Abstract
We have introduced the LTR-retrotransposon MAGGY into a naive genome of Magnaporthe grisea and estimated the copy number of MAGGY in a cell by serial isolation of fungal protoplasts at certain time intervals. The number of MAGGY elements rapidly increased for a short period following introduction. However, it did not increase geometrically and reached equilibrium at 20–30 copies per genome, indicating that MAGGY was repressed or silenced during proliferation. De novo methylation of MAGGY occurred immediately following invasion into the genome but the degree of methylation was constant and did not correlate with the repression of MAGGY. 5-Azacytidine treatment demethylated and transcriptionally activated the MAGGY element in regenerants but did not affect transpositional frequency, suggesting that post-transcriptional suppression, not methylation, is the main force that represses MAGGY proliferation in M.grisea. Support for this conclusion was also obtained by examining the methylation status of MAGGY sequences in field isolates of M.grisea with active or inactive MAGGY elements. Methylation of the MAGGY sequences was detected in some isolates but not in others. However, the methylation status did not correlate with the copy numbers and activity of the elements.
INTRODUCTION
DNA methylation has been implicated in a wide range of cellular processes, including embryogenesis (1), X-chromosome inactivation (2), genomic imprinting (3) and repression of transposable elements (4). It remains unknown, however, whether DNA methylation is the mechanism responsible for these processes. The role of DNA methylation in repressing or taming transposable elements in the mammalian genome is also a matter of considerable debate (5,6). A series of findings support the idea that the activity of transposons is repressed by methylation of the transposon promoters in plants (7–10). The observation that the majority of the 5-methylcytosine in mammalian DNA resides in transposable elements is also consistent with this idea (4). In certain fungal species, repeated sequences are silenced or inactivated by processes called repeat-induced point mutation (RIP) and methylation-induced premeiotically (MIP), which are characterized by hypermethylation (MIP) and both point mutations and DNA methylation (RIP) during a specific period of the sexual cycle (11,12).
On the other hand, several contradictory facts have also arisen. The transposase of a plant DNA transposon, Ac, preferentially interacts with hemimethylated rather than unmethylated transposon DNA (13). In the genome of the invertebrate chordate, transposable elements were found to be unmethylated even though cellular genes were predominantly methylated (14). In addition, Drosophila and Caenorhabditis elegans maintain transposable elements within a certain range of copy numbers despite the fact that these organisms lack DNA methylation. In fact, homology-dependent gene silencing was shown to be involved in taming transposable elements in Drosophila (15).
The LTR-retrotransposon is one of the transposable elements that transpose via a process termed retrotransposition. Retrotransposition involves a series of intracellular processes: transcription of an RNA intermediate of the element, reverse transcription of the intermediate, synthesis of dsDNA using the resulting single-stranded cDNA as a template and integration of the dsDNA into the host genome. One cycle of retrotransposition results in one additional replica of the element in the genome. Thus, how the genome copes with the increase in copy number of LTR-retrotransposons is an open question. MAGGY is a gypsy-like LTR-retrotransposon found in the rice blast fungus Magnaporthe grisea (16). This element has been shown to be active not only in original hosts (17,18) but also in naive genomes of other fungal isolates, including heterologous species, when it is introduced by polyethylene glycol (PEG)-mediated transformation (19). MAGGY transposes at a high frequency after introduction to a naive isolate of M.grisea, but at a low frequency when introduced into Colletotrichum lagenarium (19).
Because of the difficulty of direct examination of transposition events, the activity of transposable elements has mostly been assessed by their transcriptional activities or indirectly by observations of phenotypic changes caused by transposition, in most cases by excision of the element. So far, there is no report chasing actual frequency of transpositions in a cell after the introduction of a transposable element into a new host. Thus, we monitored the dynamics of the number of MAGGY transpositions in a fungal cell over time using Southern analysis. This approach enabled us to accurately examine the relationship between the activity of MAGGY and DNA methylation within it. We conclude that methylation is not the mechanism responsible for repression of the MAGGY element in M.grisea.
MATERIALS AND METHODS
Transformation of fungi
pMGY70, a plasmid containing a copy of MAGGY, was introduced into a M.grisea isolate possessing no MAGGY (a wheat isolate, Br48) by a PEG-mediated transformation method using the plasmid pSH75 as described previously (19). pSH75 carries a hygromycin B phosphotransferase gene as a selective marker.
Extraction of fungal genomic DNA and Southern blot hybridization
Fungal genomic DNA was extracted as described previously (19). Southern blot analysis was performed using the dioxetane chemiluminescence system, Gene Images™ (Amersham). Genomic DNA was digested with appropriate restriction enzymes. The digests were fractionated on a TAE agarose gel and transferred to Hybond N+ (Amersham). The hybridization and detection procedures were performed according to the manufacturer’s instructions.
