Abstract
Allograft rejection in sensitized recipients remains the major problem in clinical organ transplantation. We have developed a donor-type skin-sensitized mouse cardiac allograft model (BALB/c→C57BL/6) in which both rejection (<5 days) and alloreactive CD8 activation are resistant to CD154 blockade. First, we attempted to elucidate why CD154 blockade fails to protect cardiac grafts in sensitized recipients. The gene array analysis has revealed that treatment with anti-CD154 mAb (MR1) had distinctive impact on host immunity in naive vs sensitized animals. Unlike in naive counterparts, host sensitization mitigated the impact of CD154 blockade on critical immune signaling pathways. Indeed, we identified 3234 genes in cardiac grafts that were down-regulated by MR1 in naive (at least 5-fold), but remained unaffected in sensitized hosts. Moreover, MR1 treatment failed to prevent accumulation of CD4 T cells in cardiac allografts of sensitized recipients. Then, to determine the role of CD4 help in CD154 blockade-resistant immune response, we used CD4-depleting and CD4-blocking Ab, in conjunction with MR1 treatment. Our data revealed that CD154 blockade-resistant CD8 activation in sensitized mice was dependent on CD4 T cells. In the absence of CD4 help, CD154 blockade prevented differentiation of alloreactive CD8 T cells into CTL effector/memory cells and abrogated acute rejection (cardiac graft survival for >30 days), paralleled by selective target gene depression at the graft site. These results provide the rationale to probe potential synergy of adjunctive therapy targeting CD4 and CD154 to overcome graft rejection in sensitized recipients.
Transplant patients become sensitized to a broad range of donor Ags through multiple blood transfusions, previous failed transplants, or pregnancies. These sensitized patients experience an increased rate of early rejection episodes, which are difficult to control with current immunosuppressive agents (1–3). The historical concept that rejection in sensitized individuals is driven primarily by the humoral component of host immunity led to cellular responses receiving relatively little attention. However, current concepts emphasize the interdependence of the two pathways and recognize the key role for T cells in controlling the process. It is now well recognized that alloreactive memory T cells generated from previous exposure to alloantigens or from heterologous immunity of the cross-reactive antimicrobial T cells represent the major barrier in transplant tolerance induction (4–6). Importantly, memory T cells are resistant to the majority of newly developed immune modulatory therapies (7, 8).
We have long been interested in elucidating the mechanisms of and developing novel approaches to manage sensitized transplant recipients (1, 2). More recently, we focused on the immunological mechanisms of CD154-CD40 T cell costimulation blockade in a model of cardiac allograft rejection in skin-sensitized mice (BALB/c to C57BL/6). We have shown that CD8 T cells represent the key mediator of cardiac allograft rejection in this sensitized model (9, 10). Moreover, CD154 blockade was ineffective and failed to suppress alloreactive CD8 T cell activation or to prolong cardiac allograft survival in sensitized recipients (11). This conclusion derived from effector vs memory cell-mediated rejection models in which the first allogeneic donor-type skin graft was used to activate recipient immune system at either 10 or >40 days before cardiac graft challenge. Furthermore, CD8 T cells were responsible for costimulation-resistant rejection, as their depletion was required to facilitate graft survival after CD154 blockade. Interestingly, CD4-deficient mice fail to develop memory-type rejection response, because CD8 T cell activation and cardiac graft rejection in CD4 knockout-sensitized recipients remain CD154 blockade sensitive (12). We have also documented that alloreactive CD8 T cells become activated via CD4-dependent and -independent pathways and that both pathways in naive mice remain CD154 blockade sensitive (13). Thus, the question arises as to whether alloreactive memory CD8 activation in sensitized recipient proceed via CD4-dependent and/or -independent pathways and whether these pathways are CD154 costimulation independent.
In this study, we first analyzed why CD154 blockade fails to protect cardiac allografts in sensitized recipients. We used the gene array approach and focused on the immune pathways differentially affected by the treatment in naive vs sensitized recipients. Then, we determined whether simultaneous targeting of CD4 and CD154 might synergize to promote long-term graft survival in sensitized recipients. We analyzed the role of CD4 T cells in CD154 blockade-resistant alloreactive CD8 T cell activation and intragraft immune activation.
Materials and Methods
Animals and grafting techniques
Wild-type BALB/c (B/c; H-2d) and C57BL/6 (B6; H-2b) mice were obtained from The Jackson Laboratory and housed in the University of California, Los Angeles animal facilities under pathogen-free conditions. Orthotopic full-thickness skin grafts (~0.5 cm in diameter) from B/c donors were sutured bilaterally onto the flanks of prospective B6 recipients. At 60–80 days later, these skin-sensitized recipients were challenged with intra-abdominal vascularized B/c hearts. Graft survival was assessed daily by palpation of ventricular activity. Cardiac allografts were harvested at defined time points and tissue RNA was isolated for gene expression analysis. The tail vein blood was taken at several time points for the measurement of CTL activation.
Ab therapy
Anti-CD154 (MR1) and anti-CD4 (GK1.5, depleting) Abs were purchased from BioExpress; anti-CD4 mAb (YTS177, blocking) was a generous gift from Dr. H. Waldman (Oxford University, Oxford, U.K.). Abs were administered i.v. at the time (MR1) or at day −1 and +1 (CD4) of transplantation (0.5 mg/mouse). Control recipients were treated with relevant doses of isotype Ig.
