, Abassi YA. The application of cell-based label-free technology in drug discovery. Biotechnol J. 2008;3:484–495. doi: 10.1002/biot.200800020. [DOI] [PubMed] [Google Scholar]
- 67.Andersson H, van den Berg A. Microtechnologies and nanotechnologies for single-cell analysis. Curr Opin Biotechnol. 2004;15:44–49. doi: 10.1016/j.copbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 68.Sia SK, Whitesides GM. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis. 2003;24:3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 69.Svahn HA, van den Berg A. Single cells or large populations? Lab Chip. 2007;7:544–546. doi: 10.1039/b704632b. [DOI] [PubMed] [Google Scholar]
- 70.Manz A, Dittrich PS. Lab-on-a-chip: Microfluidics in drug discovery. Nat Rev Drug Discov. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 71.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–372. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 72.Sims CE, Allbritton NL. Analysis of single mammalian cells on-chip. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 73.Wlodkowic D, Skommer J, Faley S, Darzynkiewicz Z, Cooper JM. Dynamic analysis of apoptosis using cyanine SYTO probes: From classical to microfluidic cytometry. Exp Cell Res. 2009;315:1706–1714. doi: 10.1016/j.yexcr.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takeda K, Jimma F. Maintenance free biosafety flowcytometer using disposable microfluidic chip (FISHMAN-R) Cytometry B Clin Cytom. 2009;76B:405–406. [Google Scholar]
- 75.Takao M, Jimma F, Takeda K. Expanded applications of new-designed microfluidic flow cytometer (FISHMAN-R) Cytometry B Clin Cytom. 2009;76B:405. [Google Scholar]
- 76.Wlodkowic D, Faley S, Zagnoni M, Wikswo JP, Cooper JM. Microfluidic single-cell array cytometry for the analysis of tumor apoptosis. Anal Chem. 2009;81:5517–5523. doi: 10.1021/ac9008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Faley SL, Copland M, Wlodkowic D, Kolch W, Seale KT, Wikswo JP, Cooper JM. Microfluidic single cell arrays to interrogate signalling dynamics of individual, patient-derived hematopoietic stem cells. Lab Chip. 2009;9:2659–2664. doi: 10.1039/b902083g. [DOI] [PubMed] [Google Scholar]
- 78.Jaiswal JK, Simon SM. Potential and pitfalls of fluorescent quantum dots for biological imaging. TRENDS Cell Biol. 2004;14:497–504. doi: 10.1016/j.tcb.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 79.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotech. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 80.Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup RA, de Rosa SC, Bruchez MP, Roederer M. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 81.Hanshaw RG, Lakshmi C, Lambert TN, Johnson JR, Smith BD. Fluorescent detection of apoptotic cells by zinc coordination complexes with a selective affinity for membrane surfaces enriched with phosphatidylserine. ChemBioChem. 2005;6:2214–2220. doi: 10.1002/cbic.200500149. [DOI] [PubMed] [Google Scholar]
- 82.Koulov AV, Stucker KA, Lakshmi C, Robinson JP, Smith BD. Detection of apoptotic cells using a synthetic fluorescent sensor for membrane surfaces that contain phosphatidylserine. Cell Death Differ. 2003;10:1357–1359. doi: 10.1038/sj.cdd.4401315. [DOI] [PubMed] [Google Scholar]
- 83.Laakko T, King L, Fraker P. Versatility of merocyanine 540 for flow cytometric detection of apoptosis in human and murine cells. J Immunol Methods. 2002;261:129–139. doi: 10.1016/s0022-1759(01)00562-2. [DOI] [PubMed] [Google Scholar]
- 84.Idziorek T, Estaquier J, De Bels F, Ameisen JC. YOPRO-1 permits cytofluorometric analysis of programmed cell death (apoptosis) without interfering with cell viability. J Immunol Methods. 1995;185:249–258. doi: 10.1016/0022-1759(95)00172-7. [DOI] [PubMed] [Google Scholar]
- 85.Cankurtaran-Sayar S, Sayar K, Ugur M. P2×7 receptor activates multiple selective dye-permeation pathways in RAW 264.7 and human embryonic kidney 293 cells. Mol Pharmacol. 2009;76:1323–1332. doi: 10.1124/mol.109.059923. [DOI] [PubMed] [Google Scholar]