Regeneration of fungi from a single protoplast
A piece of fungal potato dextrose agar (PDA) culture was transferred to CM broth (0.3% Casamino acids, 0.3% yeast extract, 0.5% sucrose) and incubated at 26°C for a week. Fungal protoplasts were produced by incubating mycelia in a digestion buffer (10 mM Na2HPO4, 1.2 M Mg2SO4) containing 5 mg/ml lysing enzymes (Sigma) and 3.5 mg/ml Zymolyase 20T (Seikagaku, Japan) for 3 h. Serial 10-fold dilutions of protoplast suspension were made with STC (1 M sorbitol, 50 mM Tris–HCl pH 8.0, 50 mM CaCl2) and mixed with regeneration agar medium, then poured onto regeneration agar plates containing 400 µg/ml of hygromycin B. After 3–5 days, single colonies of regenerants were picked and transferred to PDA slant media.
RNA isolation and northern blot analysis
Total RNA was extracted from mycelia using the RNeasy plant mini kit (Qiagen). Total RNA (20 µg) was separated on a 1.2% denaturing agarose gel. After electrophoresis, RNA was transferred to Hybond N+ (Amersham). Hybridization and detection were performed using the dioxetane chemiluminescence system (Gene Images™). RNA probes were labeled with UTP-fluorescein using T3 and T7 transcriptional system (Roche Diagnostics). A SalI–BamHI fragment of MAGGY (nucleotides 885–1445) was used as a template for the labeling after digesting with an appropriate restriction enzyme. Hybridization was performed in 5× SSC, 5% (w/v) SDS, 5% (w/v) dextran sulfate and 5% (v/v) liquid block (Amersham) at 65°C overnight. After hybridization, membranes were washed twice in 1× SSC containing 0.1% SDS for 15 min at 65°C and twice in 0.1× SSC containing 0.1% SDS for 15 min at 65°C. Loading of total RNA was estimated by ethidium bromide staining. Transcript sizes were determined by comparison with RNA molecular-weight marker (Toyobo).
RESULTS
Monitoring the copy number of MAGGY in a fungal cell
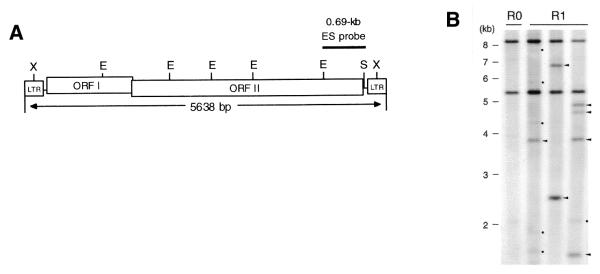
The plasmid pMGY70 containing a copy of MAGGY was introduced into a naive genome of M.grisea (Br48) by PEG-mediated transformation. The copy number of MAGGY in transformants was monitored by isolation of a single protoplast from each cultured transformant over time, followed by propagation of the mycelia and Southern analysis of the mycelial DNA. We designated the population of the original MAGGY transformants as R0, the population that regenerated from a single protoplast of R0 as R1, the one regenerated from a single protoplast of R1 as R2, etc. It took 2–3 weeks to generate a new population of regenerants. Southern analysis was performed on genomic DNAs digested with EcoRI using a 0.69 kb EcoRI–SmaI fragment (ES) of MAGGY (Fig. 1A) as a probe. In this way, the length of detectable fragments varies depending on the proximity of the closest EcoRI site in the genomic sequence outside the transposable element, enabling estimation of MAGGY copy number as total number of bands detected with the ES probe. The Southern blots of an R0 transformant and three of its protoplast regenerants (R1) are shown in Figure 1B. In addition to MAGGY bands in R0, newly appearing bands were observed independently in each R1 regenerant. The timing of MAGGY transposition was estimated by the intensity of the signals appearing in the Southern blots. A MAGGY insert that had already appeared in the genome of the isolated single protoplast would be detected as a strong band in the Southern blots (Fig. 1B, arrowheads), since the mycelial cells are supposed to share the MAGGY insert. On the other hand, a MAGGY insert that appeared during the culture of mycelia after the protoplast isolation would not be detected as a band in most cases. The amount of DNA molecules sharing the MAGGY insert would be too small to be detected in the population since MAGGY transposition occurs independently in each cell. Nevertheless, a MAGGY insert appearing at a very early stage of the culture could be detected as a faint band (Fig. 1B, circles). We took account of only the strong bands and estimated the copy number of MAGGY in the single cell at protoplast isolation. Because of the nature of retrotransposition, the copy number of the element increases one per transposition. Thus, the number of increases of the element in a cell fundamentally represents the number of transpositions that have occurred in the cell.
