Abstract
Genes of the immune system are generally considered to evolve rapidly due to host–parasite coevolution. They are therefore of great interest in evolutionary biology and molecular ecology. In this study, we manually annotated 144 avian immune genes from the zebra finch (Taeniopygia guttata) genome and conducted evolutionary analyses of these by comparing them with their orthologs in the chicken (Gallus gallus). Genes classified as immune receptors showed elevated dN/dS ratios compared with other classes of immune genes. Immune genes in general also appear to be evolving more rapidly than other genes, as inferred from a higher dN/dS ratio compared with the rest of the genome. Furthermore, ten genes (of 27) for which sequence data were available from at least three bird species showed evidence of positive selection acting on specific codons. From transcriptome data of eight different tissues, we found evidence for expression of 106 of the studied immune genes, with primary expression of most of these in bursa, blood, and spleen. These immune-related genes showed a more tissue-specific expression pattern than other genes in the zebra finch genome. Several of the avian immune genes investigated here provide strong candidates for in-depth studies of molecular adaptation in birds.
Keywords: genomics, bird, immunogenetics, next-generation sequencing, digital transcriptomics, Taeniopygia guttata
Introduction
Genes of the immune system have been found to show signatures of positive selection in genome-wide scans (Nielsen et al. 2005; Wang et al. 2006; Axelsson et al. 2008). The selection has generally been attributed to an evolutionary arms race between parasites and hosts. However, the kind of selection observed will often differ between different classes of immune genes and within individual loci (Mukherjee et al. 2009). Gene duplications that ultimately result in large multigene families are also common in genes involved in disease resistance, such as major histocompatibility (MHC) genes, toll-like receptors, and antimicrobial peptides (Nei et al. 1997; Wong et al. 2007; Hughes and Piontkivska 2008), again implying a significant role of selection in shaping immune genes. These genes have therefore received considerable attention in evolutionary biology and molecular ecology as candidates for local adaptation and for studying functionally important polymorphism.
In recent years, the focus of molecular ecology and conservation genetics has shifted from classical studies of selectively neutral variation to studies of functionally important genes (Sommer 2005; Piertney and Webster 2010). Many studies that have used such a candidate gene approach to study these topics in vertebrates have investigated variation in MHC genes and have especially focused on the antigen peptide–binding exons of MHC class I and class IIβ genes (see e.g., Edwards et al. 2000; Sommer et al. 2002; Aguilar et al. 2004; Kurtz et al. 2004; Ekblom et al. 2007; Westerdahl 2007; Alcaide et al. 2008; Babik et al. 2008; Burri et al. 2008; Hale et al. 2009). MHC genes are important in vertebrate immune defense and have been investigated in order to answer a range of ecologically important questions. However, it has been argued that focusing on only one or a few specific immune genes (like the MHC) may not give a very complete picture of genetic variation of the complex vertebrate immune system (Acevedo-Whitehouse and Cunningham 2006). One of the strategies suggested to change this picture is to survey variation in a large number of immune candidate genes; for example, in mosquitoes (Waterhouse et al. 2007), salmon (Tonteri et al. 2010), and Drosophila (Obbard et al. 2009). However, multicandidate gene studies have been unrealistic in birds due to the lack of comparative genomic information (Edwards 2007); until recently the domestic chicken (Gallus gallus) was the only bird with a characterized genome (International Chicken Genome Sequencing Consortium 2004). Recently, the second bird genome, that of the zebra finch (Taeniopygia guttata), was released (Warren et al. 2010), opening up this field for avian research (Ellegren 2007; Clayton et al. 2009). This resource, together with advances in sequencing technology (Hudson 2008; Morozova and Marra 2008), now facilitates the multicandidate gene approach for studying avian immune genes.
Complementary to the molecular evolution approach is the idea of examining variation in gene expression between tissues, individuals, populations, and even species. Traditionally, gene expression studies were conducted using microarrays and were largely restricted to genetic model species (see e.g., Nuzhdin et al. 2004; Rottscheidt and Harr 2007). The use of massively parallel pyrosequencing means that it is now feasible to get data on both gene sequence and gene expression from transcriptomes of nonmodel organisms very quickly and at a reasonable cost (Vera et al. 2008). This high-throughput digital transcriptomics approach, known as RNA-Seq (Wang et al. 2009) has received a lot of attention in recent years, and its application will increase further with new technological advances in sequencing chemistry and bioinformatics.
Immune genes differ extensively in their expression pattern with certain genes, for example, MHC class II, being expressed only in specialized immune tissues, whereas others, such as MHC class I, have equal expression in most tissues (Roitt 1997). Some genes are expressed at all times, whereas others are only turned on or are upregulated following a specific infection (Wang et al. 2006). Variation in expression of specific genes between individuals may contribute substantially to resistance or susceptibility to infection (Schadt et al. 2005; Dixon et al. 2007).
Here, we investigate molecular evolution on avian immune genes by comparing chicken and zebra finch coding sequences (CDSs). The rate of nonsynonymous (dN) and synonymous (dS) nucleotide substitutions of immune genes is contrasted against the genome-wide rate, and substitution rates are also compared between different classes of immune genes, to find out what categories of genes involved in different parts of the immune response are most rapidly evolving. There may be tendency for dN/dS ratios to become unreliable over large evolutionary distances (such as our zebra finch–chicken comparison) due to saturation of the dS (Smith JM and Smith NH 1996). There is also a risk for dN/dS to be underestimated for distant comparisons if rapidly evolving genes are systematically excluded from the analyses due to the difficulty of correctly aligning and annotating them. However, such methods to infer signatures of selection have been widely used in the past (Ellegren 2008) and have been shown to yield informative results (e.g., Künstner et al. 2010). Furthermore, simulations have shown that likelihood tests (such as used here) to detect selection in deep lineages of vertebrates are generally robust against this problem (Studer et al. 2008). We also describe tissue-specific expression patterns of zebra finch immune genes using a next-generation digital transcriptomic approach; RNA-Seq (Nagalakshmi et al. 2008; Wang et al. 2009).