Microarray and data analysis
Total tissue RNA from cardiac allografts was prepared using TRIzol reagent (Life Technologies). Three individual samples from each experimental group were pooled by mixing equal amounts of RNA. The pooled samples were subjected to cDNA microarray analysis performed by the University of California, Los Angeles Microarray Core using an Affymetrix GeneChip Mouse Genome 430 2.0 Array. Data were uploaded to GeneSifter.Net and normalized by samples’ overall expression mean. Pairwise analysis of control vs MR1-treated samples was performed to identify differentially expressed genes, followed by pathway analysis of the gene list. Gene expression pattern analysis was also performed with the four groups of samples. The relevance of biological pathways with a defined gene list was evaluated by z-scores, calculated as described in the Genesifter.net web site.
CTL effector differentiation in vivo
RBC-free lymphocytes were used for Ab staining in ice-cold PBSA (PBS with 1% BSA). Cells were first incubated with 1μg of normal rat IgG to block Fc binding sites. After washing, cells were stained with 0.5–1 μg of rat anti-mouse CD8α-FITC (clone 53-6.7), CD62 ligand (CD62L)-R-PE (clone MEL-14), and CD44-CyChrome (clone IM7) (BD Pharmingen). After washing, three-color flow cytometry was performed on a FACSCan cytometer (BD Biosciences). CD8-positive lymphocytes were gated and analyzed for their CD62L and CD44 expression. CTL effectors were identified as CD8+CD62LlowCD44high population.
Quantitative RT-PCR
RNA (2.5 μg) was reverse-transcribed into cDNA using SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative PCR was performed using the DNA Engine with a Chromo 4 Detector (MJ Research). In a final reaction volume of 25 μl, the following were added: 1× Super-Mix (Platinum SYBR Green qPCR Kit; Invitrogen), cDNA, and 0.5 mM of each primer. Amplification conditions were: 50°C (2 min), 95°C (5 min) followed by 50 cycles of 95°C (15 s) and 60°C (30 s). Primers used to amplify a specific gene fragments are the same as published previously (12). Gene inductions were calculated as the following: individual gene levels were calibrated by their ratios to the housekeeping gene HPRT and the ratios in allografts were compared with those in isografts.
Statistical analysis
The results are shown as mean ± SD. Statistical analyses were performed using Student’s t test with p < 0.05 considered as significant.
Results
CD154 blockade fails to affect critical immune signaling pathways in sensitized hosts
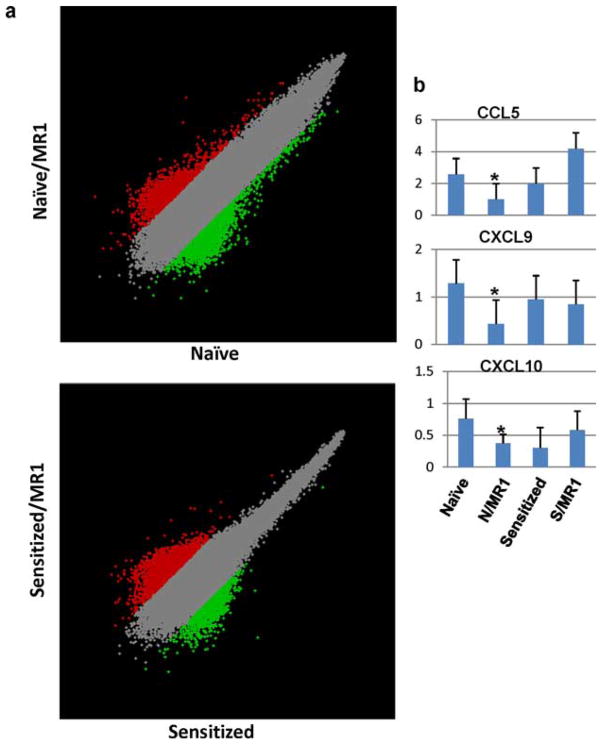
We have shown that sensitized B6 recipients (challenged with B/c skin at day −60) are resistant to CD154 blockade, as evidenced by prompt (<5 day) rejection of B/c cardiac grafts comparable to Ig-treated controls (11). To determine the immunological basis of CD154 blockade-resistant rejection, we harvested heart grafts at the peak of rejection response from age-matched naive (day 6) or sensitized (day 3) recipients in four experimental groups: 1) naive plus control Ig; 2) naive plus MR1; 3) sensitized plus control Ig; and 4) sensitized plus MR1, followed by gene expression microarray analysis. Samples from three individual grafts were pooled in each group. Differentially expressed genes between control and MR1-treated samples in naive, as well as sensitized, hosts were identified by pairwise analysis with a 5-fold difference as the arbitrarily assigned threshold. Intragraft expression of 3839 genes in naive and 3388 genes in sensitized recipients was either up-regulated or down-regulated following CD154 blockade, as shown by scatter plots (Fig. 1a). To determine the functional significances of these differentially expressed genes, we did the biological pathway analysis of the gene lists using the Kyoto Encyclopedia of Genes and Genomes (KEGG) source. The pathways that were most frequently associated with these genes (z-score, ≥1.9) are listed in Table I. Clearly, distinctive pathways were associated with gene lists derived from naive vs sensitized mice, indicating that CD154 blockade had distinct impact in the two types of recipients. In particular, TCR signaling pathway, cell adhesion molecules, NK-mediated cytotoxicity, and cell communication were affected most significantly by CD154 blockade in naive animals. However, these pathways became largely irrelevant (z-score negative) or affected only marginally in sensitized animals. This alteration occurs specifically in the immunology relevant pathways, since other pathways such as neuroactive ligand-receptor interaction, maturity onset diabetes of the young, or γ-hexachlorocyclohexane degradation were equally targeted with similar z-scores in the two recipient groups. Moreover, MR1 Ab treatment targets some novel pathways selectively in sensitized recipients, e.g., basal cell carcinoma, metabolism of xenobiotics by cytochrome P450, androgen, and estrogen metabolism, of which the biological significance is currently unclear. Thus, unlike in naive animals, host sensitization mitigates the impact of CD154 blockade on critical immunological pathways.