Figure 1.
Southern blot analysis of a MAGGY transformant of M.grisea (R0) and its protoplast regenerants (R1). (A) Restriction map of MAGGY and the ES fragment used for probing the Southern blots. (B) Southern blot of DNA samples that were extracted from R0 and R1 mycelia, digested with EcoRI and probed with the ES fragment. Arrowheads, MAGGY bands presumed to have already occurred in the isolated single protoplast; circles, MAGGY bands presumed to have appeared during the culture of mycelia after the protoplast isolation (see text). E, EcoRI; S, SmaI; X, XhoI.
Copy number dynamics of MAGGY after introduction into a virgin genome of M.grisea
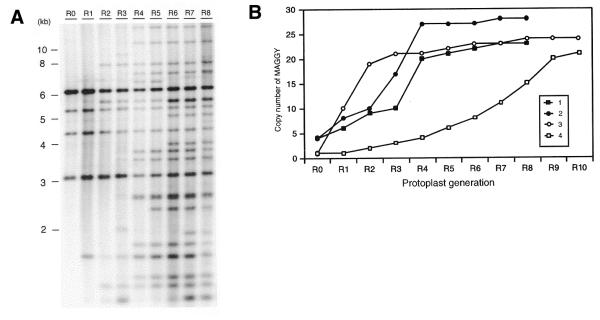
We analyzed the copy number dynamics of MAGGY in four series of protoplast regenerants from R0–R8 or R10. A typical example of the Southern blots (series 1) is given in Figure 2A. In this case, there appeared to be four integrations of the MAGGY plasmid as shown in R0. The copy number of MAGGY gradually increased up to 20 copies by R4 or R5 (2–3 months after the introduction of MAGGY), i.e. five times the original copy number of the MAGGY plasmid integrated in the genome. However, little further increase in copy number appeared thereafter. A similar increase in copy number was observed in two of the other three series of regenerants (Fig. 2B). Series 4 showed distinct dynamics in copy number increase. The copy number of MAGGY in this series increased more slowly, but it eventually reached about 20 at R9 and the rate of the increase seemed to be reduced thereafter. If the frequency of transposition of each MAGGY copy were constant, the number of MAGGY copies in the genome would increase geometrically. However, Figure 2B shows that the copy number of MAGGY seemed to attain equilibrium after propagation of the element to 20–30 copies per genome. This indicated that transposition frequency was repressed at this stage by some mechanism, which would emerge to counter the proliferation of the element but would not pre-exist or appear promptly after the invasion of MAGGY.
Figure 2.
Southern blot analysis of protoplast regenerants of MAGGY transformants. Protoplasts were repeatedly isolated from MAGGY transformants at certain time intervals to establish four series of regenerants (R0–R8 or R10). Southern blot analyses of the regenerants were performed by digesting genomic DNA with EcoRI and probing with the ES fragment. An example (series 1) is given in (A). The copy number of MAGGY in each regenerant series estimated by Southern blot analysis is shown in (B). Closed square, regenerant series 1; closed circle, regenerant series 2; open circle, regenerant series 3; open square, regenerant series 4.
It is noteworthy that some MAGGY bands were lost after several generations (data not shown). This indicated that excision of the element or rearrangement of genomic sequence flanking the element sometimes took place. In addition, amplification of transgenes is often reported after transformation in filamentous fungi. Thus, we estimated the rate of sub-genomic MAGGY copies that were independent of transposition by probing with flanking plasmid sequence. In fact, >95% of the sub-genomic MAGGY copies did not carry any plasmid sequence, indicating that most of them were derived from transposition events.