- 86.Ormerod MG, Sun XM, Snowden RT, Davies R, Fearhead H, Cohen GM. Increased membrane permeability of apoptotic thymocytes: A flow cytometric study. Cytometry. 1993;14:595–602. doi: 10.1002/cyto.990140603. [DOI] [PubMed] [Google Scholar]
- 87.Schmid I, Uittenbogaart C, Jamieson BD. Live-cell assay for detection of apoptosis by dual-laser flow cytometry using Hoechst 33342 and 7-amino-actinomycin D. Nat Protoc. 2007;2:187–190. doi: 10.1038/nprot.2006.458. [DOI] [PubMed] [Google Scholar]
- 88.Antczak C, Takagi T, Ramirez CN, Radu C, Djaballah H. Live-cell imaging of caspase activation fort high-content screening. J Biomol Screen. 2009;14:956–969. doi: 10.1177/1087057109343207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Packard BZ, Komoriya A. Intracellular protease activation in apoptosis and cell mediated cytotoxicity characterized by cell-permeable fluorogenic protease substrates. Cell Res. 2008;18:238–247. doi: 10.1038/cr.2008.17. [DOI] [PubMed] [Google Scholar]
- 90.Cen H, Mao F, Aronchik I, Fuentes RJ, Firestone GL. DEVD-NucView488: A novel class of enzyme substrates for real-time detection of caspase-3 activity in live cells. FASEB J. 2008;22:2243–2252. doi: 10.1096/fj.07-099234. [DOI] [PubMed] [Google Scholar]
- 91.Gorman AM, Hirt UA, Zhivotovsky B, Orrenius S, Ceccatelli S. Application of a fluorometric assay to detect caspase activity in thymus tissue undergoing apoptosis in vivo. J Immunol Methods. 1999;226:43–48. doi: 10.1016/s0022-1759(99)00054-x. [DOI] [PubMed] [Google Scholar]
- 92.Hug H, Los M, Hirt W, Debatin KM. Rhodamine 110-linked amino acids and peptides as substrates to measure caspase activity upon apoptosis induction in intact cells. Biochemistry. 1999;38:13906–13911. doi: 10.1021/bi9913395. [DOI] [PubMed] [Google Scholar]
- 93.Telford WG, Komoriya A, Packard BZ. Detection of localized caspase activity in early apoptotic cells by laser scanning cytometry. Cytometry Part A: J Intl Soc Anal Cytol. 2002;47A:81–88. doi: 10.1002/cyto.10052. [DOI] [PubMed] [Google Scholar]
- 94.Lee BW, Johnson GL, Hed SA, Darzynkiewicz Z, Talhouk JW, Mehrotra S. DEVDase detection in intact apoptotic cells using the cell permeant fluorogenic substrate (z-DEVD)2 -cresyl violet. Biotechniques. 2003;35:1080–1085. doi: 10.2144/03355pf01. [DOI] [PubMed] [Google Scholar]
- 95.Alnemri ES, Livingston DI, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J. Human ICE/CED-4 protease nomenclature. Cell. 1996;87:171–173. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 96.Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 97.Li X, Darzynkiewicz Z. Cleavage of poly(ADP-ribose) polymerase measured in situ in individual cells: Relationship to DNA fragmentation and cell cycle position during apoptosis. Exp Cell Res. 2000;255:125–132. doi: 10.1006/excr.1999.4796. [DOI] [PubMed] [Google Scholar]
- 98.Pop C, Salvesen GS. Human caspases: Activation, specificity, and regulation. J Biol Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pozarowski P, Huang X, Halicka DH, Lee B, Johnson G, Darzynkiewicz Z. Interactions of fluorochrome-labeled caspase inhibitors with apoptotic cells. A caution in data interpretation. Cytometry Part A: J Intl Soc Anal Cytol. 2003;55A:50–60. doi: 10.1002/cyto.a.10074. [DOI] [PubMed] [Google Scholar]
- 100.Bedner E, Smolewski P, Amstad P, Darzynkiewicz Z. Activation of caspases measured in situ by binding of fluorochrome-labeled inhibitors of caspases (FLICA): Correlation with DNA fragmentation. Exp Cell Res. 2000;259:308–313. doi: 10.1006/excr.2000.4955. [DOI] [PubMed] [Google Scholar]
- 101.Van Noorden CJF. The history of Z-VAD-FMK, a tool for understanding the significance of caspase inhibition. Acta Histochem. 2001;103:241–251. doi: 10.1078/0065-1281-00601. [DOI] [PubMed] [Google Scholar]