FIGURE 1.
a, Scatter plots of pairwise analysis results. Gene expression data were normalized to samples’ global mean and compared between control Ig and anti-CD154 mAb (MR1)-treated samples from naive (upper panel) vs sensitized (lower panel) recipients. b, Quantitative RT-PCR results of CCL5, CXCL9, and CXCL10 gene expression at the peak of cardiac allograft rejection in naive (day 6) vs sensitized (day 3) recipients. Averages of three individual samples and SD are shown; p < 0.05 when naive vs N/MR1 are compared; p = NS when sensitized vs S/MR1 are compared.
Table I.
Differential impact of CD154 blockade on KEGG pathways in naive vs. sensitized cardiac allograft recipients
| KEGG Pathway | Array | Naive |
Sensitized |
||
|---|---|---|---|---|---|
| List | z-score | List | z-score | ||
| TCR signaling pathway | 11 | 16 | 1.94 | 3 | −2.13 |
| Cell adhesion molecules | 129 | 26 | 3.77 | 11 | −0.49 |
| NK cell-mediated cytotoxicity | 112 | 22 | 2.32 | 0 | −0.13 |
| Cell communication | 108 | 21 | 2.0 | 14 | 1.13 |
| Calcium signaling pathway | 100 | 26 | 2.42 | 28 | 3.14 |
| Olfactory transduction | 29 | 12 | 5.21 | 5 | 1.36 |
| Neuroactive ligand-receptor interaction | 26.8 | 54 | 4.89 | 60 | 7.21 |
| Maturity onset diabetes of the young | 25 | 7 | 2.7 | 10 | 5.1 |
| γ-Hexachlorocyclohexane degradation | 23 | 6 | 2.29 | 5 | 1.94 |
| Basal cell carcinoma | 55 | 6 | −0.05 | 15 | 4.4 |
| Metabolism of xenobiotics by cytochrome P450 | 59 | 7 | −0.05 | 15 | 4.48 |
| Androgen and estrogen metabolism | 38 | 7 | 1.44 | 9 | 2.9 |
| Wnt signaling pathway | 14.2 | 14 | −0.48 | 22 | 2.34 |
To further examine molecular mechanisms of CD154 blockade-resistant rejection, we performed a gene expression pattern analysis of the two sets of samples and identified 3234 genes in cardiac grafts that were down-regulated by MR1 in naive (at least 5-fold), but failed to respond in sensitized recipients. The KEGG pathway analysis of this gene list revealed several immune relevant pathways (Table II), such as cell adhesion molecules, NK-mediated cytotoxicity, and TCR signaling pathway. These were also identified by pairwise analysis of the naive set of samples (Table I), indicating that they might represent putative therapeutic targets for CD154 blockade in transplant recipients. Among the genes clustered in these immune pathways (Table III lists 50 highly expressed of a total 114, levels >1), CCL5, CXCL9, and CXCL10 are T cell targeted chemokines, whereas lymphotoxin B, IL-1β, and IL-6 are tissue proinflammatory cytokines. The expression levels of CCL5, CXCL9, and CXCL10 in individual cardiac allografts of distinct experimental groups were determined by quantitative RT-PCR, and the results were in agreement with the microarray analysis data (Fig. 1b; p < 0.05 between naive (N) vs N/MR1; p = NS between sensitized (S) vs S/MR1). Consistent with T cells as the major targets of CD154 blockade, intragraft CD4 and CD8 expression levels in naive MR1-treated recipients were markedly lower compared with those of controls (CD4: 0.0064 vs 0.019; CD8: 0.64 vs 3.52), whereas in sensitized MR1-treated mice, their levels remained as high as (or even higher than) those in controls (CD4: 0.046 vs 0.021; CD8: 3.03 vs 3.02; Table III, see CD8 Ag, α-chain and CD4 Ag).
Table II.
KEGG pathway analysis of the gene list sensitive to CD154 blockade in naive recipients, but resistant to CD154 blockade in sensitized recipients
| KEGG Pathway | List | Array | z-score |
|---|---|---|---|
| Cell adhesion molecules | 34 | 129 | 4.38 |
| NK cel-mediated cytotoxicity | 27 | 112 | 3.36 |
| BCR signaling pathway | 17 | 64 | 3.11 |
| One carbon pool by folate | 6 | 15 | 3.02 |
| Hematopoietic cell lineage | 19 | 76 | 2.99 |
| Type I diabetes mellitus | 13 | 47 | 2.88 |
| Leukocyte transendothelial migration | 25 | 111 | 2.85 |
| Cytokine-cytokine receptor interaction | 42 | 220 | 2.53 |
| TCR signaling pathway | 20 | 92 | 2.36 |
| Apoptosis | 18 | 81 | 2.34 |
| Vascular endothelial growth factor signaling pathway | 16 | 70 | 2.33 |
| Fcε RI signaling pathway | 17 | 76 | 2.31 |
| Chronic myeloid leukemia | 16 | 74 | 2.08 |
Table III.