DNA methylation in the MAGGY sequences during proliferation in the transformants
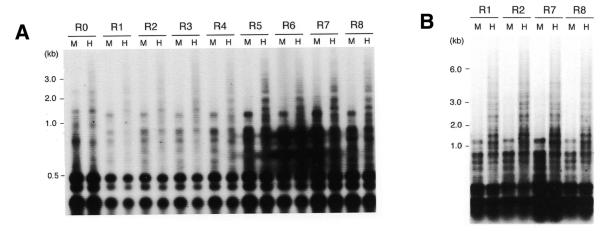
Since DNA methylation has been suggested as a mechanism for repression of transposable elements or repeated sequence in a wide range of organisms including mammals, plants and fungi (4,7,8,10,12,20), we tested whether DNA methylation was involved in the repression of MAGGY in the M.grisea transformants. Four series of regenerants were examined by Southern hybridization using a pair of restriction endonuclease isochizomers, HapII and MspI. HapII is sensitive to cytosine methylation in the recognition site, but MspI is not. HapII and MspI digests were separated in an agarose gel and hybridized with a 5.4 kb XhoI fragment of MAGGY. DNA methylation was detected at HapII sites in the MAGGY sequences just after transformation (R0) and was maintained up to R8 (Fig. 3A). Occurrence of DNA methylation was observed at R0 in all series of regenerants we tested (data not shown). Based on the relative intensity of the hybridizing high (uncut) and low (cut) molecular weight bands, the degree of methylation was low in all the series, and it was almost constant during the whole period. Higher molecular weight bands caused by DNA methylation were more intense in the latter stages, but this can be attributed to increased MAGGY copy number.
Figure 3.
Restriction endonuclease analysis to determine methylation status of MAGGY sequences in R0–R8 regenerants (A). Genomic DNA extracted from the regenerants was digested with isoschizomers HapII (H) and MspI (M). HapII is sensitive to cytosine methylation in the recognition sequence CCGG but MspI is not. HapII and MspI digests were separated in an agarose gel and hybridized to a 5.4 kb XhoI fragment of MAGGY (Fig. 1A). Complete digestion with the enzymes was checked by reprobing with pEBA18 containing a LINE-like retrotransposon, MGR583 (24) (data not shown). (B) For accurate comparison of the degree of methylation in the MAGGY sequence, the concentrations of genomic DNA of R1, R2, R7 and R8 were adjusted according to the copy numbers of MAGGY they possessed and subjected to analysis as in (A).
To clarify the last point, the concentrations of genomic DNA of R1, R2, R7 and R8 were adjusted according to the copy numbers of MAGGY they possessed. They were then subjected to the Southern analysis using the isochizomers. Almost the same degree of methylation was observed in all the samples tested (Fig. 3B). If DNA methylation was the main force repressing MAGGY transposition, the occurrence of methylation could be expected to correlate with the MAGGY repression occurring at the latter stages of MAGGY proliferation, but this was not the case. Thus, these results suggest that the repression of MAGGY in M.grisea does not directly depend on DNA methylation.
Effect of 5-azacytidine (5AC) treatment on the transcriptional and transpositional activities of MAGGY
To examine other aspects of the involvement of methylation in the repression of MAGGY, the effect of demethylation on the transcriptional and transpositional activities of MAGGY was assessed by treatment of the MAGGY transformant with 5AC, which inhibits cytosine methylation. A regenerant with methylated MAGGY was cultured in CM medium with or without 20 µM 5AC for a week. Aliquots of the cultures were harvested to examine the methylation status of MAGGY sequences and the expression level of MAGGY RNA, respectively. Genomic DNA was subjected to Southern analysis with the endonuclease isochizomers HapII and MspI. As expected, detectable cytosine methylation in MAGGY sequences was drastically reduced in the 5AC-treated cells (Fig. 4A), indicating that most of the MAGGY sequences were demethylated. Furthermore, total RNA was extracted and subjected to northern blot analysis. MAGGY RNA was observed at a position of 5.4 kb, which corresponds to the full length of MAGGY (Fig. 4B). More MAGGY RNA was detected in the 5AC-treated cells than in the untreated cells, suggesting that demethylation up-regulated transcription of MAGGY. The northern analysis was repeated three times. In every experiment, up-regulation of MAGGY RNA was observed with 5AC treatment.
Figure 4.
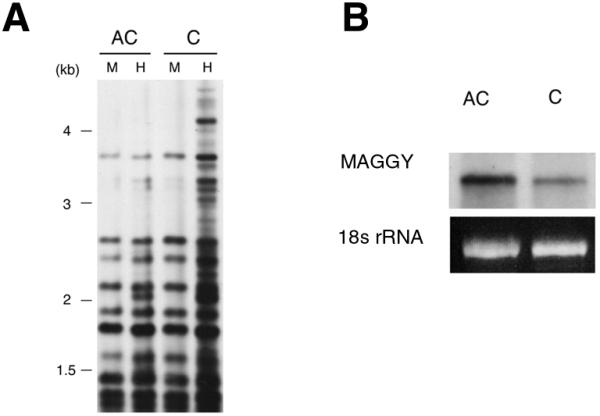
Effect of 5AC treatment on methylation status and transcriptional activity of MAGGY. An R8 regenerant with methylated MAGGY was cultured in CM medium with (AC) or without (C) 20 µM 5AC for 1 week. Aliquots of the cultures were harvested and subjected to Southern (A) and northern (B) blot analyses to examine the methylation status of MAGGY sequences and the expression of MAGGY RNA, respectively. Southern blot analysis was carried out with isoschizomers HapII (H) and Msp I (M). HapII is sensitive to cytosine methylation but MspI is not. Genomic DNA digested with the isoschizomers was run on a 0.8% agarose gel and probed with a 5.4 kb XhoI fragment of MAGGY. With northern blot analysis, total RNA (20 µg) extracted from 5AC-treated and untreated cells was run on a 1.0% formaldehyde agarose gel and probed with a SalI–BamHI fragment of MAGGY. The18S rRNA stained with ethidium bromide and photographed before blotting is shown below the blot.