These analyses are performed on a data set of 144 manually annotated immune genes. Because the nomenclature of immune genes can sometimes be particularly confusing with several names being used for a single gene, we provide an extensive list of alternative names of these genes in addition to the official symbol (supplementary Appendix 1, Supplementary Material online). Manual annotation is a slow process, which is not feasible for a whole genome (Curwen et al. 2004). Instead, genomes are primarily annotated via automated systems, and the accuracy of these predictions is important for downstream analysis (Altenhoff and Dessimoz 2009). The incorrect alignment of genes increases the risk of falsely detecting positive selection (Hughes and Friedman 2008; Mallick et al. 2009; Schneider et al. 2010). Therefore, we also compare the results obtained from the analysis of manually annotated genes with data from the same genes generated by automated gene predictions available in the Ensembl database.
Materials and Methods
Compiling a List of Avian Immune Genes
We made an extensive search of the scientific literature for genes that have been described as being important to the bird immune response and disease resistance. Most of these studies have been performed on chicken, but a few have also investigated immune genes in other species (for references see supplementary Appendix 1, Supplementary Material online), such as duck (Anas platyrhynchos) and quail (Coturnix sp.). In addition, we also searched the NCBI Entrez Gene database for chicken genes containing gene ontology terms associated with immune response (“immune response,” “inflammatory,” “resistance,” “B cell,” and “T cell”).
Search for Homologs of Chicken Immune Genes in the Zebra Finch Genome
The zebra finch genome sequence (version 3.2.4) was downloaded from the Washington University Genome Sequencing Center web site (http://genome.wustl.edu/). The nucleotide CDS and protein amino acid sequence for each of the identified chicken immune genes were downloaded from the NCBI gene site (http://www.ncbi.nlm.nih.gov/gene/). The zebra finch genome was searched for homologues to these sequences using version 2.2.18 of stand-alone BlastN and TBlastN (Altschul et al. 1997). For rapidly evolving multigene families (like MHC), we aligned the CDS for several species of birds and mammals. Conserved regions within each of the exons were then used to search (TBlastN) for the orthologous regions of the zebra finch genome.
Aligning and Annotating Zebra Finch Immune Genes
Regions of the zebra finch genome (usually 20 kbp surrounding the location of the Blast hit) with significant Blast hits (e value < 1 × 10−10) against chicken immune genes were aligned to each exon of the chicken gene using ClustalW (Thompson et al. 1994). The alignments were then carefully manually checked using BioEdit (Hall 1999), for example, exon–intron boundaries were verified (and shifted where appropriate) using the GT-AG rule (in the cases where this criterion was fulfilled in the chicken equivalent); start and stop codons were verified in the first and last exon; individual exons were removed if there was no clear match in the zebra finch genome sequence; and frame shifting gaps were corrected if possible and exons with frame shifting gaps that could not be corrected were removed. The zebra finch CDS was then constructed by combining the different identified exons. This was Blasted back against the chicken genome, and the principle of best reciprocal Blast hit was used to determine if the two sequences were orthologs (Overbeek et al. 1999). For each confirmed orthologous pair of chicken and zebra finch genes, the CDSs were then aligned using ClustalW and checked manually (in a few cases, gaps were manually introduced or removed at this stage to improve the alignment) before performing downstream analyses. In total, 144 zebra finch–chicken orthologs were annotated in this study. For 25 of the genes, automated zebra finch gene predictions were downloaded directly from the NCBI gene database and used as guidance (e.g., to locate exon–intron boundaries) in the annotation process (note that these genes were not used in the analysis comparing manual annotation and Ensembl gene prediction).
Evolutionary Analyses
The Codeml application in PAML4 (Yang 2007), run using the IDEA interface (Egan et al. 2008), was used to perform evolutionary analysis of immune genes. For pairwise analyses (using only data from zebra finch and chicken), runmode was set to −2 and NSsites to 0. The ω values (dN/dS) were then averaged over all identified immune genes and compared with the genome-wide ω value obtained by downloading data on zebra finch–chicken orthologs genes from Ensembl (version 57) BioMart (http://www.ensembl.org/biomart/martview/). dN/dS data from 14,800 zebra finch–chicken orthologs (the most similar zebra finch ortholog for each chicken gene) were used for this comparison. We also compared our ω values with those obtained in a whole-transcriptome analysis of zebra finch brain expressed sequence tags (ESTs) (Axelsson et al. 2008). Note that the brain EST data set may be biased toward evolutionary conserved genes, if these are more likely to be expressed in brain. There may also be a slight similar bias in the genome-wide BioMart data set if conserved genes are more likely to be annotated in both chicken and zebra finch compared with fast evolving genes.
For genes where we had sequence data from more than two species of birds, we also performed Codeml analyses using runmode 0 and NSsites models 1, 2, 7, and 8. NSsites 1 and 7 represent models of neutral evolution, whereas models 2 and 8 allow for positive selection (ω > 1) on parts of the gene. To test for signs of positive selection, a likelihood ratio test (LRT) was performed (using IDEA) between models 1 and 2 and between models 7 and 8. If evidence of positive selection was found in a gene (model 2 or 8 better fitting the data compared with model 1 or 7), sites within that gene under selection (using the best model) were identified using the Bayes empirical Bayes approach in PAML (Yang 2007). In addition, sites evolving under positive selection were also identified in genes with sequences from three or more species using the random effects likelihood method implemented in the HyPhy software (Pond et al. 2005), through the Datamonkey web interface (Pond and Frost 2005). Codon-wise ω and posterior probabilities of positive selection across genes were inferred using omegaMap version 0.5 (Wilson and McVean 2006) and plotted using R 2.7.2 (R Development Core Team 2008). The analysis was run twice for each gene for 75,000 generations. After discarding the first 5,000 generations as “burn-in,” the two independent runs of each locus were assessed manually for convergence and then combined.
