Abstract
A growing body of evidence suggests that DNA methylation is functionally divergent among different taxa. The recently discovered functional methylation system in the honeybee Apis mellifera presents an attractive invertebrate model system to study evolution and function of DNA methylation. In the honeybee, DNA methylation is mostly targeted toward transcription units (gene bodies) of a subset of genes. Here, we report an intriguing covariation of length and epigenetic status of honeybee genes. Hypermethylated and hypomethylated genes in honeybee are dramatically different in their lengths for both exons and introns. By analyzing orthologs in Drosophila melanogaster, Acyrthosiphon pisum, and Ciona intestinalis, we show genes that were short and long in the past are now preferentially situated in hyper- and hypomethylated classes respectively, in the honeybee. Moreover, we demonstrate that a subset of high-CpG genes are conspicuously longer than expected under the evolutionary relationship alone and that they are enriched in specific functional categories. We suggest that gene length evolution in the honeybee is partially driven by evolutionary forces related to regulation of gene expression, which in turn is associated with DNA methylation. However, lineage-specific patterns of gene length evolution suggest that there may exist additional forces underlying the observed interaction between DNA methylation and gene lengths in the honeybee.
Keywords: honeybee, DNA methylation, gene lengths, gene expression
Introduction
DNA methylation is phylogenetically widespread and likely to have an ancient evolutionary origin (Colot and Rossignol 1999; Ponger and Li 2005). Although DNA methylation has been studied extensively in mammalian model systems, its function in other taxa, especially in invertebrate animals, is poorly understood. In the last few years, it has become apparent that the patterns of genomic DNA methylation differ greatly between vertebrates and invertebrates (Suzuki et al. 2007; Elango and Yi 2008; Elango et al. 2009; Wang and Leung 2009; Feng et al. 2010; Zemach et al. 2010). Accordingly, the functions of DNA methylation are also likely to vary significantly between taxa (Kucharski et al. 2008; Elango et al. 2009; Foret et al. 2009; Yi and Goodisman 2009). Thus, investigating patterns of genomic DNA methylation in diverse taxa provides fundamental information on evolution of epigenetic regulation.
The majority of vertebrate genomes are methylated, with the only exceptions being short regions of high CpG dinucleotide frequencies, the so-called “CpG islands” (Suzuki and Bird 2008; Illingworth and Bird 2009). Genomic distributions of DNA methylation in invertebrates appear to be diametrically different from this vertebrate pattern (Suzuki et al. 2007; Elango and Yi 2008; Suzuki and Bird 2008). For instance, studies of the sea squirt Ciona intestinalis, pea aphid Acyrthosiphon pisum as well as honeybee Apis mellifera demonstrate that DNA methylations in these species are targeted to “gene bodies” or transcriptional units rather than nongenic regions (Wang et al. 2006; Suzuki et al. 2007; Elango et al. 2009; Feng et al. 2010; Walsh et al. 2010; Zemach et al. 2010). Furthermore, only subsets of genes are methylated in these species (Suzuki et al. 2007; Elango et al. 2009; Foret et al. 2009; Wang and Leung 2009; Walsh et al. 2010).
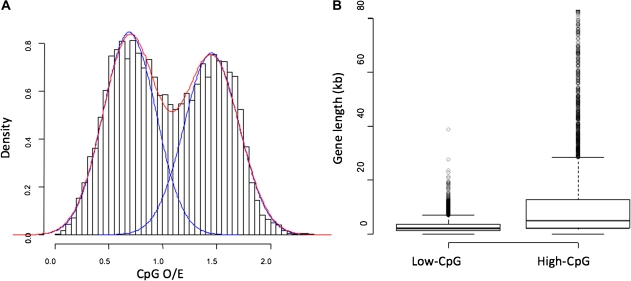
Such a pattern of “partial” DNA methylation can be represented by genomic patterns of CpG depletion, which is a good proxy for the level of DNA methylation. Briefly, DNA methylation in animal genomes predominantly targets CpG dinucleotides. Because methylated CpG dinucleotides are readily converted to TpG dinucleotides via spontaneous deamination, methylated regions gradually lose CpG dinucleotides (Duncan and Miller 1980). In other words, regions with low CpG dinucleotides indicate high levels of DNA methylation and vice versa (Bird 1980). CpG O/E corresponds well to the actual level of DNA methylation observed in experimental studies in mammals (e.g., Weber et al. 2007) as well as in the honeybee (Foret et al. 2009) and the silkworm (Xiang et al. 2010). According to the CpG depletion profile, it is clear that honeybee genes can be divided into two distinctive groups, namely low-CpG O/E and high-CpG O/E classes (henceforth referred to as low-CpG and high-CpG, respectively), representing hyper and hypomethylated genes in germlines (fig. 1, also shown in the aforementioned references). Here, we report that these two epigenetic classes of honeybee genes exhibit another dramatic difference in their characteristics.
FIG. 1.
(A) The distribution of CpG O/E in Apis mellifera genes. A mixture of two distributions (represented by two blue curves) fit the observed distribution of CpG O/E (the red curve represents the sum of the two distributions). Accordingly, honeybee genes are classified into low- and high-CpG O/E genes (see text). (B) Length differences of low- and high-CpG O/E genes are represented in the boxplot. Note that gene length diagrams are shown only up to 80 kbps for display purposes. Analyzing exons and introns separately leads to similar patterns of bimodal distributions (supplementary material, Supplementary Material online).