- 102.Darzynkiewicz Z, Pozarowski P. All that glitters is not gold: All that binds FLICA is not caspase. A caution in data interpretation- and new opportunities. Cytometry Part A: J Intl Soc Anal Cytol. 2007;71A:536–537. doi: 10.1002/cyto.a.20425. [DOI] [PubMed] [Google Scholar]
- 103.Jones J, Heim R, Hare E, Stack J, Pollok BA. Development and application of a GFP-FRET intracellular caspase assay for drug screening. J Biomol Screen. 2000;5:307–318. doi: 10.1177/108705710000500502. [DOI] [PubMed] [Google Scholar]
- 104.Kawai H, Suzuki T, Kobayashi T, Sakurai H, Ohata H, Honda K, Momose K, Namekata I, Tanaka H, Shigenobu K, Nakamura R, Hayakawa T, Kawanishi T. Simultaneous real-time detection of initiator- and effector-caspase activation by double fluorescence resonance energy transfer analysis. J Pharmacol Sci. 2005;97:361–368. doi: 10.1254/jphs.fp0040592. [DOI] [PubMed] [Google Scholar]
- 105.Luo KQ, Yu VC, Pu Y, Chang DC. Measuring dynamics of caspase-8 activation in a single living HeLa cells during THF-α-induced apoptosis. Biochem Biophys Res Commun. 2003;304:217–222. doi: 10.1016/s0006-291x(03)00559-x. [DOI] [PubMed] [Google Scholar]
- 106.Tawa P, Tam J, Cassady R, Nicholson DW, Xanthoudakis S. Quantitative analysis of fluorescent caspase substrates cleavage in intact cells and identification of novel inhibitors of apoptosis. Cell Death Differ. 2001;8:30–37. doi: 10.1038/sj.cdd.4400769. [DOI] [PubMed] [Google Scholar]
- 107.Gorczyca W, Gong J, Darzynkiewicz Z. Detection of DNA strand breaks in individual apoptotic cells by the in situ terminal deoxynucleotidyl transferase and nick translation assays. Cancer Res. 1993;53:1945–1951. [PubMed] [Google Scholar]
- 108.Halicka HD, Huang X, Traganos F, King MA, Dai W, Darzynkiewicz Z. Histone H2AX phosphorylation after cell irradiation with UV-B: Relationship to cell cycle phase and induction of apoptosis. Cell Cycle. 2005;4:339–345. [PubMed] [Google Scholar]
- 109.Huang X, Okafuji M, Traganos F, Luther E, Holden E, Darzynkiewicz Z. Assessment of histone H2AX phosphorylation induced by DNA topoisomerase I and II inhibitors topotecan and mitoxantrone and by DNA crosslinking agent cisplatin. Cytometry Part A: J Intl Soc Anal Cytol. 2004;58A:99–110. doi: 10.1002/cyto.a.20018. [DOI] [PubMed] [Google Scholar]
- 110.Darzynkiewicz Z, Crissman HA, Jacobberger JW. Cytometry of the cell cycle. Cycling through history. Cytometry Part A: J Intl Soc Anal Cytol. 2004;58A:21–32. doi: 10.1002/cyto.a.20003. [DOI] [PubMed] [Google Scholar]
- 111.Godell MA, Brose K, Paradis G, Conner AS, Mullican RC. Isolation and functional properties of mutine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Durand RE, Olive PL. Cytotoxicity, mutagenicity and DNA damage by Hoechst 33342. J Histochem Cytochem. 1982;30:111–116. doi: 10.1177/30.2.7061816. [DOI] [PubMed] [Google Scholar]
- 113.Singh S, Dwarakanath BS, Mathew TL. DNA ligand Hoechst-33342 enhances UV induced cytotoxicity in human glioma cells. J Photochem Photobiol B. 2004;77:45–54. doi: 10.1016/j.jphotobiol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 114.Smith PJ, Blunt N, Wiltshire M, Hoy T, Teesdale-Spittle P, Craven MR, Watson JV, Amos WB, Errington RJ, Patterson LH. Characteristics of a novel deep red/infrared fluorescent cell permeant DNA probe. DRAQ5, in intact human cells analyzed by flow cytometry and multiphoton microscopy. Cytometry. 2000;40:280–291. doi: 10.1002/1097-0320(20000801)40:4<280::aid-cyto4>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 115.Wojcik K, Dobrucki JW. Interaction of a DNA intercalator DRAQ5, and a minor groove binder SYTO17, with chromatin in live cells—Influence on chromatin organization and histone-DNA interactions. Cytometry Part A: J Intl Soc Anal Cytol. 2008;73A:555–562. doi: 10.1002/cyto.a.20573. [DOI] [PubMed] [Google Scholar]