Functional Categories of Immune Genes
To compare signatures of selection and expression between different categories of immune genes, our manually annotated genes were classified according to their specific function. Information on functional categories was extracted from the IRIS database (Kelley et al. 2005). This database places each immune gene into one or more of the following 22 functional categories: Functions in innate immunity, inflammation, coagulation, complement, phagocytosis, innate killing (including natural killer [NK] cell function), chemotaxis/cell adhesion, cytokine/chemokine, adaptive immunity, cellular immunity (including immune-related apoptosis), humoral immunity (including antibody-related genes and B cell function), antigen processing, pathways or signaling that result in expression of immune molecules, development of immune system (including receptor formation), hematopoiesis (and maturation/selection), induced by immunomodulator, expressed primarily in immune tissues, involved in immunodeficiency, involved in autoimmunity, associated with other disease, immune receptor, and other putative immune function. Testing for differences of ω between genes in the various categories were performed using a linear regression model (lm function in R), with ranked pairwise ω as response variable and fitting presence or absence in each of the functional categories for all genes as separate effects.
Expression Profiling of Zebra Finch Immune Genes
Expression levels of the manually annotated zebra finch immune genes were analyzed using a digital transcriptomics approach (RNA-Seq; Wang et al. 2009). Roche 454 sequencing data from cDNA libraries of eight different tissues (blood, bursa, embryo, liver, muscle, skin, spleen, and testes) were trimmed and assembled using NGen 2.0 (DNASTAR, Inc.). In total, 2,020,514 sequence reads were entered into the assembly. The cDNA came from pools of six different adult zebra finch individuals form the captive population at the University of Sheffield (Stapley et al. 2008), except for the blood which was derived from only one individual and the bursa which came from a pool of four 18-day-old chicks. For details about library construction and sequence assembly, see Ekblom et al. (2010). The sequence reads entered into the assembly are available from the NCBI trace archive (http://www.ncbi.nlm.nih.gov/Traces/home/).
The nucleotide CDSs for each of the manually annotated zebra finch immune genes were Blasted (BlastN) against all contigs (n = 49,606) and singletons (n = 1,298,597) from the 454 sequence assembly. The best hit (with an e value smaller than 1 × 10−10) for each contig and singleton were kept as immune gene candidates and Blasted back (reciprocal BlastN) against the full data set of zebra finch gene predictions (ftp://ftp.ncbi.nih.gov). Ninety-four contigs and 1,182 singletons gave significant (e < 1 × 10−10) best reciprocal Blast hits against one of the immune genes examined here, representing a total of 106 different genes.
For each gene, the numbers of reads from contigs and singletons were counted for each tissue separately. Expression of each gene in every tissue, calibrated for transcript length was defined as the number of reads per million per kilobase (RPKM), according to the method in Mortazavi et al. (2008). The index of tissue specificity of expression (τ) (Yanai et al. 2005) was calculated according to Mank et al. (2008). Theoretically, τ ranges from 0 to 1 with low numbers indicating even expression over the sampled tissues and high numbers being obtained for genes expressed in only one tissue. The τ index for immune genes was contrasted against τ calculated from all zebra finch genes, from Ekblom et al. (2010). Only expression data from the six tissues investigated by Ekblom et al. (2010) were used in this comparison. The tissue of maximal expression was defined as the tissue with the highest RPKM for the gene in question.
Comparison of Manual Annotation and Ensembl Data
To evaluate the automatic gene annotation in Ensembl, the ω values for zebra finch–chicken comparisons of immune genes were compared with our manually calculated omega values for the same genes. For the zebra finch immune genes that were annotated without using a priori information from automated zebra finch gene predictions, we downloaded dN and dS values from the Ensembl database (release 54) using the BioMart web interface (http://www.ensembl.org/biomart/martview). For genes with more than one zebra finch ortholog on Ensembl, the pair with least sequence difference (minimum dN + dS) was selected.
Statistical Analyses
Because the dN/dS ratios of genes were generally not normally distributed, nonparametric statistics were used in tests including this parameter. Statistical analyses and the handling of large data files were performed using R 2.7.2 (R Development Core Team 2008).
Results
Evolutionary Analyses of Zebra Finch Immune Genes
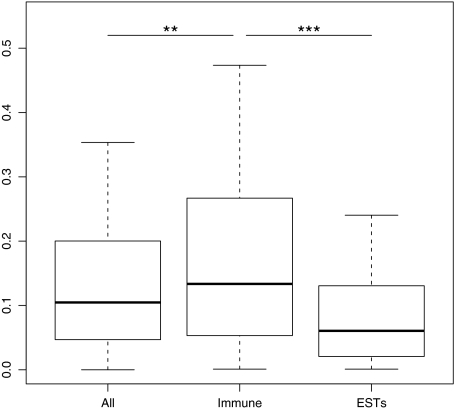
A total of 144 chicken–zebra finch immune genes orthologs were found using our manual annotation (Appendix 1 and 2). Pairwise dN/dS (ω) values varied from 0.0001 to 1.5, with a median value of 0.134. This was significantly higher than the ω obtained for all genes in the genome (median = 0.105, Mann–Whitney U (MWU) test, W = 93,6134, P = 0.01, fig. 1). The ω values of immune genes were also higher than the whole-genome values calculated from zebra finch brain ESTs (Axelsson et al. 2008) (median = 0.061, MWU test, W = 24,7158, P < 0.0001, fig. 1). The median dS value for all immune genes was 0.52 with the third quartile also below one suggesting that substitution saturation between chicken and zebra finch is probably not a big problem for analyses of most of the genes studied here.
FIG. 1.—
Box-and-whisker plot of dN/dS (ω) values between zebra finch and chicken orthologs for immune genes and whole-genome comparison (All genes). Whole-brain transcriptome comparisons (EST) between chicken and zebra finch (Axelsson et al. 2008) are also included as reference.