Materials and Methods
Genome Sequences and Annotations
The genome sequences and annotations of Drosophila melanogaster were downloaded from the University of California, Santa Cruz genome browser, RefSeq Genes Track (April 2006 assembly, dm3). Genome sequences of C. intestinalis were downloaded them from Ensembl 55 (JGI2). We extracted annotations of C. intestinalis by application program interfaces code from Ensembl. Annotations from the A. mellifera genome assembly 4.0 were downloaded from the beebase (http://www.beebase.org/). For Ac. pisum, the annotations are downloaded from the aphidbase (http://www.aphidbase.com/aphidbase). Only the Refseq gene model was used for analyses.
We used honeybee recombination rate estimates obtained by Beye et al. (2006). Local recombination rates were estimated by comparing genetic distances between markers with physical distance in 125-kb nonoverlapping windows.
Ortholog Identification
To identify orthologous genes among D. melanogaster, C. intestinalis, and A. mellifera, we utilized the Roundup database of orthologs (DeLuca et al. 2006), which identifies orthologous proteins using the Reciprocal Smallest Distance algorithm. We first downloaded the protein clusters containing the three pairwise orthologous proteins separately with the default parameter setting. If the same A. mellifera protein ID appeared in all three clusters, we took the combined protein clusters as the 1 to 1 to 1 orthologs among D. melanogaster, C. intestinalis, and A. mellifera. To identify four-way orthologs between Ac. pisum, D. melanogaster, C. intestinalis, and A. mellifera, we performed BlastP comparisons of complete protein sequence sets between Ac. pisum and other species with a cutoff value of 1 × 10−5, to identify reciprocal best hits. Once the four-way orthologs were identified, all protein GI identifiers were converted to RNA nucleotide accessions using the gene2refseq database from the National Center for Biotechnology Information ftp site (http://www.ncbi.nlm.nih.gov/ftp/). We identified a total of 2,026 four-way orthologs.
Measurement and Classification of CpG O/E Distribution
CpG O/E or “normalized CpG content” measures depletion of CpG dinucleotides for certain regions of interest. It is defined as
 |
where PCpG, Pc, and PG are the frequencies of CpG dinucleotides, C nucleotides, and G nucleotides, respectively.
Alternatively, CpG O/E can be also calculated as
where G + C content is calculated as total number of G and C divided by the total number of nucleotides. The values calculated by these methods are nearly identical, as expected under the Chargaff’s rule (Chargaff 1951; Rudner et al. 1968).
In the honeybee, the distributions of CpG O/E from exons, introns, and exons + introns (referred to as gene bodies) are not unimodal but mixtures of distributions (fig. 1, supplementary fig. 1, Supplementary Material online). We estimated the number of components in those mixture distributions using a model-based clustering. The “mclust” package in R package (www.r-project.org) was used to estimate the number of components under the Gaussian mixture model.
Gene Ontology Enrichment Analysis
Due to the limitation of the gene ontology (GO) annotation in A. mellifera, we only used the orthologs in D. melanogaster for GO biological process term analysis. Enrichment of specific GO terms was compared with the background (all D. melanogaster orthologs used) using the DAVID tools (Dennis et al. 2003). A Benjamini multiple-testing correction of the EASE score (a modified Fisher exact test) was used to determine the significance of gene enrichment.
Results
Dramatic Length Difference between Low- and High-CpG Genes of A. mellifera
The distributions of CpG O/E from honeybee exons, introns, and exons + introns (referred to as gene bodies) are best explained by “bimodal” distributions (fig. 1, supplementary fig. 1, Supplementary Material online), as previously described (Elango et al. 2009). Based upon this observation, we have previously proposed that honeybee genes can be divided into two distinctive epigenetic classes. Namely, we proposed low- and high-CpG genes in honeybee represent hyper and hypomethylated genes in the germlines. Newly available experimental data on genomic methylation patterns provide supports to this hypothesis (Zemach et al. 2010).
Here, we report that in addition to the distinctive CpG depletion profile, low- and high-CpG genes in the honeybee differ greatly in their lengths (fig. 1, table 1). On average, high-CpG genes are over five times longer than low-CpG genes (P < 2 × 10−16, table 1). The difference is most pronounced in introns: introns of high-CpG genes are approximately an order of magnitude longer than those from low-CpG genes (table 1). The average numbers of introns in low- and high-CpG groups are similar (6.2 and 6.4 for low- and high-CpG genes, respectively: supplementary table 1, Supplementary Material online), suggesting that the observed pattern cannot be explained by preferential insertions of new exons and/or introns into high-CpG genes. Rather, high-CpG genes exhibit greater variance than low-CpG genes in terms of lengths (fig. 1 and table 1). In other words, some particularly long genes exist in the high-CpG class.
Table 1.