Fifty highly expressed immune-related genes in cardiac allografts altered by CD154 blockade in naive, but not sensitized recipients
| Gene | Expression Levels |
|||
|---|---|---|---|---|
| 1 (N) | 2 (N/MR1) | 3 (S) | 4 (S/MR1) | |
| Chemokine (C-C motif) ligand 5 | 16.49 | 5.14 | 19.59 | 21.94 |
| Fc receptor, IgE, high-affinity I, γ-polypeptide | 15.03 | 7.53 | 17.74 | 18.42 |
| Integrin β2 | 12.02 | 4.61 | 13.04 | 14.39 |
| Histocompatibility 2, Q region locus 8 | 11.83 | 5.05 | 10.93 | 10.83 |
| Chemokine (CXC motif) ligand 10 | 10.93 | 3.47 | 8.29 | 7.80 |
| Fc receptor, IgG, low-affinity IV | 10.14 | 4.01 | 12.65 | 12.11 |
| IL-2R, γ-chain | 9.56 | 3.33 | 8.27 | 8.47 |
| Fc receptor, IgG, low-affinity III | 8.42 | 4.73 | 11.06 | 11.57 |
| Killer cell lectin-like receptor, subfamily D, member 1 | 8.22 | 1.51 | 6.46 | 6.29 |
| RAS-related C3 botulinum substrate 2 | 6.89 | 2.00 | 5.01 | 5.04 |
| Selectin, platelet (P-selectin) ligand | 5.64 | 1.79 | 4.27 | 4.07 |
| Integrin αL | 5.02 | 1.77 | 4.77 | 4.41 |
| Killer cell lectin-like receptor subfamily K, member 1 | 4.86 | 1.16 | 4.29 | 4.31 |
| Lymphotoxin B | 4.75 | 1.22 | 3.24 | 3.33 |
| IL-18R1 | 4.71 | 0.99 | 4.52 | 4.83 |
| Fc receptor, IgG, high-affinity I | 4.63 | 2.00 | 5.49 | 5.42 |
| Integrin α4 | 4.57 | 1.12 | 3.14 | 3.37 |
| Histocompatibility 2, class II, locus Mb2 | 4.35 | 2.39 | 4.90 | 5.32 |
| Lymphocyte cytosolic protein 2 | 4.20 | 1.64 | 4.11 | 4.02 |
| FBJ osteosarcoma oncogene | 3.59 | 1.28 | 4.70 | 4.93 |
| CD8 Ag, α-chain | 3.52 | 0.64 | 3.02 | 3.03 |
| Chemokine (CXC motif) ligand 9 | 3.21 | 1.12 | 3.48 | 3.90 |
| IL-1β | 2.98 | 0.96 | 3.40 | 5.48 |
| Histocompatibility 2, class II, locus Mb1 | 2.86 | 1.38 | 3.70 | 2.86 |
| Casitas B-lineage lymphoma b | 2.48 | 0.80 | 1.72 | 1.99 |
| Chemokine (C-C motif) receptor 5 | 2.46 | 0.74 | 2.73 | 2.87 |
| Chemokine (C-C motif) receptor 1 | 2.44 | 0.72 | 3.14 | 3.27 |
| IL-6 | 2.41 | 0.49 | 2.98 | 3.07 |
| TNFR superfamily, member 1b | 2.27 | 1.09 | 2.47 | 2.65 |
| PI3K catalytic δ-polypeptide | 2.24 | 0.64 | 1.54 | 1.85 |
| CD40 Ag | 2.18 | 0.81 | 1.69 | 1.71 |
| CD44 Ag | 2.03 | 0.63 | 2.10 | 2.47 |
| Selectin, lymphocyte | 2.03 | 0.54 | 1.49 | 1.49 |
| Programmed cell death 1, ligand 2 | 1.94 | 0.20 | 1.90 | 2.02 |
| Fas ligand (TNF superfamily, member 6) | 1.67 | 0.24 | 1.32 | 1.47 |
| Integrin β7 | 1.59 | 0.50 | 1.29 | 1.66 |
| CD38 Ag | 1.58 | 0.35 | 1.84 | 1.51 |
| BH3-interacting domain death agonist | 1.55 | 0.65 | 1.41 | 1.28 |
| CD86 Ag | 1.53 | 0.83 | 2.29 | 1.68 |
| RAS guanyl-releasing protein 1 | 1.49 | 0.67 | 1.26 | 1.37 |
| IL-18R accessory protein | 1.44 | 0.35 | 1.71 | 1.53 |
| Bone morphogenetic protein 2 | 1.41 | 0.48 | 2.09 | 1.35 |
| PTK2 protein tyrosine kinase 2β | 1.39 | 0.53 | 1.33 | 1.27 |
| Colony-stimulating factor 2 receptor, β 2, low affinity | 1.24 | 0.57 | 1.84 | 2.22 |
| Syndecan 4 | 1.21 | 0.68 | 1.92 | 1.82 |
| CD1d1 Ag | 1.17 | 0.50 | 1.35 | 1.63 |
| Colony-stimulating factor 3 receptor (granulocyte) | 1.13 | 0.56 | 1.61 | 1.85 |
| Vav 1 oncogene | 1.08 | 0.32 | 0.92 | 0.68 |
| IL-10Rα | 1.02 | 0.53 | 1.26 | 1.02 |
| CD6 Ag | 1.01 | 0.29 | 0.73 | 0.86 |
| CD4 Ag | 0.019 | 0.0064 | 0.021 | 0.046 |
CD4 T cells are instrumental in CD154 blockade-resistant graft rejection in sensitized recipients
Although CD8 T cells represent the primary target of CD154 blockade in our model, CD4 T cells play an important role in CD8 activation (10, 13). Particularly, by using CD4-deficient mice, we have previously shown that alloreactive CD8 T cells do require CD4 help to develop the CD154 blockade-resistant memory response (12). Thus, an obvious option to improve the efficacy of CD154 blockade is to eliminate the CD4 T cell help. In this study, CD4 T cells were either depleted (by GK1.5) or blocked (by YTS177) in concert with CD154 blockade (by MR1) in sensitized mice. Six experimental groups were tested: 1) control Ig, 2) MR1 at day 0, 3) MR1 at day 0 plus GK1.5 at day −1/+1, 4) MR1 at day 0 plus YTS177 at day −1/+1, 5) GK1.5 at day −1/+1, or 6) YTS177 at day −1/+1. As shown in Fig. 2, single blockade with either MR1 or anti-CD4 mAb failed to prevent graft rejection. In groups 1 (control Ig), 2 (MR1), or 5 (GK1.5), all grafts were rejected promptly in 4–8 days. In contrast, simultaneous blockade of CD154 costimulation and CD4 help by either depleting (group 3) or blocking CD4 (group 4) effectively prolonged cardiac allograft survival for up to 30 days. However, such an adjunctive CD4-targeted one-dose therapy around the time of transplantation failed to protect transplants long term, with the majority of grafts rejected ultimately by day 40.