The other aliquots of cultures were further transferred to CM medium without 5AC. After an additional 2 weeks of culture, seven protoplasts were isolated from the 5AC-treated mycelia and seven from the untreated mycelia. The average number of MAGGY transpositions in the protoplasts was assessed by Southern analysis of regenerated mycelia (Table 1). The average numbers of MAGGY transpositions in the 5AC-treated and untreated cells were 2.86 and 2.57, respectively. No significant difference was found in the two averages by a t-test (0.05%). An independent experiment was carried out in the same way as described above. Again, no significant difference was observed in the average numbers of MAGGY transposition between the 5AC-treated (average: 2.20) and untreated cells (average: 2.40). Evidence that treatment with 5AC reduced methylation drastically in MAGGY sequences and up-regulated transcription of MAGGY RNA indicated that the treatment was effective. Surprisingly, however, it did not affect the transpositional activity of MAGGY. This means that the main force repressing MAGGY transposition is not methylation even though it reduces transcriptional background, and also suggests involvement of some post-transcriptional mechanism in the prevention of MAGGY proliferating in the M.grisea genome.
Table 1. Effect of 5AC treatment on the frequency of MAGGY transposition in M.grisea with repressed MAGGY.
| Number of MAGGY transpositions | ||
| Regenerant |
20 µM
5AC |
Control |
| 1 | 4 | 3 |
| 2 | 3 | 2 |
| 3 | 3 | 2 |
| 4 | 1 | 4 |
| 5 | 1 | 0 |
| 6 | 6 | 4 |
| 7 | 2 | 3 |
| Total | 20 | 18 |
| Average | 2.86a | 2.57a |
aNo significant difference by a t-test (5%)
Survey of the methylation status in MAGGY sequences in various isolates of M.grisea
MAGGY-related sequences have been reported in several M.grisea isolates from various monocot plants (16,21,22). We analyzed the methylation status of MAGGY-related sequences in nine M.grisea isolates from rice, foxtail millet, Panicum miliaceum and Panicum repens. The presence of active copies of MAGGY was reported in the rice and foxtail millet isolates (17,18), whereas only an inactive copy of MAGGY that had been degenerated by a RIP-like process was found in the P.miliaceum isolate (23). Isolates from P.repens seemed not to possess an active MAGGY copy, because restriction fragments conserved in active MAGGY elements were not detected in them, and the apparent copy numbers of the element were also low (one to two copies) (21,23).
Genomic DNA from these isolates was digested with the isochizomers HapII and MspI and subjected to Southern analysis. DNA methylation at HapII sites in the MAGGY-related elements was found in all of the rice isolates and the P.miliaceum isolate, but not in any of the foxtail millet or the P.repens isolates (Fig. 5A). As a control, the blot was stripped and reprobed with pEBA18 that contained a LINE-like retrotransposon, MGR583 (24). No difference was detected in the hybridization patterns between the isoschizomers in any isolates tested, which indicated that genomic DNA was completely digested and that no CCGG sites were methylated in the MGR583 homologs derived from various M.grisea isolates (Fig. 5B).
Figure 5.