Length Difference between Low- and High-CpG Genes in Honeybee
| Apis mellifera (N = 9,159) | Acyrthosiphon pisum (N = 10,248) | Ciona intestinalis (N = 14,497) | ||||||||||
| Low-CpG | High-CpG | Ratioa | P Value | Low-CpG | High-CpG | Ratioa | P Value | Low-CpG | High-CpG | Ratioa | P Value | |
| Gene body | 2,815 (35.5) | 15,118 (497.1) | 5.37 | <2.2 × 10−16 | 5,301 (65.9) | 13,495 (265.6) | 2.54 | <2.2 × 10−16 | 4,962 (66.6) | 4,961 (70.9) | 1.00 | 0.99 |
| Exons | 1,626 (21.9) | 1,837 (23.5) | 1.13 | 2.9 × 10−16 | 2,001 (19.5) | 1,849 (18.5) | 0.92 | 1.4 × 10−8 | 1,204 (13.3) | 971 (14.2) | 0.81 | 0.0065 |
| Introns | 1,189 (62.1) | 13,281 (400.7) | 11.12 | <2.2 × 10−16 | 3,300 (54.1) | 11,646 (261.4) | 3.53 | <2.2 × 10−16 | 3,758 (56.7) | 3,990 (63.9) | 1.06 | <2.2 × 10−16 |
NOTE.—Mean lengths in basepairs in each class are presented (standard errors are shown in parentheses). Significance values are assessed using a t-test. Data from C. intestinalis and Ac. pisum are also shown for comparison.
Ratio of high-CpG/low-CpG genes.
This pattern is persistent regardless of whether exons, introns, or gene bodies are used for classifying low- and high-CpG genes (supplementary table 1, Supplementary Material online). Note that the difference in gene lengths between the two groups does not influence the bimodal distribution of CpG depletion: when we assess the distribution of CpG O/E while controlling for gene lengths, the bimodality persists (supplementary fig. S2, Supplementary Material online).
We investigated if a similar length difference is also present in two invertebrate outgroups where similar bimodal distributions of CpG O/E have been reported, namely the pea aphid Ac. pisum (Walsh et al. 2010) and the sea squirt C. intestinalis (Suzuki et al. 2007). The same trend among these species would suggest that gene length difference is a common theme of invertebrate gene methylation. Note that low- and high-CpG classes in the honeybee, the pea aphid, and the sea squirt are independently assessed based upon species-specific distributions of CpG depletion.
In Ac. pisum, gene bodies of high-CpG genes are longer than those of low-CpG genes (table 1). However, when divided into exons and introns, a conflicting pattern emerges: exons of low- and high-CpG genes in Ac. pisum exhibit the opposite pattern to that in A. mellifera: low-CpG exons are significantly longer than high-CpG exons. Most length difference in the gene bodies thus comes from the difference in the intron lengths. However, the length difference in the pea aphid is much less pronounced than in the honeybee. Although the introns of high-CpG genes in honeybee are on average over an order of magnitude longer than those of low-CpG genes, in Ac. pisum, the difference is less than 4-fold (table 1).
Intriguingly, in the sea squirt, lengths of gene bodies show little difference between the two groups (table 1). Again, the exons in C. intestinalis exhibit the opposite pattern to that in A. mellifera (low-CpG exons are 24% longer). High-CpG introns, in contrast, are on average 6% longer than low-CpG introns (table 1). Thus, lengths of exons and introns exhibit a complex, potentially lineage-specific variation in terms of their relation to the epigenetic status.
Length difference Is Not Caused by G + C Content, Recombination Rates, or Preferential Accumulation of Repetitive Sequences
What accounts for this dramatic difference in gene length in the honeybee genes in general and intron length, in particular?
The honeybee genome paper reported a positive correlation between G + C content and the length of genes (HoneyBee Genome Sequencing Consortium 2006). Interestingly, G + C contents and CpG O/E tend to be correlated in a variety of taxa (Duret and Galtier 2000; Fryxell and Zuckerkandl 2000; Elango et al. 2008), even though CpG O/E is “normalized” for G + C content (see Fryxell and Zuckerkandl [2000] and Elango et al. [2008] for discussions on the potential causes for this phenomenon). Thus, we investigated whether the observed covariation of CpG O/E and gene length in the honeybee genome is caused indirectly due to the underlying relationship between G + C content and CpG O/E.
We first examined whether the correlation between CpG O/E and gene length is confounded by the effect of G + C content. We used the partial correlation method (Kim and Yi 2007). The correlation between CpG O/E and gene length is highly significant (Spearman’s r = 0.47, P < 2 × 10−16). The partial correlation between CpG O/E and gene length, after controlling for G + C content, is only slightly decreased (Spearman’s r for CpG O/E ∼ gene length|G + C content = 0.45, P < 2 × 10−16). Thus, G + C content appears to have little influence on the relationship between CpG O/E and gene length. Second, we divided genes into four equal-sized bins according to their G + C content and examined whether we observe significant length difference between low- and high-CpG groups in each bin. If the observed length difference between G + C content and CpG O/E is due to the confounding effect of G + C content, then there should be little difference between the two CpG O/E groups within each bin. In contrast, we observe highly significant differences between low- and high-CpG genes in all bins examined (supplementary fig. 3, Supplementary Material online). Finally, it should be noted that unlike CpG O/E, gene G + C content does not exhibit bimodal distribution (supplementary fig. 4, Supplementary Material online, also see Elango et al. [2009]). Therefore, the observed length difference between low- and high-CpG groups of genes is not caused by the correlation between G + C content and gene length.
Another possibility is that recombination plays a role in modulating gene length. Regions of low recombination are known to harbor longer introns in some taxa because natural selection may act against long introns in highly recombining regions (Carvalho and Clark 1999). Alternatively, long introns may be favored by natural selection in regions of low recombination: low recombining regions suffer from decreased efficiency of natural selection due to the interference between linked loci (Yi and Charlesworth 2000; Betancourt et al. 2009). It is proposed that long introns may help dilute effects of interference by increasing chances of recombination (Comeron and Kreitman 2000). Thus, it is possible that high-CpG genes of honeybee reside in low-recombination environment and accumulate longer introns than low-CpG genes.