For 27 immune genes, sequence information was available from three or more bird species. In these cases, we also tested for positive selection acting on one or more codons in each gene. Six of these genes (21%) showed evidence for selection in the PAML analysis (LRT, P < 0.05, table 1), with the number of codons under selection varying from 1 to 10. In the HyPhy analysis, an additional four genes were identified as having sites under positive selection with the number of identified codons ranging between 2 and 16 (table 1). For plots of codon-wise ω and posterior probability of positive selection across the length of these genes, see supplementary Additional figures 1–10 (Supplementary Material online).
Table 1.
Genes with Data from At Least Three Bird Species that Were Identified as Targets of Positive Selection
| Gene Symbol | Number of Bird Species | χ2 | P value | Number (and identity) of Positively Selected Sites, from PAML | Number (and identity) of Positively Selected Sites, from HyPhy |
| BLB2 (MHCIIB) | 9 | 118.605 | <0.0001 | 10 (34–37, 81, 91, 94, 95, 97, 101) | 7 (36, 38, 39, 64, 83, 97, 104) |
| BF2 (MHCI) | 7 | 37.888 | <0.0001 | 6 (66, 79, 86, 124, 126, 165) | 4 (81, 103, 184, 366) |
| B-NK | 4 | 16.576 | 0.0003 | 3 (5, 6, 60) | 6 (83, 84, 85, 86, 98, 156) |
| IL1B | 8 | 16.666 | 0.0002 | 2 (47, 192) | 7 (50, 53, 60, 76, 77, 224, 255) |
| MX1 | 6 | 12.291 | 0.0021 | 3 (212, 388, 436) | 16 (20, 130, 230, 241, 265, 318, 345, 357, 406, 454, 486, 506, 537, 538, 539, 585) |
| IFNG | 6 | 6.974 | 0.0306 | 1 (111) | 2 (111, 159) |
| CD9 | 8 | 5.534 | 0.0629 | 0 | 8 (39, 92, 152, 169, 174, 179, 186) |
| B2M | 3 | 3.950 | 0.139 | 0 | 3 (37, 73, 113) |
| CD44 | 3 | 1.187 | 0.552 | 0 | 11 (127, 184, 242, 245, 268, 277, 282, 285, 292, 299, 393) |
| PIK3AP1 | 3 | 0.804 | 0.669 | 0 | 8 (55, 62, 81, 166, 508, 664, 752, 760) |
Codons with a posterior probability greater than 95% for being under positive selection (calculated in PAML and HyPhy) are indicated. For plots of codon-wise ω and posterior probability of positive selection across the length of these genes, see Additional figs. 1–10.
Analysis of Functional Categories of Immune Genes
There was information on the functional category available in the IRIS database for 103 immune genes identified in the zebra finch. Genes in the category “immune receptor” had significantly higher dN/dS ratios than other immune genes included in this study (Table 2). There was also a tendency for genes in the category “expressed primarily in immune tissues” to have higher dN/dS ratios, whereas genes in the categories “cytokines and chemokines” and “transcription factor” tended to have lower than average dN/dS ratios (Table 2).
Table 2.
Regression Model of ω Values for the Different Functional Categories of Immune Genes
| Category | Median ω | N | Model Estimate | Model Standard error | t | P |
| Intercept | 35.3924 | 7.5845 | 4.666 | 1.18 × 10−5*** | ||
| Receptor | 0.260 | 23 | 19.5739 | 8.198 | 2.388 | 0.0193* |
| Antigen processing | 0.236 | 12 | 10.7259 | 11.3136 | 0.948 | 0.3459 |
| Humoral response | 0.219 | 8 | 13.4626 | 13.8312 | 0.973 | 0.3332 |
| Development of immune system | 0.219 | 6 | 7.3719 | 13.4588 | 0.548 | 0.5854 |
| Involved in autoimmunity | 0.210 | 3 | 4.402 | 18.3264 | 0.24 | 0.8108 |
| Innate NK killing | 0.200 | 7 | −6.1044 | 15.3811 | −0.397 | 0.6925 |
| Expressed primarily in immune tissues | 0.200 | 25 | 12.2644 | 7.3473 | 1.669 | 0.0989† |
| Inflammation | 0.190 | 20 | 17.8269 | 11.6274 | 1.533 | 0.1291 |
| Innate immunity | 0.189 | 46 | 6.8735 | 8.7715 | 0.784 | 0.4355 |
| Adaptive immunity | 0.178 | 26 | −5.0489 | 10.6001 | −0.476 | 0.6351 |
| Related to disease | 0.177 | 24 | 0.6073 | 7.2483 | 0.084 | 0.9334 |
| Immune pathway or signaling | 0.174 | 47 | 10.2255 | 6.2947 | 1.624 | 0.1081 |
| Involved in immunodeficiencies | 0.165 | 10 | 5.6751 | 10.546 | 0.538 | 0.5919 |