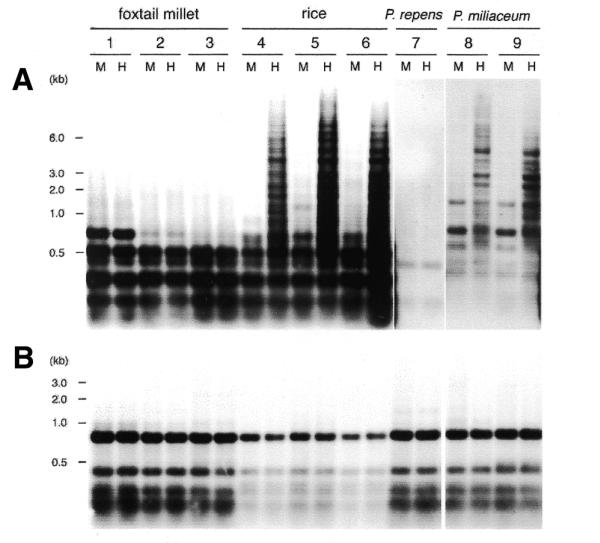
Restriction endonuclease analysis to determine methylation status of MAGGY (A) and MGR583 (B) sequences in M.grisea isolates from various monocot plants. Genomic DNA was extracted from mycelia, digested with isoschizomers HapII (H) and Msp I (M), and hybridized with a 5.4 kb XhoI fragment of MAGGY. HapII is sensitive to cytosine methylation but MspI is not. Southern blots of isolates 7, 8 and 9 were overexposed in (A) due to the low copy numbers of MAGGY-related sequences in them. 1, IN77-16-1-1 (isolated from foxtail millet in India); 2, NNSI2-1-1 (foxtail millet in Japan); 3, GFSI1-7-2 (foxtail millet in Japan); 4, IN77-46-1-3 (rice in India); 5, 1601-3 (rice in Japan); 6, 1836-3 (rice in Japan); 7, IN77-28-1-1 (P.repens in India); 8, STPM4-3-3 (P.miliaceum in Japan); 9, YNPM2-2-1 (P.miliaceum in Japan).
To further examine methylation status in the foxtail millet isolates, Southern analysis was carried out using another set of isochizomers, MboI and Sau3AI, both of which digest GATC but show different sensitivities to cytosine methylation. The results indicated that no methylation occurred at Sau3AI sites in MAGGY sequences of the isolates either (data not shown).
In conclusion, methylation of MAGGY-related sequence did occur in some isolates of M.grisea but showed no correlation with the copy numbers or activity of the elements. Evidence that even the foxtail millet isolates with active, unmethylated copies of MAGGY could restrict MAGGY copy number to less than that in the rice isolates (22) indicated methylation-independent repression of MAGGY in this fungus.
DISCUSSION
Introduction of a transposable element into a naive genome could take place by horizontal transfer or sexual hybridization. A typical example is the P-element that invaded the genome of Drosophila melanogaster by horizontal transfer from Drosophila willistoni and has rapidly spread through the natural population of D.melanogaster, probably within the twentieth century (25). In Drosophila the introduction of new transposable elements by hybridization sometimes results in a syndrome termed hybrid dysgenesis, which was characterized by high rates of mutation, chromosomal rearrangements, female sterility, etc. (26). An important question is how the eukaryotic genome tames the incoming elements and maintains their copy numbers.
Association of methylation with the repression of transposable elements has been reported in a wide range of organisms. In fungi, many transposable elements or repeated sequences in the genome were found to be methylated (27). In some cases methylation of the transposon promoter was shown to reduce transcriptional activity of the elements (7–9). It is unclear, however, to what extent methylation is responsible for the repression of transposon proliferation, i.e. is methylation the regulatory mechanism for the repression of transposition or just an accompanying phenomenon? Our study addressed this question. We examined in detail the relationship between copy number kinetics of the incoming transposon and methylation of the transposon by using the filamentous fungus M.grisea and the retrotransposon MAGGY as a model. The results indicated that methylation was associated with repression of the incoming transposon and reduced transcriptional activity of MAGGY to a certain degree but could not have been responsible for the repression of MAGGY in M.grisea. Several methylation-independent mechanisms that repress the transposable elements have been discovered in Drosophila, which lacks DNA methylation, e.g. homology-dependent gene silencing (15), transcriptional repression by a particular gene (28), overproduction inhibition (29), etc. One or more of these mechanisms could be involved in the repression of MAGGY observed in our experiments. Especially, homology-dependent gene silencing seems to be a good candidate to explain the MAGGY repression since this kind of gene silencing exists in a broad range of eukaryotes, including insects, nematode, plants and filamentous fungi (ex, co-suppression, quelling, etc.) (30–32). In addition, these gene silencing processes do not necessarily involve methylation and some of them are apparently post-transcriptional (33).
Besides the epigenetic regulation of transposable elements described above, repeated sequences in certain fungal species are genetically and irreversibly modified in a specific period of the sexual cycle by RIP (11). Certain M.grisea isolates, exemplified by the P.miliaceum isolate, seem to have succeeded in preventing MAGGY from proliferating in their genome by degenerating it through a RIP-like process (23). Since association of DNA methylation with RIP or RIP-like processes has been reported in several fungi (23,34), de novo methylation on MAGGY that we observed might indicate some role in a RIP-like process rather than a direct involvement in repression of the element.