To test this hypothesis, we analyzed empirically determined recombination data from the honeybee (Beye et al. 2006). We found a weak and significant negative correlation between recombination rates and intron lengths (Spearman’s r = −0.09, P = 5 × 10−5, see supplementary material, Supplementary Material online for other relations between recombination rates and genomic traits). However, the mean recombination rates of low-CpG and high-CpG genes are not significantly different from each other (26.0 cM/Mb and 26.5 cM/Mb, respectively; Mann–Whitney test, P = 0.7449). Furthermore, recombination rates are not significantly correlated with CpG O/E (P > 0.05). Thus, difference in recombination rates cannot account for the observed length difference between high- and low-CpG genes.
We also investigated whether preferential accumulation of repetitive sequences in high-CpG genes may account for the dramatic length difference between the two classes. Because there is extremely limited number of annotated transposable elements in the honeybee genome (only 11 mapped onto the assembly, according to the HoneyBee Genome Sequencing Consortium [2006]), we focused on simple repeats and interspersed repeats. We found that less than 1% of genes harbor these repetitive sequences. Moreover, the proportions of coding sequence lengths accounted by repetitive sequences are negligible in both classes: less than 1% of sequences in each class are occupied by repetitive sequences (0.61% and 0.48% of sequences in low- and high-CpG classes, respectively). However, we note that this aspect of honeybee genome needs to be revisited with improved annotation, as it is possible that there are honeybee transposable elements currently unbeknownst to us. Nevertheless, our analyses indicate that we can rule out simple and interspersed repeats as the main cause of length difference between low- and high-CpG genes in A. mellifera.
Comparative Analyses of Gene Lengths Indicate That Historically Long Genes Are Now Preferentially Found in High-CpG Genes
Length difference between low- and high-CpG genes in the honeybee appears to have a deep evolutionary origin. Gene lengths of A. mellifera are highly correlated with those from other invertebrate outgroups. The strength of correlation follows nicely with the proposed phylogenetic relationship among the four species: the correlation between gene lengths of the two closest species, the honeybee and the fruitfly, is the strongest (Spearman’s r = 0.69, P < 2 × 10−16), followed by that between the honeybee and the pea aphid (Spearman’s r = 0.58, P < 2 × 10−16) and between the honeybee and the sea squirt (Spearman’s r = 0.49, P < 2 × 10−16). These observations indicate that gene lengths in A. mellifera are determined largely by ancestral gene lengths.
Indeed, in all three outgroup species, orthologs of genes belonging to high-CpG class in A. mellifera are longer than those belonging to low-CpG class in A. mellifera (table 2). The pattern is consistent in exons, introns, and gene bodies, although the effect is weaker in exons than in introns. We asked how likely it is that we observe such pronounced length difference in randomly separated groups of genes, by simulation. We randomly grouped honeybee genes into two groups, of the same sample sizes as those observed (1,503 and 526 for low-CpG and high-CpG genes, respectively), and then assessed length difference between the two groups of genes. We repeated this procedure by 100,000 times. Similar experiments were performed for D. melanogaster, Ac. pisum, and C. intestinalis genes. We found that the actual length differences between these two classes are far greater than those from random simulation in all three species. In fact, we never observe length difference identical or greater than the observed difference, leading to empirically determined P values of <10−5 in all cases (supplementary fig. 5, Supplementary Material online).
Table 2.
Gene Length Distribution among the 1:1:1:1 Orthologs between the Honeybee, the Fruitfly, the Pea Aphid, and the Sea Squirt
| Apis mellifera | Drosophila melanogaster | Acyrthosiphon pisum | Ciona intestinalis | |||||||||
| Low-CpG | High-CpG | P Value | Ortholog Low-CpGa | Ortholog High-CpGb | P Value | Ortholog Low-CpGa | Ortholog High-CpGb | P Value | Ortholog Low-CpGa | Ortholog High-CpGb | P Value | |
| Gene body | 3,034 (55.4) | 19,046 (1847.2) | <2.2 × 10−16 | 3,646 (124.7) | 11,459 (3,646.5) | <2.2 × 10−16 | 6,216 (149.3) | 15,293 (812.6) | <2.2 × 10−16 | 5,559 (135.1) | 6,644 (264.3) | 0.0003 |
| Exons | 1,792 (29.6) | 2,014 (67.9) | 0.0027 | 2,220 (36.6) | 2,753 (92.5) | 1.1 × 10−7 | 2,123 (31.0) | 2,178 (72.8) | 0.487 | 1,603 (23.2) | 1,721 (50.0) | 0.032 |
| Introns | 1,242 (34.3) | 17,032 (1838.1) | <2.2 × 10−16 | 1,426 (109.6) | 8,706 (775.1) | <2.2 × 10−16 | 4,093 (136.8) | 13,115 (795.1) | <2.2 × 10−16 | 3,956 (120.4) | 4,923 (230.4) | 0.0002 |
NOTE.—Mean values for each class are presented (standard errors are shown in parentheses). T-test was used for test of significance (N = 2,026).
Orthologs of low-CpG genes in A. mellifera.
Orthologs of high-CpG genes in A. mellifera.
The fact that we can observe length difference in C. intestinalis orthologs indicates that some of the observed gene length difference traces back to the split of chordates and arthropods. Thus, genes that were historically short and long are clustered to low- and high-CpG classes in the honeybee.