| Chemotaxis | 0.157 | 16 | 2.9733 | 10.3082 | 0.288 | 0.7737 |
| Induced by immunomodulator | 0.149 | 21 | 3.8621 | 7.8428 | 0.492 | 0.6237 |
| Coagulation | 0.149 | 6 | −17.9895 | 15.0288 | −1.197 | 0.2348 |
| Phagocytosis | 0.142 | 3 | −13.6846 | 19.0216 | −0.719 | 0.4739 |
| Cellular response | 0.138 | 12 | 20.7341 | 12.8684 | 1.611 | 0.111 |
| Cytokines and chemokines | 0.122 | 38 | −12.4645 | 6.8252 | −1.826 | 0.0715† |
| Transcription factor | 0.080 | 6 | −24.2605 | 13.1965 | −1.838 | 0.0696† |
***P < 0.001; *P < 0.05; †P < 0.10.
Expression Profiling of Immune Genes
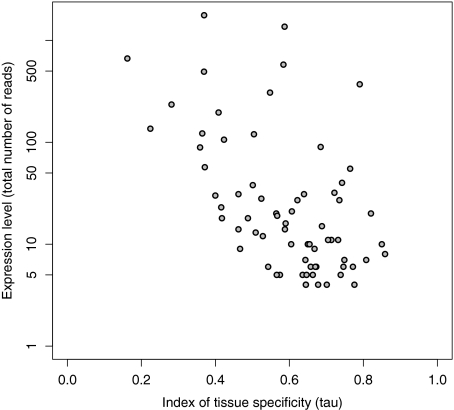
Expression levels of immune genes were estimated using RNA-Seq data from eight different tissues. The average contig length was only 150 bp but still about 65% of the zebra finch transcriptome was covered (Ekblom et al. 2010). We found evidence for expression of 106 of the 144 genes related to the immune system investigated in this study. Immune genes were expressed in a more tissue-specific manner (mean τ = 0.54) than other genes (mean τ = 0.49) in the genome (t = 2.35, degrees of freedom [df] = 60.2, P = 0.022). Most immune genes were mainly expressed in bursa, blood, and spleen, whereas only a few immune genes had primary expression in muscle, embryo, and skin (Table 3). The index of tissue specificity of gene expression (τ) was negatively correlated with (log) total expression level (rP = −0.541, df = 67, P < 0.0001, fig. 2). Thus, genes with high tissue specificity in expression (indicating a more specific function) generally appeared to be expressed at a lower level than genes with more even expression levels across the sampled tissues (indicating a more general function).
Table 3.
Numbers of Immune Genes with Primary Expression in Each of the Studied Tissues
| Tissue | Number of Genes | Mean ω | Mean τ |
| Bursa | 28 | 0.28 | 0.63 |
| Blood | 28 | 0.17 | 0.60 |
| Spleen | 19 | 0.26 | 0.63 |
| Testes | 11 | 0.09 | 0.48 |
| Liver | 8 | 0.26 | 0.61 |
| Skin | 6 | 0.22 | 0.46 |
| Embryo | 5 | 0.22 | 0.53 |
| Muscle | 1 | 0.13 | — |
Mean values of ω (dN/dS ratio) and τ (index of expression specificity) for genes with primary expression in each of the tissues are also reported.
FIG. 2.—
Relationship between total expression levels and tissue specificity in expression (τ) for zebra finch immune genes.
Comparison between Manual Annotation and Ensembl Automatic Annotation
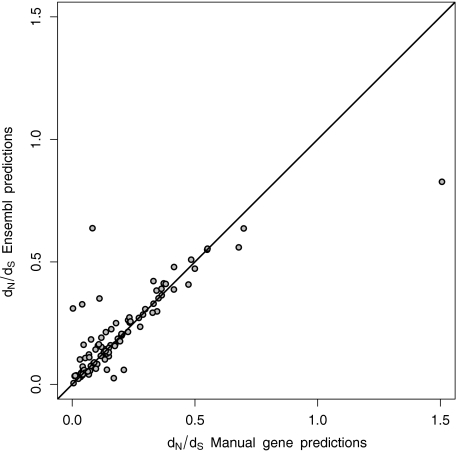
A total of 119 zebra finch–chicken orthologs for immune genes were identified without using any a priori information from automated zebra finch gene predictions. For 95 of these zebra finch–chicken orthologs, gene pairs had also been identified by the automated Ensembl gene prediction pipeline. We found a very strong positive correlation between ω values obtained from the manual annotation and those from automated gene prediction (rS = 0.811, df = 93, P < 0.0001, fig. 3). However, ω values calculated from manually annotated gene pairs (median = 0.137) tended to be lower than the corresponding values downloaded from the Ensembl database (median = 0.160, Wilcoxon's test for matched pairs, T = 1805, df = 94, P = 0.08). In only a few cases were there conspicuous differences between the two different annotations (fig. 3). In particular, the gene coding for “integrin-associated protein (IAP, CD47)” showed higher ω for the manually annotated transcript, whereas the gene coding for “interleukin 2 receptor, gamma” had a much higher ω value as calculated from the Ensembl gene prediction. In both these cases, the discrepancy arises from the fact that a large part of the CDS was left out in the manual annotation compared with the automated ENSEMBL annotation.
FIG. 3.—
Correlation between dN/dS (ω) ratios of automated and manually annotated genes. Automated dN/dS ratios were downloaded from Ensembl BioMart, whereas the manually annotated ratios were calculated in PAML4. The line represents x = y. The outlier to the right (with Manual ω = 1.51 and Ensembl ω = 0.83) is the gene for “integrin-associated protein (IAP, CD47)” and the outlier to the left (with Manual ω = 0.08 and Ensembl ω = 0.64) is the gene for interleukin 2 receptor, gamma.
Discussion
We found higher values of ω (dN/dS) between the chicken and the zebra finch immune genes than for the rest of the genome. This suggests that these genes are in general rapidly evolving and that host–parasite interaction is an important selective force on avian genomes. Positive selection is often evoked to explain such high ω values; note, however, that relaxed evolutionary constraints may have the same effect and these two mechanisms cannot be distinguished with our data. In particular, immune receptors had higher ω values compared with other categories of immune genes. Because many such gene products might be involved directly in antigen recognition, it seems reasonable that these should be the primary target for host–pathogen coevolution (Borghans et al. 2004). This category includes several toll-like receptors and interleukin receptors as well as genes linked to the MHC region. MHC genes in particular are well known to be affected by balancing selection from previous studies (Sommer 2005; Westerdahl 2007). There was also a tendency for genes expressed primarily in immune tissues to have high ω values. Genes that have a more specific immune function may therefore be less constrained than genes with a more general expression. This pattern has also been observed in genome-wide tests of selection in a wide array of organisms (Axelsson et al. 2008; Larracuente et al. 2008; Ekblom et al. 2010) and is thus not restricted to genes of the immune system.