MAGGY carriers occurring in the natural population of M.grisea possess 35–40 copies of MAGGY (22). This implies that the copy number of MAGGY in our transformants (20–30 copies) had not attained, by R8, the equilibrium that has been established in the natural evolution of M.grisea and MAGGY. Actually, the copy numbers of MAGGY in most transformants grew to greater than 30 by 18 months after transformation (data not shown). Leaky transposition contributing to this gradual rise may ultimately be stopped at the equilibrium observed in nature by a distinct, stronger force of repression. This might include genetic modification of the element by RIP and/or an increase of excision frequency of the element in response to increase in copy number.
It is reported that mammals, plants and filamentous fungi differ in both 5-methylcytosine content in genomic DNA and preferred target sequences for cytosine methylation (34–36), which might indicate that DNA methylation plays distinct or multiple functions in different eukaryotic systems. Actually, our data showed variability in methylation status of MAGGY sequences even among field isolates of M.grisea. Furthermore, in the same fungal isolate, e.g. rice isolates or MAGGY transformants, one retrotransposon (MAGGY) was methylated and the other (MGR583) was apparently not. The variability discussed here, and our observed lack of correlation between DNA methylation and suppression of MAGGY, render DNA methylation unlikely as a responsive mechanism for protection of the genome against rampant transposition of mobile elements. Our results imply that post-transcriptional and methylation-independent gene silencing can be the general and main mechanism repressing transposons not only in organisms without methylation but also in some of those with methylation.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Hajime Kato, the former professor of Kobe University, for valuable suggestions, and Dr Adam J.Bogdanove for critical reading of the manuscript and suggestions for improvement. This work was partly supported by a grant from the Ministry of Education, Science, Sports and Culture of Japan (No. 10660049).
References
- 1.Macleod D., Charlton,J., Mullins,J. and Bird,A. (1994) Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev., 8, 2282–2292. [DOI] [PubMed] [Google Scholar]
- 2.Singer-Sam J. and Riggs,A. (1993) In Jost,J. and Saluz,H. (eds), DNA Methylation: Molecular Biology and Biological Significance. Birkhauser Verlag, Boston, MA, pp. 358–384.
- 3.Li E., Beard,C. and Jaenisch,R. (1993) Role for DNA methylation in genomic imprinting. Nature, 366, 362–365. [DOI] [PubMed] [Google Scholar]
- 4.Yoder J., Walsh,C. and Bestor,T. (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet., 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 5.Bird A. (1997) Does DNA methylation control transposition of selfish elements in the germline? Trends Genet., 13, 469–470. [DOI] [PubMed] [Google Scholar]
- 6.Bestor T., Walsh,C. and Yoder,J. (1997) Reply. Trends Genet., 13, 470–472. [DOI] [PubMed] [Google Scholar]
- 7.Chandler V. and Walbot,V. (1986) DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl Acad. Sci. USA, 83, 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomet P., Wessler,S. and Dellaporta,S. (1987) Inactivation of the maize transposable element Activator (Ac) is associated with DNA modification. EMBO J., 6, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banks J., Masson,P. and Fedoroff,N. (1988) Molecular mechanisms in the developmental regulation of the maize suppressor-mutator transposable element. Genes Dev., 2, 1364–1380. [DOI] [PubMed] [Google Scholar]
- 10.Hirochika H., Okamoto,H. and Kakutani,T. (2000) Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell, 12, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cambareri E.B., Jensen,B.C., Schabtach,E. and Selker,E.U. (1989) Repeat-induced G-C to A-T mutations in Neurospora. Science, 244, 1571–1575. [DOI] [PubMed] [Google Scholar]
- 12.Goyon C. and Faugeron,G. (1989) Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol. Cell. Biol., 9, 2818–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Heinlein,M. and Kunze,R. (1996) Methylation pattern of Activator transposase binding sites in maize endosperm. Plant Cell, 8, 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmen M., Leitgeb,S., Charlton,J., Jones,S., Harris,B., Clark,V. and Bird,A. (1999) Nonmethylated transposable elements and methylated genes in a chordate genome. Science, 283, 1164–1167. [DOI] [PubMed] [Google Scholar]