Honeybee Epigenetic Status Explains Additional Variation in Gene Lengths
We now focus on the gene length difference between the two epigenetic classes in A. mellifera that cannot be explained by the shared evolutionary histories between flies and bees. In figure 2A, we depict a linear regression model where gene lengths of A. mellifera are dependent variables and those of D. melanogaster are independent variables (log-transformed to improve normality). As expected from the highly significant correlation between these two variables, this regression model can explain substantial amount of the observed length variation (the R2 of this model is 0.41).
FIG. 2.
(A) Gene lengths between Apis mellifera and Drosophila melanogaster are highly correlated. Ortholog length in D. melanogaster can explain 41% of observed variation in A. mellifera gene lengths in a linear regression model (see text). Note that the lengths are log-transformed to improve normality. (B) Residuals remaining from the regression model in figure 2A. Residuals from low-CpG genes (red triangles) tend to be negative, whereas those from the high-CpG genes (green circles) tend to be positive, demonstrating that low-CpG genes are shorter and high-CpG genes are longer than expected from the linear regression model alone. Note that there exists a subset of high-CpG genes with particularly large residuals (denoted as darker green circles). These genes include those related to specific developmental functions (see text).
The residuals from this regression model (fig. 2B) represent the amount of variation in A. mellifera gene lengths that cannot be explained by the evolutionary relationship alone. Note that the residuals are not uniformly distributed. Rather, residuals in low- and high-CpG genes tend to be negative and positive, respectively: the mean residuals are −0.107 and 0.188 from low- and high-CpG genes, which are highly significantly different from each other (P < 2 × 10−16, Mann–Whitney test). In other words, on average, low-CpG genes tend to be shorter and high-CpG genes tend to be longer than expected, based solely upon phylogenetic relationships. This trend is stronger for high-CpG genes, where a subset of genes appear clearly longer than expected under evolutionary relationships alone (upper left corner in fig. 2A).
We then asked whether we could explain some variation remaining in the residual by the current epigenetic profile of A. mellifera genes (in other words, if there exists additional gene length variation in the honeybee lineage, related to their DNA methylation status). We investigated this by assessing the relationship between the residuals in figure 2B and the CpG O/E measures, which is the proxy of methylation status in A. mellifera. We performed this analysis separately for low- and high-CpG genes. CpG O/E is highly significantly positively correlated with the residuals for both low- and high-CpG genes (Spearman’s r = 0.19 and 0.42, for low- and high-CpG classes. P < 2.2 × 10−16 in both cases). Therefore, CpG O/E explains substantial amount of gene lengths evolution in the honeybee lineage, and this effect is stronger for high-CpG genes. In particular, a subset of high-CpG genes is markedly longer than predicted by the phylogenetic relationship alone (dark green circles, fig. 2B).
Discussion
Causes of Length Difference between Hyper and Hypomethylated Honeybee Genes: Expression Provides a Partial Answer
We have established that hyper and hypomethylated genes in A. mellifera also differ greatly in their lengths. We have excluded G + C content, recombination, and repetitive sequences as main causes of length differences, although the importance of the latter two factors needs to be revisited with improved annotations of the honeybee genome. What can we say about the causes of length difference between the two epigenetic groups with the data in hand?
We hypothesize that length difference between hypo and hypermethylated genes in the honeybee is at least partially mediated by selection related to regulation of gene expression. In mammals, genes that are constitutively expressed in many tissues or “housekeeping genes” are shorter than tissue-specific genes (Eisenberg and Levanon 2003). Natural selection may prefer compact housekeeping genes because it is beneficial for efficient transcription and translation (Eisenberg and Levanon 2003; Urrutia and Hurst 2003).
In the honeybee, low-CpG genes are broadly expressed, whereas high-CpG genes tend to exhibit tissue-specific expression (Foret et al. 2009). Computational studies also reveal that low-CpG genes in honeybee are enriched in housekeeping functions (Elango et al. 2009; Wang and Leung 2009). Therefore, natural selection toward compactness of broadly expressed housekeeping genes may underlie the observed length difference. Furthermore, the fact that high-CpG genes tend to be narrowly expressed (Foret et al. 2009) raises the possibility that high-CpG genes may have the propensity to accumulate weakly deleterious insertion mutations because they are under reduced selective constraints (Duret and Mouchiroud 2000).
We investigated the relationship between expression breadth and gene lengths using data from six different tissues (Foret et al. 2009). In accord to the idea that gene expression plays a significant role in determining gene lengths, we observe that honeybee gene length decreases as the number of tissues where it is expressed increases (fig. 3A); the most broadly expressed genes are the shortest. Thus, the observed gene length difference in the honeybee follows well the general trend between gene lengths and gene expression.
FIG. 3.
(A) Gene lengths decrease as expression breadths increase in honeybee genes. (B) Experimentally determined levels of CG methylation increase with expression breadths in honeybee genes. Data on expression breadths are obtained from Foret et al. (2009), who combined microarray profiling of six tissues: antennae, brain, larvae, ovary, thorax, and hypopharyngeal gland. Data on experimental verified CpG methylation are from Zemach et al. (2010). Gene lengths are shown in kilobases.