We identified ten of 27 tested immune genes (for which sequence information was available from at least three bird species) that showed evidence of being under positive selection in birds. It is likely that further sequencing in other bird species (enabling this kind of analysis for a larger number of genes) in the future will reveal more immune genes that are evolving under positive selection. In a study of 136 immunity genes in multiple Drosophila species Obbard et al. (2009) concluded that the rate of adaptive evolution were higher (and more variable) in these compared with nonimmune genes. In particular, genes belonging to certain immunological pathways were found to be very rapidly evolving. Similarly, in mosquitoes, different classes of immune genes are evolving at different rates (Waterhouse et al. 2007). In atlantic salmon (Salmo salar), microsatellites linked to immune genes were showing signs of adaptive evolution more often than other microsatellites (Tonteri et al. 2010). These results together with the conclusions from our study indicate that parasites may be an important factor driving molecular evolution in many different systems. And that a large number of immune-related genes may be influenced by adaptive evolution.
Of the ten loci where specific amino acid residues were identified as candidates for being under positive selection, MHC class I and IIB genes have already been under detailed investigation elsewhere. The general pattern is that selection seems to favor polymorphism in residues that deal specifically with antigen binding (Edwards and Hedrick 1998). The codons identified as evolving under positive selection in our analysis of avian MHC class IIB genes overlap considerably with regions that have previously been identified as important to antigen binding (Brown et al. 1993; Tong et al. 2006), especially in the beginning (codons 34–39) and middle (codons 82–115) of the second exon of this gene. Similarly, a large region around codon number 100 of the MHC class I gene was identified as having a high ω value in our sliding window analysis. This region, situated in exon 2 has previously been shown to contain many codons directly involved in antigen binding (Westerdahl et al. 1999). However, our analyses failed to identify the peptide-binding regions of exon 3 as evolving under positive selection.
The B-NK gene (also known as Blec2 or natural killer like receptor) is also linked to the MHC region in chicken, but seems to be located on the Z chromosome in zebra finch (Balakrishnan et al. 2010). The exact function of this gene in birds is not known but receptors on NK cells are generally involved in MHC class I recognition and cell-mediated cytotoxicity (Lanier 1998). Interleukin 1β (IL1B) is an important cytokine (a small secreted protein that carries signals between immune cells) produced by macrophages and involved in the inflammatory response. Some sequence variation exists in mammalian IL1B genes, and this has been shown to be related to particular disease (Bird et al. 2002). Interestingly, many of the aligned bird sequences seem to have gaps extending over traditionally conserved amino acid residues, involved in receptor binding such as the histidine at position 141 (146 in the human protein) and valine at position 250 (the last position of the IL-1 family signature). The MX1 gene has been extensively studied in birds due to its function in protection against avian flu. There are several polymorphisms in the avian MX1 genes and in particular a transition from aspargine to serine at amino acid position 631 (position 563 in our incomplete alignment) seem to mediate antiviral activities (Li et al. 2006). Our analyses revealed several codons with a signature of positive selection and a peak of ω in the region just upstream from this position. An avian-specific region in the beginning of the MX1 gene has also been found to be rich in codons affected by positive selection (Berlin, Qu, Li, et al. 2008). Unfortunately, we were not able to identify this part of the coding region in our manual annotation of the zebra finch MX1 homolog. The CD9 gene is immunologically important for platelet activation and aggregation but has mainly been studied because of its role in gamete fusion during fertilization in animals (Clark et al. 2006). Our results on this gene are similar to other studies showing a hotspot of positively selected sites around codons 150–186 (Swanson et al. 2003; Berlin, Qu, and Ellegren 2008).
In general, zebra finch immune genes were found to have a more tissue-specific expression pattern than other genes in the zebra finch genome. A majority of the immune-related genes were primarily expressed in bursa, blood, and spleen. Similar patterns of immune gene expression have also been reported in other organisms (Kocabas et al. 2002; Chan et al. 2009). As has previously been reported from whole-genome analyses (Axelsson et al. 2008; Ekblom et al. 2010), the index of specificity of genes (τ) was negatively correlated with overall expression. Thus, genes with a highly specific function seem to have low expression levels compared with genes with more general functions. Another implication of this is that genes that are involved in adaptive evolution may be the ones that are hardest to study as these are less universally expressed. Great care is thus needed at the planning stage of a study to ensure sampling of RNA from the right tissue and at the right time if the objective is to identify such genes (Chintapalli et al. 2007).
Our study also provides evidence that dN/dS ratios calculated from automated gene annotation are similar to those calculated from manually annotated genes. This result is useful, as the large amount of data available following a genome sequencing project makes it unrealistic to manually annotate all genes, and it is important to know that the data generated in this automated way are reliable (Smedley et al. 2009). Note, however, that mis-assemblies or sequencing errors, even at a very low rate, may lead to false positive signals of positive selection (Mallick et al. 2009). This analysis assumes that the chicken predictions are annotated correctly as the annotated chicken genome was used as a reference in the manual annotations of the zebra finch. Generally, there is a risk of stepwise deterioration in the quality of genome annotation as an annotation error made in one organism will be carried forward to another (Artamonova et al. 2005).
The immune genes studied here will provide a good starting point as a list of candidate genes for further studies of avian immunogenetics and molecular ecology. Several of these loci appear to evolve under positive selection and many are also likely to exhibit functionally important polymorphism and local adaptation. This study therefore provides a significant step away from the hitherto dominant strategy of using only one or a few genes (mainly MHC) when investigating host–parasite coevolution in vertebrates. The nomenclature of genes can be rather messy with several different names for the same gene, and this is especially obvious for immune-related genes. To facilitate future studies, we therefore provide both the official gene symbols as well as common alternative gene names for many of the genes in Appendix 1. With CDSs now being available for at least two very divergent bird taxa (chicken and zebra finch), it is also likely that conserved regions that are suitable for primer design in these genes will be identifiable, something that will enable studying these loci in novel bird species.