FIGURE 2.
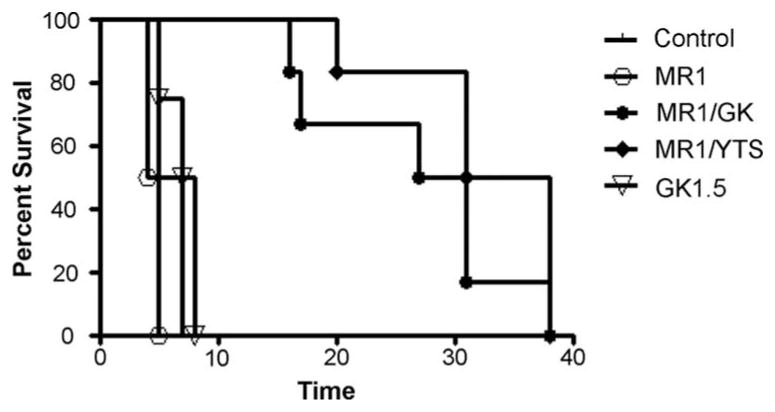
Kaplan-Meier survival curve of cardiac allografts in sensitized recipients. Groups of skin-sensitized mice (day −60) were treated with different CD154- and/or CD4-targeting regimens (n = 4–6/group) around the time of cardiac engraftment as described in Materials and Methods. Graft survival was monitored daily by palpation.
CD4 T cells are required for CD154 blockade-resistant CD8 activation in sensitized hosts
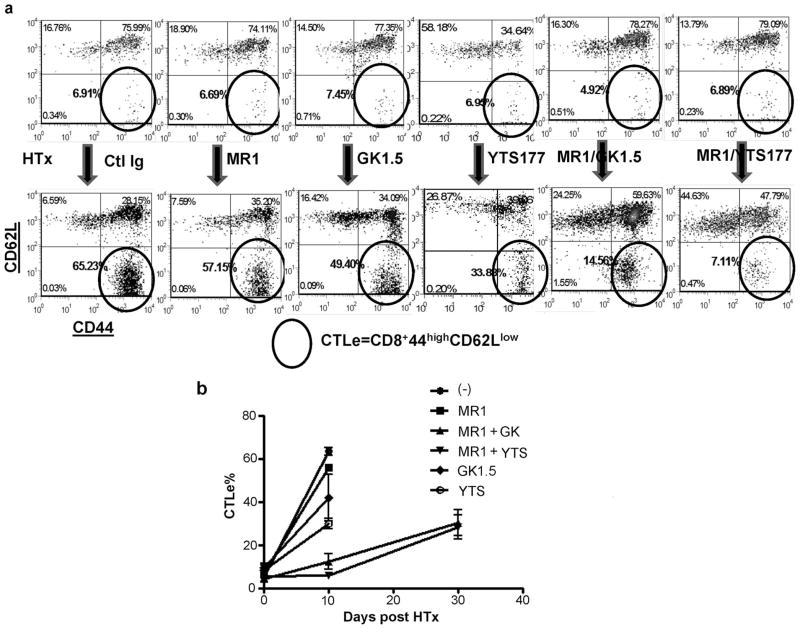
Because alloreactive CD8 T cells represent the key effectors that are resistant to CD154 blockade in sensitized recipients (11, 12), we then profiled their activation in the absence of CD4 help (days 10 and 30 after transplant). Before cardiac engraftment, numbers of CTL effectors (CTLe4; CD8+CD44highCD62Llow) in sensitized recipients stabilized at <10% of total CD8 T cells (Fig. 3a, upper panel). By day 10 after cardiac engraftment, CTL activation peaked in the control group (group 1), with CTLe increasing from 6 to 60% (Fig. 3). Treatment with MR1 or anti-CD4 mAb alone failed to prevent the activation process, as compared with controls. In marked contrast, simultaneous targeting of CD154 and CD4 (GK1.5 or YTS177) effectively suppressed alloreactive CD8 T cell activation at day 10 (7–14%; Fig. 3). However, the frequency of CTLe eventually increased by day 30 (Fig. 3b). Thus, the kinetics of CD8 T cell activation correlated well with cardiac graft survival (Fig. 2) in sensitized recipients.
FIGURE 3.
a, Representative density plots of FACS analysis of CD8 CTLs. PBLs were harvested at day 10 after cardiac transplant and stained with fluorochrome-labeled Abs against CD8, CD44, and CD62L as described in Materials and Methods. Gated CD8 cells were analyzed for CD44 and CD62L expression. CD8+CD44highCD62Llow cells are circled and their percentages in total CD8 are shown. b, Kinetics of alloreactive CD8 activation in different experimental groups were plotted by averaging percentages of CD8+CD44highCD62Llow cells in PBLs of individual recipients within the same experimental group (n = 4–6/group).