However, some questions still remain before we can conclude that gene expression is the sole determinant underling the relationship between CpG O/E and gene lengths in honeybee genes. First, if the length difference between the two epigenetic groups of honeybee genes represents a general trend between gene expression and length, why do the patterns exhibit lineage-specific variation? For example, in Ac. pisum, exons of low-CpG genes were significantly longer than those of high-CpG genes. In C. intestinalis, gene bodies of low- and high-CpG genes are of similar lengths (table 1). Second, we assessed the influence of gene expression breadths on the correlation between CpG O/E and gene lengths, using partial correlation (Kim and Yi 2006, 2007). As discussed, CpG O/E and gene lengths are strongly positively correlated (Spearman’s r = 0.52, P < 2 × 10−16). Partial correlation between these two variables after removing the effect of gene expression breadths is still highly significant (Spearman’s r = 0.44, P < 2 × 10−16), although the amount of variation explained by this relationship is substantially reduced (i.e., r2 decreased by 27%).
Note, however, that the data on gene expression in honeybee are still quite limited: the data on gene expression breadths we used are from only six different tissues, and a comprehensive data set on gene expression levels of honeybee genes, normalized for different castes, is currently lacking. Furthermore, recent studies reveal that the relationship between gene length and expression is rather complex rather than a linear pattern (i.e., highly expressed genes are not necessarily shorter than lowly expressed genes: Vinogradov 2006a; Carmel and Koonin 2009).
Regulatory Complexity and Specific Functional Enrichment of Long Honeybee Genes
It has been proposed that gene lengths indicate regulatory complexity (Vinogradov 2004, 2006a, 2006b): according to this theory, longer introns represent greater amount of regulatory sequences within them, required for more complex regulation and chromatin-mediated suppression of these genes. Likewise, longer exons may be related to more complex protein functional architectures (Vinogradov 2004). High-CpG genes in A. mellifera include those that are differentially expressed between different castes (Elango et al. 2009), as well as expressed in specific sets of tissues (Foret et al. 2009). Accumulation of regulatory sequences to facilitate tissue-specific expressions of some high-CpG genes could then cause the observed length difference.
Furthermore, length increases of high-CpG genes appear to be related to specific functions. High-CpG genes are enriched with GO terms belonging to development and regulation in A. mellifera (Elango et al. 2009). For example, the top five GO categories overrepresented in high-CpG genes of A. mellifera included organ development, cell communication, and system development (Elango et al. 2009). In diverse taxa, genes belonging to developmental processes tend to be longer than the rest of genes in the genome (Yi S, unpublished data). Therefore, enrichment of long developmental genes in high-CpG class of A. mellifera may partially account for the observed length difference between the two classes.
To further investigate this hypothesis, we determined whether high-CpG genes that show particular length increase in the honeybee compared with flies (fig. 2B) are enriched in specific functional categories. Table 3 shows enrichment of specific GO terms for genes within top 100 of residuals from the regression in figure 2. These genes exhibit a striking overrepresentation of GO terms for development, particularly functions related to imaginal disc and appendage, and postembryonic development.
Table 3.
Genes with the Greatest Deviations in Length from Associations Predicted by Phylogenetic Analysis (Top 100 Residuals Genes in fig. 2B) Are Enriched in Specific GO Terms
| GO Biological Process Term | Accession | Fold Enrichment | Significancea |
| Postembryonic development | GO:0009791 | 4.23 | 1.00 × 10−04 |
| Imaginal disc development | GO:0007444 | 4.15 | 3.27 × 10−04 |
| Appendage morphogenesis | GO:0035107 | 5.44 | 4.92 × 10−04 |
| Imaginal disc-derived appendage morphogenesis | GO:0035114 | 5.44 | 4.92 × 10−04 |
| Appendage development | GO:0048736 | 5.37 | 5.73 × 10−04 |
| Imaginal disc-derived appendage development | GO:0048737 | 5.37 | 5.73 × 10−04 |
| Postembryonic organ development | GO:0048569 | 4.84 | 7.04 × 10−04 |
Significance is denoted by a Benjamini correction for multiple testing.
Insights into the Conserved and Derived Roles of DNA methylation in Animal Genomes
One of the prevailing ideas posits that DNA methylation evolved mainly to suppress deleterious transpositions of repetitive elements (Yoder et al. 1997). This is not likely to be universal (Simmen et al. 1999) and certainly not supported by our observation. If genes harboring transposable elements are primary targets of DNA methylation, we should observe longer hypermethylated genes than hypomethylated genes, exactly the opposite pattern to what we have demonstrated in this paper.
Our results, together with recent comparative analyses of DNA methylation, emphasize the importance of evolutionary perspective on understanding functional aspects of DNA methylation. In mammals, genes harboring hypermethylated promoters are silenced in most tissues, whereas those with hypomethylated promoters exhibit broad expression (Antequera 2003; Saxonov et al. 2006; Weber et al. 2007; Elango and Yi 2008). It is well accepted that promoter methylation silences gene expression. In contrast, in the honeybee and the silkworm, where DNA methylation is mainly targeted to gene bodies rather than promoters, the levels of DNA methylation are positively correlated with the breadths and levels of gene expression (fig. 3B, also Foret et al. 2009; Xiang et al. 2010). Likewise, in C. intestinalis, hypermethylated genes represent broadly expressed housekeeping genes (Suzuki et al. 2007).
Although these observations at first appear at odds with the well-established principle from the mammalian studies, newly available data on DNA methylation from diverse animals indicate that promoter methylation and subsequent silencing of gene expression actually represent a derived pattern and function of genomic DNA methylation. Comparative analyses show that promoter methylation is a vertebrate-specific feature (Elango and Yi 2008). In several invertebrate and vertebrate taxa, high levels of gene body methylation consistently manifests in moderate levels of gene expression (Feng et al. 2010; Zemach et al. 2010). Taken together, it appears that gene body methylation, which does not suppress but rather promote gene expression, likely to represent a conserved, ancestral function of DNA methylation.