Supplementary Material
Supplementary Additional figs. 1–10 and Appendix 1 and 2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We thank Erik Axelsson for sharing his data on dN/dS values for zebra finch ESTs, and Dave Burt for general discussions and suggestions regarding analyses. Tim Birkhead kindly provided the birds and performed the tissue preparations. Scott Edwards and four anonymous reviewers provided valuable feedback on previous versions of this manuscript. Owen Petchey and Alexie Papanicolaou assisted with computing. R.E. was supported by a Marie Curie Transfer of Knowledge project (MAERO) awarded to J.S. and T.B. by the European Commission.
References
- Acevedo-Whitehouse K, Cunningham AA. Is MHC enough for understanding wildlife immunogenetics? Trends Ecol Evol. 2006;21:433–438. doi: 10.1016/j.tree.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Aguilar A, et al. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc Natl Acad Sci U S A. 2004;101:3490–3494. doi: 10.1073/pnas.0306582101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaide M, Edwards SV, Negro JJ, Serrano D, Tella JL. Extensive polymorphism and geographical variation at a positively selected MHC class II B gene of the lesser kestrel (Falco naumanni) Mol Ecol. 2008;17:2652–2665. doi: 10.1111/j.1365-294X.2008.03791.x. [DOI] [PubMed] [Google Scholar]
- Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artamonova II, Frishman G, Gelfand MS, Frishman D. Mining sequence annotation databanks for association patterns. Bioinformatics. 2005;21:iii49–iii57. doi: 10.1093/bioinformatics/bti1206. [DOI] [PubMed] [Google Scholar]
- Axelsson E, et al. Natural selection in avian protein-coding genes expressed in brain. Mol Ecol. 2008;17:3008–3017. doi: 10.1111/j.1365-294X.2008.03795.x. [DOI] [PubMed] [Google Scholar]
- Babik W, Pabijan M, Radwan J. Contrasting patterns of variation in MHC loci in the Alpine newt. Mol Ecol. 2008;17:2339–2355. doi: 10.1111/j.1365-294X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- Balakrishnan C, et al. Gene duplication and fragmentation in the zebra finch major histocompatibility complex. BMC Biol. 2010;8:29. doi: 10.1186/1741-7007-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S, Qu L, Ellegren H. Adaptive evolution of gamete-recognition proteins in birds. J Mol Evol. 2008;67:488–496. doi: 10.1007/s00239-008-9165-6. [DOI] [PubMed] [Google Scholar]
- Berlin S, Qu L, Li X, Yang N, Ellegren H. Positive diversifying selection in avian Mx genes. Immunogenetics. 2008;60:689–697. doi: 10.1007/s00251-008-0324-0. [DOI] [PubMed] [Google Scholar]
- Bird S, et al. Evolution of interleukin-1β. Cytokine Growth Factor Rev. 2002;13:483–502. doi: 10.1016/s1359-6101(02)00028-x. [DOI] [PubMed] [Google Scholar]
- Borghans JAM, Beltman JB, De Boer RJ. MHC polymorphism under host-pathogen coevolution. Immunogenetics. 2004;55:732–739. doi: 10.1007/s00251-003-0630-5. [DOI] [PubMed] [Google Scholar]
- Brown JH, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Burri R, Hirzel HN, Salamin N, Roulin A, Fumagalli L. Evolutionary patterns of MHC Class II B in owls and their implications for the understanding of avian MHC evolution. Mol Biol Evol. 2008;25:1180–1191. doi: 10.1093/molbev/msn065. [DOI] [PubMed] [Google Scholar]
- Chan ET, et al. Conservation of core gene expression in vertebrate tissues. J Biol. 2009;8:33. doi: 10.1186/jbiol130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Clayton DF, Balakrishnan CN, London SE. Integrating genomes, brain and behavior in the study of songbirds. Curr Biol. 2009;19:R865–R873. doi: 10.1016/j.cub.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwen V, et al. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Edwards S. Genomics and ornithology. J Ornithol. 2007;148:27–33. [Google Scholar]
- Edwards SV, Hedrick PW. Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol Evol. 1998;13:305–311. doi: 10.1016/s0169-5347(98)01416-5. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Nusser J, Gasper J. Characterization and evolution of Major histocompatibility complex (MHC) genes in non-model organisms, with examples from birds. In: Baker AJ, editor. Molecular methods in ecology. Oxford: Blackwell Science; 2000. pp. 168–207. [Google Scholar]
- Egan A, et al. IDEA: interactive display for evolutionary analyses. BMC Bioinformatics. 2008;9:524. doi: 10.1186/1471-2105-9-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblom R, Balakrishnan CN, Burke T, Slate J. Digital gene expression analysis of the zebra finch genome. BMC Genomics. 2010;11:219. doi: 10.1186/1471-2164-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblom R, et al. Spatial pattern of MHC class II variation in the great snipe (Gallinago media) Mol Ecol. 2007;16:1439–1451. doi: 10.1111/j.1365-294X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Molecular evolutionary genomics of birds. Cytogenet Genome Res. 2007;117:120–130. doi: 10.1159/000103172. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Comparative genomics and the study of evolution by natural selection. Mol Ecol. 2008;17:4586–4596. doi: 10.1111/j.1365-294X.2008.03954.x. [DOI] [PubMed] [Google Scholar]
- Hale ML, Verduijn MH, Møller AP, Wolff K, Petrie M. Is the peacock's train an honest signal of genetic quality at the major histocompatibility complex? J Evol Biol. 2009;22:1284–1294. doi: 10.1111/j.1420-9101.2009.01746.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hudson ME. Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol Ecol Notes. 2008;8:3–17. doi: 10.1111/j.1471-8286.2007.02019.x. [DOI] [PubMed] [Google Scholar]