CD4 T cells are required for CD154 blockade-resistant intragraft gene induction in sensitized recipients
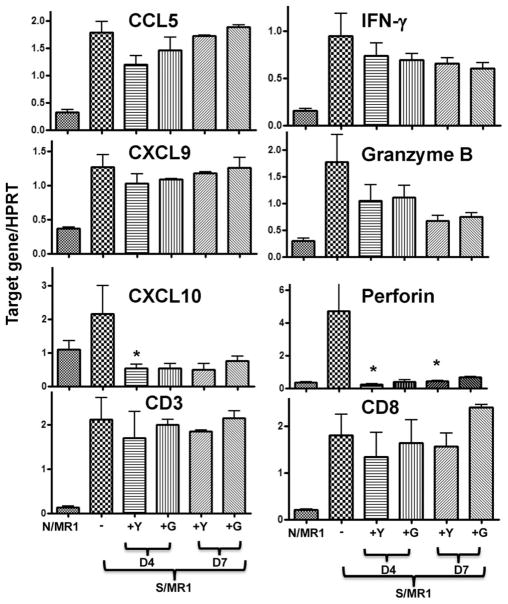
To analyze the effect of simultaneous CD154 and CD4 blockade on intragraft immune response, cardiac allografts were harvested from groups of sensitized recipients (days 4 and 7). Grafts from naive MR1-treated hosts served as controls. As shown in Fig. 4, CD4 suppression in conjunction with CD154 blockade selectively inhibited intragraft CXCL10 expression (p < 0.05), with little effect on CCL5 or CXCL9. Intragraft perforin expression was inhibited profoundly throughout (p < 0.05), while granzyme B expression was minimally suppressed and IFN-γ levels remained unchanged. Interestingly, the combined therapy did not affect graft CD3 and CD8 levels, indicating impairment of the CD8 function rather than their accumulation in the graft itself. Thus, adjunctive CD4 suppression inhibited CD154 blockade-resistant intragraft immune response on selective gene targets.
FIGURE 4.
Intragraft gene expression after CD154 blockade in the following experimental groups: N/MR1, naive recipients; S/MR1, sensitized recipients. The latter group received no adjunctive (−) or adjunctive CD4-blocking (+Y) or CD4-depelting (+G) Ab. Cardiac allografts were harvested at day 4 or 7 and tissue RNA samples were subjected to quantitative RT-PCR as described in Materials and Methods. Target gene expressions were calibrated by their ratios against HPRT levels of the same sample. Average expression ratios in different treatment groups were plotted. n = 3–6/group;*, p < 0.05.
Discussion
The principal findings of this study are as follows: 1) systemic analysis of differentially expressed genes in cardiac allografts has revealed that CD154 blockade has a distinctive impact on host immunity in naive vs sensitized recipients; 2) CD4 T cells play a critical role in the resistance of sensitized recipients to CD154 blockade; 3) simultaneous targeting of CD154 and CD4 synergize to prolong cardiac allograft survival, inhibit alloreactive CD8 activation, and intragraft immune response in sensitized hosts; and 4) the immune suppression after combined therapy is selective, as evidenced by diminished intragraft induction of CXCL10 and perforin, but not CXCL9/CCL5 or granzyme B/IFN-γ. Although clinical and experimental data document the importance of humoral immune responses (1, 9, 14), in this study we focused on T cells, which do play a critical role in the accelerated form of rejection in sensitized recipients (9). From our array analysis, the relevance of humoral response in CD154 blockade-resistant rejection became obvious, because the BCR signaling was one of the major pathways that escaped the treatment in sensitized recipients (Table II). Additionally, our adjunctive T cell-targeted therapies markedly prolonged the survival but failed to achieve permanent cardiac graft acceptance. To ameliorate humoral immune response, we plan to concomitantly target the CD27-CD70 costimulation pathway in CD154 Ab-treated sensitized hosts. Indeed, CD70 signaling is required not only for alloreactive memory CD8 T cell activation but also for B cell activation and Ig production (15).
To the best of our knowledge, this study is the first to analyze in a comprehensive manner gene programs associated with transplant rejection in sensitized recipients, particularly those that remain CD154 blockade resistant. Not surprisingly, our current finding that CD154 blockade fails to control immune-related KEGG pathways, which otherwise associate with prolonged graft survival in naive mice, provides compelling evidence that the rejection response in sensitized recipients is distinctive from that in naive counterparts. Furthermore, the direction of changes in gene profiles altered by MR1 Ab was distinct in sensitized vs naive recipients. For example, 53 genes clustered in the TCR signaling pathway were mostly down-regulated by at least 5-fold after CD154 blockade in naive recipients. In sensitized recipients, however, we identified only 20 such genes; these remained mostly up-regulated (data not shown). Interestingly, CD154 blockade targeted several pathways, such as basal cell carcinoma or metabolism of xenobiotics by cytochrome P450, in sensitized but not naive recipients. Although the significance of these novel pathways remains unclear, the genes listed in the basal cell carcinoma pathway relate to Wnt/Hedgehog signaling, whereas those in the metabolism of xenobiotics by cytochrome P450 pathway exhibit the oxidoreductase activity. Based on their involvement in the innate immunity, the CD154 blockade may preferentially impart innate immune responses in sensitized recipients.