DNA Methylation and Recombination on Nucleotide Content Heterogeneity of the Honeybee Genome
The honeybee genome exhibits unique characteristics that are distinct from other insect genomes. First is the presence of relatively homogeneous nucleotide “domains,” reminiscent of the classical “isochores” in genomes of mammals and birds (HoneyBee Genome Sequencing Consortium 2006). Similar to the observations in isochores, gene G + C contents are strongly correlated with domain G + C contents (HoneyBee Genome Sequencing Consortium 2006; Jørgensen et al. 2006). For example, in our data, gene G + C contents are strongly correlated with the G + C contents of surrounding genomic regions (when defined as 20 kb adjacent each gene, the Spearman’s correlation coefficient r is 0.57, P < 2 × 10−16). Jørgensen et al. (2006) performed an extensive analysis of codon and amino acid usage as well as nucleotide substitutions of honeybee genes from heterogeneous G + C domains and concluded that the presence of low G + C content domains in the honeybee genome could be explained by a distinctive AT-biased mutational process.
The nature of such mutational bias remained unknown to Jørgensen et al. (2006). With the knowledge on functional DNA methylation in the honeybee and its mutational property toward AT nucleotides, it is tempting to hypothesize that DNA methylation may lie at the origin of the heterogeneous G + C domains in the honeybee genome. It is worthwhile to note that a parallel argument exists regarding the origin of mammalian isochores: Fryxell and Zuckerkandl (2000) hypothesized that the mutagenetic property of DNA methylation and its relationship to DNA melting can explain the evolution of isochores in warm-blooded vertebrates. However, the fact that DNA methylation is only targeted toward gene bodies in the honeybee genome suggests that there may exist additional mechanisms that can explain the extension of nucleotide heterogeneity outside of genic regions.
The honeybee genome is also outstanding in its excess of CpG dinucleotides (HoneyBee Genome Sequencing Consortium 2006). This is reflected in the high CpG O/E value throughout the honeybee genome, as well as in the majority of the honeybee genes (Elango et al. 2009). We have previously proposed that biased gene conversion may partially explain this phenomenon (Elango et al. 2009). Because G + C contents and CpG O/E are significantly positively correlated in many taxa (Fryxell and Zuckerkandl 2000; Elango et al. 2008), we posited that high-CpG genes of honeybee might undergo increased biased gene conversion events and hence increase CpG O/E indirectly (Elango et al. 2009).
According to the hypothesis that biased gene conversion increases the CpG contents of high-CpG genes specifically, recombination rates of high-CpG genes should be higher than those of low-CpG genes. However, the average recombination rates of low-CpG and high-CpG genes are, not different from each other (26.0 cM/Mb and 26.5 cM/Mb, respectively, Mann–Whitney test, P = 0.7449), and there was no correlation between recombination rates and CpG O/E (P > 0.05). Thus, there is no support for a major role of biased gene conversion responsible for the excess of CpG dinucleotides of high-CpG genes. The causes of the excess CpG dinucleotides in the honeybee genome remains to be resolved.
Conclusions
The honeybee A. mellifera is an emerging model system to study molecular and evolutionary aspects of invertebrate DNA methylation. Here, we report an intriguing covariation between several genomic traits and the evolutionary signature of DNA methylation of the honeybee genes. We demonstrate that long genes are found preferentially in hypomethylated class, whereas hypermethylated genes are short. Comparative analyses indicate that the length distinction between the two classes of genes has a deep evolutionary origin, tracing back well beyond to the split of Diptera and Hymenoptera. We demonstrate that several factors, including selection for transcription efficiency, functional loads, regulatory complexity as potential mechanisms underlying the covariation between genomic traits and DNA methylation. Thus, DNA methylation may play critical regulatory roles and influence genome evolution in distinctive ways. With the anticipated additional data on genomic and transcriptomic profiles from several Hymenopteran outgroups (e.g., Smith et al. 2008), we can elucidate the dynamics of genome evolution in relation to epigenetic regulation of gene expression more deeply in a near future. In particular, it is of great interest to determine whether the observed covariation of gene length and epigenetic status are the ancestral pattern in arthropods.
Supplementary Material
Supplementary material, figures 1–5, and table 1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/.
Acknowledgments
We thank Martin Beye for sharing data on honeybee recombination rates, Silvain Foret for sharing data on expression breadths. Jungsun Park, Brendan Hunt, and Eddie Loh have provided valuable computational helps and discussions, and Michael Goodisman and Brendan Hunt provided comments on an earlier version of the manuscript. This study is supported by funds from the Georgia Institute of Technology and an National Science Foundation grant (MCB-0950896).