- Hughes A, Friedman R. Codon-based tests of positive selection, branch lengths, and the evolution of mammalian immune system genes. Immunogenetics. 2008;60:495–506. doi: 10.1007/s00251-008-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A, Piontkivska H. Functional diversification of the toll-like receptor gene family. Immunogenetics. 2008;60:249–256. doi: 10.1007/s00251-008-0283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Kelley J, de Bono B, Trowsdale J. IRIS: a database surveying known human immune system genes. Genomics. 2005;85:503–511. doi: 10.1016/j.ygeno.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Kocabas AM, et al. Expression profile of the channel catfish spleen: analysis of genes involved in immune functions. Mar Biotechnol. 2002;4:526–536. doi: 10.1007/s10126-002-0067-0. [DOI] [PubMed] [Google Scholar]
- Kurtz J, et al. Major histocompatibility complex diversity influences parasite resistance and innate immunity in sticklebacks. Proc R Soc Lond B Biol Sci. 2004;271:197–204. doi: 10.1098/rspb.2003.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künstner A, et al. Comparative genomics based on massive parallel transcriptome sequencing reveals patterns of substitution and selection across 10 bird species. Mol Ecol. 2010;19:266–276. doi: 10.1111/j.1365-294X.2009.04487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL. NK cell receptors. Ann Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- Larracuente AM, et al. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Li XY, Qu LJ, Yao JF, Yang N. Skewed allele frequencies of an Mx gene mutation with potential resistance to avian influenza virus in different chicken populations. Poult Sci. 2006;85:1327–1329. doi: 10.1093/ps/85.7.1327. [DOI] [PubMed] [Google Scholar]
- Mallick S, Gnerre S, Muller P, Reich D. The difficulty of avoiding false positives in genome scans for natural selection. Genome Res. 2009;19:922–933. doi: 10.1101/gr.086512.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Hultin-Rosenberg L, Zwahlen M, Ellegren H. Pleiotropic constraint hampers the resolution of sexual antagonism in vertebrate gene expression. Am Nat. 2008;171:35–43. doi: 10.1086/523954. [DOI] [PubMed] [Google Scholar]
- Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Sarkar-Roy N, Wagener DK, Majumder PP. Signatures of natural selection are not uniform across genes of innate immune system, but purifying selection is the dominant signature. Proc Natl Acad Sci U S A. 2009;106:7073–7078. doi: 10.1073/pnas.0811357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci U S A. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol. 2004;21:1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim K-W, Jiggins FM. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 2009;5:e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piertney S, Webster L. Characterising functionally important and ecologically meaningful genetic diversity using a candidate gene approach. Genetica. 2010;138:419–432. doi: 10.1007/s10709-008-9322-2. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2008. [Google Scholar]
- Roitt IM. Essential immunology. Oxford: Blackwell Science Ltd; 1997. [Google Scholar]
- Rottscheidt R, Harr B. Extensive additivity of gene expression differentiates subspecies of the house mouse. Genetics. 2007;177:1553–1567. doi: 10.1534/genetics.107.076190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, et al. Estimates of positive Darwinian selection are inflated by errors in sequencing, annotation, and alignment. Genome Biol Evol. 2010;1:114–118. doi: 10.1093/gbe/evp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, et al. BioMart—biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Smith NH. Synonymous nucleotide divergence: what ss "saturation“? Genetics. 1996;142:1033–1036. doi: 10.1093/genetics/142.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S, Schwab D, Ganzhorn JU. MHC diversity of endemic Malagasy rodents in relation to geographic range and social system. Behav Ecol Sociobiol. 2002;51:214–221. [Google Scholar]
- Stapley J, Birkhead TR, Burke T, Slate J. A linkage map of the zebra finch Taeniopygia guttata provides new insights into avian genome evolution. Genetics. 2008;179:651–667. doi: 10.1534/genetics.107.086264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer RA, Penel S, Duret L, Robinson-Rechavi M. Pervasive positive selection on duplicated and nonduplicated vertebrate protein coding genes. Genome Res. 2008;18:1393–1402. doi: 10.1101/gr.076992.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J, et al. Modeling the bound conformation of Pemphigus Vulgaris-associated peptides to MHC Class II DR and DQ Alleles. Immunome Res. 2006;2:1. doi: 10.1186/1745-7580-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonteri A, Vasemägi A, Lumme J, Primmer CR. Beyond MHC: signals of elevated selection pressure on Atlantic salmon (Salmo salar) immune-relevant loci. Mol Ecol. 2010;19:1273–1282. doi: 10.1111/j.1365-294X.2010.04573.x. [DOI] [PubMed] [Google Scholar]
- Vera JC, et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Farmer K, Hill GE, Edwards SV. A cDNA macroarray approach to parasite-induced gene expression changes in a songbird host: genetic response of house finches to experimental infection by Mycoplasma gallisepticum. Mol Ecol. 2006;15:1263–1273. doi: 10.1111/j.1365-294X.2005.02753.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerdahl H. Passerine MHC: genetic variation and disease resistance in the wild. J Ornithol. 2007;148:469–477. [Google Scholar]
- Westerdahl H, Wittzell H, von Schantz T. Polymorphism and transcription of Mhc class I genes in a passerine bird, the great reed warbler. Immunogenetics. 1999;49:158–170. doi: 10.1007/s002510050477. [DOI] [PubMed] [Google Scholar]
- Wilson DJ, McVean G. Estimating diversifying selection and functional constraint in the presence of recombination. Genetics. 2006;172:1411–1425. doi: 10.1534/genetics.105.044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JH, Xia L, Ng TB. A review of defensins of diverse origins. Curr Protein and Pept Sci. 2007;8:446–459. doi: 10.2174/138920307782411446. [DOI] [PubMed] [Google Scholar]
- Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]