- 15.Jensen S., Gassama,M. and Heidmann,T. (1999) Taming of transposable elements by homology-dependent gene silencing. Nature Genet., 21, 209–212. [DOI] [PubMed] [Google Scholar]
- 16.Farman M.L., Tosa,Y., Nitta,N. and Leong,S.A. (1996) MAGGY, a retrotransposon in the genome of the rice blast fungus Magnaporthe grisea. Mol. Gen. Genet., 251, 665–674. [DOI] [PubMed] [Google Scholar]
- 17.Eto Y., Ikeda,K., Chuma,I., Kataoka,T., Kuroda,S., Kikuchi,N., Don,L.D., Kusaba,M., Nakayashiki,H., Tosa,Y. and Mayama,S. (2001) Comparative analyses of the distribution of various transposable elements in Pyricularia and their activity during and after the sexual cycle. Mol. Gen. Genet., 264, 565–577. [DOI] [PubMed] [Google Scholar]
- 18.Shull V. and Hamer,J.E. (1996) Genetic differentiation in the rice blast fungus revealed by the distribution of the Fosbury retrotransposon. Fungal Genet. Biol., 20, 59–69. [DOI] [PubMed] [Google Scholar]
- 19.Nakayashiki H., Kiyotomi,K., Tosa,Y. and Mayama,S. (1999) Transposition of the retrotransposon MAGGY in heterologous species of filamentous fungi. Genetics, 153, 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhounim L., Rossignol,J. and Faugeron,G. (1992) Epimutation of repeated genes in Ascobolus immersus. EMBO J., 11, 4451–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusaba M., Eto,E., Tosa,Y., Nakayashiki,H. and Mayama,M. (1999) Genetic diversity in Pyricularia isolates from various hosts revealed by polymorphisms of nuclear ribosomal DNA and the distribution of the MAGGY retrotransposon. Ann. Phytopathol. Soc. Jpn, 65, 588–596. [Google Scholar]
- 22.Tosa Y., Nakayashiki,H., Hyodo,H., Mayama,S., Kato,H. and Leong,S.A. (1995) Distribution of retrotransposon MAGGY in Pyricularia species. Ann. Phytopathol. Soc. Jpn, 61, 549–554. [Google Scholar]
- 23.Nakayashiki H., Nishimoto,N., Ikeda,K., Tosa,Y. and Mayama,S. (1999) Degenerate MAGGY elements in a subgroup of Pyricularia grisea: a possible example of successful capture of a genetic invader by a fungal genome. Mol. Gen. Genet., 261, 958–966. [DOI] [PubMed] [Google Scholar]
- 24.Urashima A.S., Hashimoto,Y., Don,L.D., Kusaba,M., Tosa,Y., Nakayashiki,H. and Mayama,S. (1999) Molecular analysis of the wheat blast population in Brazil with a homologue of retrotransposon MGR583. Ann. Phytopathol. Soc. Jpn, 65, 429–436. [Google Scholar]
- 25.Daniels S., Peterson,K., Strausbaugh,L., Kidwell,M. and Chovnick,A. (1990) Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics, 124, 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bregliano J., Picard,G., Bucheton,A., Pelisson,A., Lavige,J. and L’Heritier,P. (1980) Hybrid dysgenesis in Drosophila melanogaster. Science, 207, 606–611. [DOI] [PubMed] [Google Scholar]
- 27.Selker E.U. (1997) Epigenetic phenomena in filamentous fungi: useful paradigms or repeat-induced confusion? Trends Genet., 13, 296–301. [DOI] [PubMed] [Google Scholar]
- 28.Prud’homme N., Gans,M., Masson,M., Terzian,C. and Bucheton,A. (1995) Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics, 139, 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohe A. and Hartl,D. (1996) Autoregulation of mariner transposase activity by overproduction and dominant-negative complementation. Mol. Biol. Evol., 13, 549–555. [DOI] [PubMed] [Google Scholar]
- 30.Napoli C., Lemieux,C. and Jorgensen,R. (1990) Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell, 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Krol A., Mur,L., Beld,M., Mol,J. and Stuitje,A. (1990) Flavonoid genes in Petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell, 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cogoni C., Irelan,J.T., Schumacher,M., Schmidhauser,T.J., Selker,E.U. and Macino,G. (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J., 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- 33.Cogoni C. and Macino,G. (1997) Conservation of transgene-induced post-transcriptional gene silencing in plants and fungi. Trends Plant Sci., 2, 438–443. [Google Scholar]
- 34.Selker E.U., Fritz,D.Y. and Singer,M.J. (1993) Dense nonsymmetrical DNA methylation resulting from repeat-induced point mutation in Neurospora. Science, 262, 1724–1728. [DOI] [PubMed] [Google Scholar]
- 35.Gruenbaum Y., Naveh Many,T., Cedar,H. and Razin,A. (1981) Sequence specificity of methylation in higher plant DNA. Nature, 292, 860–862. [DOI] [PubMed] [Google Scholar]
- 36.Meyer P., Niedenhof,I. and ten Lohuis,M. (1994) Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrida. EMBO J., 13, 2084–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]