References
- Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AJ, Welch JJ, Charlesworth B. Reduced effectiveness of selection caused by a lack of recombination. Curr Biol. 2009;19:655–660. doi: 10.1016/j.cub.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Beye M, et al. Exceptionally high levels of recombination across the honey bee genome. Genome Res. 2006;16:1339–1344. doi: 10.1101/gr.5680406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel L, Koonin EV. A universal nonmonotonic relationship between gene compactness and expression levels in multicellular eukaryotes. Genome Biol Evol. 2009;1:382–390. doi: 10.1093/gbe/evp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Clark AG. Intron size and natural selection. Nature. 1999;401:344. doi: 10.1038/43827. [DOI] [PubMed] [Google Scholar]
- Chargaff E. Structure and function of nucleic acids as cell constituents. Fed Proc. 1951;10:654–659. [PubMed] [Google Scholar]
- Colot V, Rossignol JL. Eukaryotic DNA methylation as an evolutionary device. Bioessays. 1999;21:402–411. doi: 10.1002/(SICI)1521-1878(199905)21:5<402::AID-BIES7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M. The correlation between intron length and recombination in Drosophila: dynamic equilibrium between mutational and selective forces. Genetics. 2000;156:1175–1190. doi: 10.1093/genetics/156.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca TF, et al. Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics. 2006;22:2044–2046. doi: 10.1093/bioinformatics/btl286. [DOI] [PubMed] [Google Scholar]
- Dennis G, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Duret L, Galtier N. The covariation between TpA deficiency, CpG deficiency, and G + C content of human isochores is due to a mathematical artifact. Mol Biol Evol. 2000;17:1620–1625. doi: 10.1093/oxfordjournals.molbev.a026261. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon E. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Elango N, Hunt BH, Goodisman MAD, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci U S A. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Kim SH NISC Sequencing Program, Vigoda E, Yi SV. Mutations of different molecular origins exhibit contrasting patterns of regional substitution rate variation. PLoS Comput Biol. 2008;4:e1000015. doi: 10.1371/journal.pcbi.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Yi SV. DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol. 2008;25:1602–1608. doi: 10.1093/molbev/msn110. [DOI] [PubMed] [Google Scholar]
- Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foret S, Kucharski R, Pittelkow Y, Lockett G, Maleszka R. Epigenetic regulation of the honeybee transcriptome: unravelling the nature of methylated genes. BMC Genomics. 2009;10:472. doi: 10.1186/1471-2164-10-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryxell KJ, Zuckerkandl E. Cytosine deamination plays a primary role in the evolution of mammalian isochores. Mol Biol Evol. 2000;17:1371–1383. doi: 10.1093/oxfordjournals.molbev.a026420. [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands-'A rough guide'. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Jørgensen FG, Schierup MH, Clark AG. Heterogeneity in regional GC content and differential usage of codons and amino acids in GC-poor and GC-rich regions of the genome of Apis mellifera. Mol Biol Evol. 2006;24:611–619. doi: 10.1093/molbev/msl190. [DOI] [PubMed] [Google Scholar]
- Kim SH, Yi SV. Correlated asymmetry between sequence and functional divergence of duplicate proteins in Saccharomyces cerevisiae. Mol Biol Evol. 2006;23:1068–1075. doi: 10.1093/molbev/msj115. [DOI] [PubMed] [Google Scholar]
- Kim SH, Yi SV. Understanding relationship between sequence and functional evolution in yeast proteins. Genetica. 2007;131:151–156. doi: 10.1007/s10709-006-9125-2. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- Ponger L, Li WH. Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol Biol Evol. 2005;22:1119–1128. doi: 10.1093/molbev/msi098. [DOI] [PubMed] [Google Scholar]
- Rudner R, Karkas JD, Chargaff E. Separation of B. subtilis DNA into complementary strands. 3. Direct analysis. Proc Natl Acad Sci U S A. 1968;60:921–922. doi: 10.1073/pnas.60.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen MW, et al. Nonmethylated transposable elements and methylated genes in a chordate genome. Science. 1999;283:1164–1167. doi: 10.1126/science.283.5405.1164. [DOI] [PubMed] [Google Scholar]
- Smith CR, Toth AL, Suarez AV, Robinson GE. Genetic and genomic analyses of the division of labour in insect societies. Nat Rev Genet. 2008;9:735–748. doi: 10.1038/nrg2429. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Kerr ARW, De Sousa D, Bird A. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 2007;17:625–631. doi: 10.1101/gr.6163007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia AO, Hurst LD. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. Compactness of human housekeeping genes: selection for economy or genomic design? Trends Genet. 2004;20:248–253. doi: 10.1016/j.tig.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. “Genome design” model and multicellular complexity: golden middle. Nucleic Acids Res. 2006a;34:5906–5914. doi: 10.1093/nar/gkl773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. Genome design” model: evidence from conserved intronic sequence in human–mouse comparison. Genome Res. 2006b;16:347–354. doi: 10.1101/gr.4318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TK, et al. A functional DNA methylation system in the pea aphid Acyrthosiphon pisum. Insect Mol Biol. 2010;19(Suppl 2):215–228. doi: 10.1111/j.1365-2583.2009.00974.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Functional CpG methylation system in a social insect. Science. 2006;314:645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- Wang Y, Leung FCC. In silico prediction of two classes of honeybee genes with CpG deficiency or CpG enrichment and sorting according to gene ontology classes. J Mol Evol. 2009;68:700–705. doi: 10.1007/s00239-009-9244-3. [DOI] [PubMed] [Google Scholar]
- Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Xiang H, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol. 2010;28:516–520. doi: 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- Yi S, Charlesworth B. Contrasting patterns of molecular evolution of genes on the new and old sex chromosomes of Drosophila miranda. Mol Biol Evol. 2000;17:703–717. doi: 10.1093/oxfordjournals.molbev.a026349. [DOI] [PubMed] [Google Scholar]
- Yi SV, Goodisman MAD. Computational approaches for understanding the evolution of DNA methylation in animals. Epigenetics. 2009;4:551–556. doi: 10.4161/epi.4.8.10345. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]