- 116.Zhao H, Traganos F, Dobrucki J, Wlodkowic D, Darzynkiewicz Z. Induction of DNA damage response by the supravital probes of nucleic acids. Cytometry Part A: J Intl Soc Anal Cytol. 2009;75A:510–519. doi: 10.1002/cyto.a.20727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wlodkowic D, Skommer J, Darzynkiewicz Z. SYTO probes in the cytometry of tumor cell death. Cytometry Part A: J Intl Soc Anal Cytol. 2008;73A:496–507. doi: 10.1002/cyto.a.20535. [DOI] [PubMed] [Google Scholar]
- 118.Kapuscinski J, Darzynkiewicz Z. Interactions of antitumor agents ametantrone and mitoxantrone (novantrone) with double-stranded DNA. Biochem Pharmacol. 1985;34:4203–4213. doi: 10.1016/0006-2952(85)90275-8. [DOI] [PubMed] [Google Scholar]
- 119.Martin RM, Leonhardt H, Cardoso MC. DNA labeling in living cells. Cytometry Part A: J Intl Soc Anal Cytol. 2005;67A:45–52. doi: 10.1002/cyto.a.20172. [DOI] [PubMed] [Google Scholar]
- 120.Telford WG, Bradford J, Godfrey W, Robey RW, Bates SE. Side population analysis using a violet-excited cell permeable DNA binding dye. Stem Cells. 2007;25:1029–1036. doi: 10.1634/stemcells.2006-0567. [DOI] [PubMed] [Google Scholar]
- 121.She JJ, Zhang PG, Wang ZM, Gan WM, Che XM. Identification of side population cells from bladder cancer cells by DyeCycle Violet staining. Cancer Biol Ther. 2008;7:1663–1669. doi: 10.4161/cbt.7.10.6637. [DOI] [PubMed] [Google Scholar]
- 122.Mathew G, Timm EA, Jr, Sotomayor P, Godoy A, Montecinos VP, Smith GJ, Huss WJ. ABCG2-mediated DyeCycle Violet efflux defined side population in benign and malignant prostate. Cell Cycle. 2009;8:1–9. doi: 10.4161/cc.8.7.8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prefetto SP, Chattopadhyay PK, Lamoreaux L, Nguyen R, Ambrozak D, Koup RA, Roederer M. Amine reactive dyes: An effective tool to discriminate live ad dead cells in polychromatic flow cytometry. J Immunol Methods. 2006;313:199–208. doi: 10.1016/j.jim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 124.Smolewski P, Grabarek J, Lee BW, Johnson GL, Darzynkiewicz Z. Kinetics of HL-60 cell entry to apoptosis during treatment with TNF-α or camptothecin assayed by the stathmo-apoptosis method. Cytometry. 2002;47:143–149. doi: 10.1002/cyto.10062. [DOI] [PubMed] [Google Scholar]
- 125.Cursio R, Colosetti P, Auberger P, Gugenheim J. Liver apoptosis following nor-mothermic ischemia-reperfusion: In vivo evaluation of casapase activity by FLIVO assay in rats. Transplant Proc. 2008;40:2038–2041. doi: 10.1016/j.transproceed.2008.05.039. [DOI] [PubMed] [Google Scholar]
- 126.Darzynkiewicz Z, Pozarowski P, Lee BW, Johnson GL. Fluorochrome-labeled inhibitors of caspases (FLICA): Convenient in vitro and in vivo markers of apoptotic cells for cytometric analysis. Meth Mol Biol. doi: 10.1007/978-1-60327-409-8_9. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Meijer AJ, Codogno P. Autophagy: Regulation and role in disease. Crit Rev Clin Lab Sci. 2009;46:210–240. doi: 10.1080/10408360903044068. [DOI] [PubMed] [Google Scholar]
- 128.Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 129.Corcelle EA, Puustinen P, Jäättelä M. Apoptosis and autophagy: Targeting autophagy signalling in cancer cells—”trick or treats”? FEBS J. 2009;276:6084–6096. doi: 10.1111/j.1742-4658.2009.07332.x. [DOI] [PubMed] [Google Scholar]
- 130.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 131.Gurusamy N, Das DK. Detection of cell death by autophagy. Methods Mol Biol. 2009;559:95–103. doi: 10.1007/978-1-60327-017-5_7. [DOI] [PubMed] [Google Scholar]
- 132.Swanlund JM, Kregel KC, Oberley TD. Investigating autophagy: Quantitative mor-phometric analysis using electron microscopy. Autophagy. 2010;6:270–277. doi: 10.4161/auto.6.2.10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4:621–628. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 134.Shvets E, Elazar Z. Flow cytometric analysis of autophagy in living mammalian cells. Methods Enzymol. 2009;452:131–141. doi: 10.1016/S0076-6879(08)03609-4. [DOI] [PubMed] [Google Scholar]
- 135.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]