It is now well recognized that allograft rejection by memory T cells is distinctive from that by their naive counterparts not only quantitatively in the kinetics and magnitude, but also qualitatively in their susceptibility to particular therapy or regulation (8, 16). The vast majority of current immune suppressive therapies, particularly those effectively targeting specific costimulation molecules in naive recipients, spare memory T cells and fail to prolong allograft survival in sensitized/primed hosts. Additionally, memory T cells are more resistant to immune down-regulation by regulatory T cells (17). Efforts aiming to improve the efficacy of co-stimulation blockade in sensitized recipients or memory T cell reconstituted recipients, included T cell depletion or simultaneous blockade of multiple costimulation pathways. We have shown that transient CD8 depletion combined with CD154 blockade could indeed effectively prolong cardiac allograft survival in presensitized hosts for up to 50 days, even when peripheral CD8 T cells were fully recovered (11). Similar deletion regimens have been reported to enhance both CD154 blockade-based therapy in primate renal allograft model by depleting CD8, as well as the rapamycin therapy in the mouse skin allograft model by a high dose of antithymocyte globulin (18, 19). We also reported the beneficial effect of adjunctive CD4 depletion to protect cardiac allografts in sensitized recipients, but to a much lesser degree, as compared with adjunctive CD8 depletion. Our current study extends this finding by analyzing CD8 activation and intragraft immune responses early posttransplant. In addition to the depletion regimen, we also tested a CD4-blocking Ab, a clinically more appealing and safer nondeletional approach. Indeed, the blockade of CD4 and CD154 signaling in concert synergistically prolonged allograft survival in sensitized recipients. Interestingly, although such an adjunctive treatment inhibited peripheral CD8 effector differentiation, it failed to abrogate intragraft immune activation, as evidenced by proinflammatory cytokine/chemokine and cytotoxic molecule profiles. In particular, perforin, but not granzyme B, levels decreased significantly in sensitized recipients after the treatment, whereas both genes remained down-regulated in the treated naive hosts. Although perforin deficiency itself does not impair the ability of alloreactive CD8 T cells to affect acute rejection cascade (20), it remains to be seen whether or not concomitant disruption of perforin signaling might benefit graft survival after CD154 blockade in sensitized recipients.
Our study has also revealed a key immunological feature of memory CD8 T cells, i.e., CD4-dependent alloreactive CD8 activation becomes CD154 costimulation independent in sensitized recipients. In other words, CD4-independent alloreactive memory CD8 T cells remain CD154 signaling dependent for their activation. Indeed, we have recently analyzed several other costimulation pathways, including ICOS, 4-1BB, and OX40 by using Abs against their ligands, alone or in combination with CD154 blockade. Interestingly, preliminary results show that none of these pathways have a significant effect on alloreactive CD8 activation or cardiac allograft survival in our sensitized transplant model (data not shown). These findings are in sharp contrast to other reports from memory T cell-reconstituted recipients (21, 22). Obviously, our skin-sensitized host with full immune repertoire represents a much more stringent model as compared with a selectively cell-reconstituted animal system. Unlike in the latter that focus at a single memory T cell subset, varieties of memory T cells, B cells, as well as other primed immune components are all present in our model. These certainly contribute to a complex rejection cascade involving not only multiple cell types, but also a plethora of cell-cell interactions. Our finding that CD4-dependent CD8 activation in sensitized recipients becomes CD154 blockade-resistant provides clear evidence for the novel feature of the memory CD8 T cell, resulting from its interaction with the memory CD4 T cell. Indeed, naive CD4-dependent CD8 activation requires CD154 costimulation (13). Further elucidation of molecular pathways that provide memory CD4 help to CD8 T cells should lead to the development of much needed and more specific strategies to optimize CD154-targeted therapy in sensitized recipients.
In summary, we present a comprehensive analysis of gene programs associated with allograft rejection in sensitized recipients, particularly those that are resistant to CD154 costimulation blockade. We also provide compelling evidence for a potentially clinically applicable adjunctive strategy targeting CD4 and CD154 to synergistically inhibit alloreactive CD8 activation and thus markedly prolong cardiac allograft survival in sensitized recipients.
Footnotes
This work was supported by National Institutes of Health Grants AI23847 and AI42223 (to J.W.K.-W.), the American Heart Association (to Y.Z.), and the Dumont Research Foundation.
Abbreviations used in this paper: CTLe, effector CTL.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Kupiec-Weglinski JW. Graft rejection in sensitized recipients. Ann Transplant. 1996;1:34–40. [PubMed] [Google Scholar]
- 2.Sablinski T, Hancock WW, Tilney NL, Kupiec-Weglinski JW. Biology of vascularized organ allograft rejection in sensitized recipients. Transplant Rev. 1990;4:108–120. [Google Scholar]
- 3.Baid S, Saidman SL, Tolkoff-Rubin N, Williams WW, Delmonico FL, Cosimi AB, Pascual M. Managing the highly sensitized transplant recipient and B cell tolerance. Curr Opin Immunol. 2001;13:577–581. doi: 10.1016/s0952-7915(00)00262-4. [DOI] [PubMed] [Google Scholar]
- 4.Burrows SR, Silins SL, Khanna R, Burrows JM, Rischmueller M, McCluskey J, Moss DJ. Cross-reactive memory T cells for Epstein-Barr virus augment the alloresponse to common human leukocyte antigens: degenerate recognition of major histocompatibility complex-bound peptide by T cells and its role in alloreactivity. Eur J Immunol. 1997;27:1726–1736. doi: 10.1002/eji.1830270720. [DOI] [PubMed] [Google Scholar]
- 5.Pantenburg B, Heinzel F, Das L, Heeger PS, Valujskikh A. T Cells primed by Leishmania major infection cross-react with alloantigens and alter the course of allograft rejection. J Immunol. 2002;169:3686–3693. doi: 10.4049/jimmunol.169.7.3686. [DOI] [PubMed] [Google Scholar]
- 6.Adams AB, Williams MA, Jones TR, Shirasugi N, Durham MM, Kaech SM, Wherry EJ, Onami T, Lanier JG, Kokko KE, et al. Heterologous immunity provides a potent barrier to transplantation tolerance. J Clin Invest. 2003;111:1887–1895. doi: 10.1172/JCI17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brook MO, Wood KJ, Jones ND. The impact of memory T cells on rejection and the induction of tolerance. Transplantation. 2006;82:1–9. doi: 10.1097/01.tp.0000226082.17507.da. [DOI] [PubMed] [Google Scholar]
- 8.Valujskikh A. The challenge of inhibiting alloreactive T-cell memory. Am J Transplant. 2006;6:647–651. doi: 10.1111/j.1600-6143.2005.01215.x. [DOI] [PubMed] [Google Scholar]
- 9.Hancock WW, Gao W, Shemmeri N, Shen XD, Gao F, Busuttil RW, Zhai Y, Kupiec-Weglinski JW. Immunopathogenesis of accelerated allograft rejection in sensitized recipients: humoral and nonhumoral mechanisms. Transplantation. 2002;73:1392–1397. doi: 10.1097/00007890-200205150-00006. [DOI] [PubMed] [Google Scholar]
- 10.Zhai Y, Shen XD, Gao F, Coito AJ, Wasowska BA, Salama A, Schmitt I, Busuttil RW, Sayegh MH, Kupiec-Weglinski JW. The CD154-CD40 T cell costimulation pathway is required for host sensitization of CD8+ T cells by skin grafts via direct antigen presentation. J Immunol. 2002;169:1270–1276. doi: 10.4049/jimmunol.169.3.1270. [DOI] [PubMed] [Google Scholar]
- 11.Zhai Y, Meng L, Gao F, Busuttil RW, Kupiec-Weglinski JW. Allograft rejection by primed/memory CD8+ T cells is CD154 blockade resistant: therapeutic implications for sensitized transplant recipients. J Immunol. 2002;169:4667–4673. doi: 10.4049/jimmunol.169.8.4667. [DOI] [PubMed] [Google Scholar]
- 12.Zhai Y, Wang Y, Wu Z, Kupiec-Weglinski JW. Defective allo-reactive CD8 T cell function and memory response in allograft recipients in the absence of CD4 help. J Immunol. 2007;179:4529–4534. doi: 10.4049/jimmunol.179.7.4529. [DOI] [PubMed] [Google Scholar]
- 13.Zhai Y, Meng L, Busuttil RW, Sayegh MH, Kupiec-Weglinski JW. Activation of alloreactive CD8+ T cells operates via CD4-dependent and CD4-independent mechanisms and is CD154 blockade sensitive. J Immunol. 2003;170:3024–3028. doi: 10.4049/jimmunol.170.6.3024. [DOI] [PubMed] [Google Scholar]
- 14.Binder J, Hancock WW, Watschinger B, Wasowska B, Sayegh MH, Kupiec-Weglinski JW. The alloantibody network following intrathymic immunomodulation of sensitized rat recipients of cardiac allografts. Transplantation. 1995;59:590–597. [PubMed] [Google Scholar]
- 15.Arens R, Nolte MA, Tesselaar K, Heemskerk B, Reedquist KA, van Lier RA, van Oers MH. Signaling through CD70 regulates B cell activation and IgG production. J Immunol. 2004;173:3901–3908. doi: 10.4049/jimmunol.173.6.3901. [DOI] [PubMed] [Google Scholar]
- 16.Valujskikh A, Li XC. Frontiers in nephrology: T cell memory as a barrier to transplant tolerance. J Am Soc Nephrol. 2007;18:2252–2261. doi: 10.1681/ASN.2007020151. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Brook MO, Carvalho-Gaspar M, Zhang J, Ramon HE, Sayegh MH, Wood KJ, Turka LA, Jones ND. Allograft rejection mediated by memory T cells is resistant to regulation. Proc Natl Acad Sci USA. 2007;104:19954–19959. doi: 10.1073/pnas.0704397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koyama I, Nadazdin O, Boskovic S, Ochiai T, Smith RN, Sykes M, Sogawa H, Murakami T, Strom TB, Colvin RB, et al. Depletion of CD8 memory T cells for induction of tolerance of a previously transplanted kidney allograft. Am J Transplant. 2007;7:1055–1061. doi: 10.1111/j.1600-6143.2006.01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minamimura K, Sato K, Yagita H, Tanaka T, Arii S, Maki T. Strategies to induce marked prolongation of secondary skin allograft survival in alloantigen-primed mice. Am J Transplant. 2008;8:761–772. doi: 10.1111/j.1600-6143.2007.02143.x. [DOI] [PubMed] [Google Scholar]
- 20.Schulz M, Schuurman HJ, Joergensen J, Steiner C, Meerloo T, Kagi D, Hengartner H, Zinkernagel RM, Schreier MH, Burki K, et al. Acute rejection of vascular heart allografts by perforin-deficient mice. Eur J Immunol. 1995;25:474–480. doi: 10.1002/eji.1830250225. [DOI] [PubMed] [Google Scholar]
- 21.Shiao SL, McNiff JM, Pober JS. Memory T cells and their costimulators in human allograft injury. J Immunol. 2005;175:4886–4896. doi: 10.4049/jimmunol.175.8.4886. [DOI] [PubMed] [Google Scholar]
- 22.Vu MD, Clarkson MR, Yagita H, Turka LA, Sayegh MH, Li XC. Critical, but conditional, role of OX40 in memory T cell-mediated rejection. J Immunol. 2006;176:1394–1401. doi: 10.4049/jimmunol.176.3.1394. [DOI] [PubMed] [Google Scholar]