Abstract
Historically, the study of bacterial catabolism of complex carbohydrates has contributed to understanding basic bacterial physiology. Recently, however, genome-wide screens of streptococcal pathogenesis have identified genes encoding proteins involved in complex carbohydrate catabolism as participating in pathogen infectivity. Subsequent studies have focused on specific mechanisms by which carbohydrate utilization proteins might contribute to the ability of streptococci to colonize and infect the host. Moreover, transcriptome and biochemical analyses have uncovered novel regulatory pathways by which streptococci link environmental carbohydrate availability to virulence factor production. Herein we review new insights into the role of complex carbohydrates in streptococcal host-pathogen interaction.
Confluence of basic bacterial physiology and microbial pathogenesis
The control of infection due to pathogenic bacteria currently is faced with great therapeutic and preventive challenges. The use of antimicrobial agents has resulted in widespread resistance to many antibiotics. At the same time, the remarkable successes of vaccines have slowed because certain important pathogens, such as Staphylococcus aureus, group A Streptococcus and Pseudomonas aeruginosa, have been recalcitrant to successful vaccine development. Therefore, we remain in need of an increased understanding of the pathogenesis of bacterial infectivity to form the basis for the development of the next generation of therapeutic and preventive agents.
The availability of many complete bacterial genomes has provided the framework for discovering novel aspects of bacterial pathogenesis, in part by facilitating hypothesis-generating, genome-wide investigations. One conventional definition of a ‘virulence factor’ is a molecule that directly causes damage in the host and is produced only by pathogenic bacteria [1]. Given that genes involved in basic physiological processes are generally present in both pathogenic and nonpathogenic organisms, such a definition traditionally has excluded such genes from being associated with virulence. In a broader view, however, central physiological processes, such as the ability of an organism to proliferate in the host, are pivotal to pathogenesis [2]. Therefore, it is not surprising that genome-wide studies, such as signature-tagged mutagenesis (STM) and expression microarray analysis, consistently have identified genes involved in basic metabolic processes, including the catabolism of complex carbohydrates, as crucial to the pathogenesis of disease caused by many bacteria. In turn these studies have led to gene-focused investigations designed to understand how genes historically not thought of as encoding virulence factors contribute to pathogenesis.
Over the past few years, the contribution of complex carbohydrate catabolism to the pathogenesis of streptococcal disease has been a particularly fruitful example of how merging basic bacterial physiology and virulence can elucidate new aspects of host-pathogen interaction. Although the role of complex carbohydrate utilization in microbial pathogenesis has long been recognized, recent investigations have suggested a far greater importance for carbohydrate catabolism in pathogen infectivity than previously appreciated [3-5]. In this review we summarize the genome-wide screens of streptococcal pathogenesis that resulted in a subsequent focus on complex carbohydrate utilization genes. We also detail gene- and/or pathway-specific investigations that have validated the findings of the genome-wide studies. Finally, we discuss recent molecular insights into how some streptococci link carbohydrate utilization and virulence factor production through regulatory systems. Owing to space limitations, this review is confined to Streptococcus pneumoniae, group A Streptococcus (GAS) and group B Streptococcus (GBS). Importantly, similar investigative pathways recently have yielded important new insights into the pathogenesis of oral streptococcal infections [6]. Moreover, the role of complex carbohydrate utilization in pathogenesis is also an active research area in non-streptococcal bacteria, mycobacteria and fungi, indicating that the findings discussed herein might have broad microbial pathogenesis implications [4,5,7].
Genome-wide studies bring focus to the role of complex carbohydrate catabolism in streptocococcal pathogenesis
Beginning in the late 1990s, a series of genome-wide experiments was performed in several streptococcal species to identify new avenues for pathogenesis research. An initial approach involved STM, in which a transposon is used to disrupt genes randomly, with the resulting pool of mutant bacteria being screened for a phenotype of interest, such as the ability to survive in the lungs of mice. By comparing the input and output pools of mutants, genes whose disruption results in decreased in vivo fitness can be identified. Three genome-wide STM screens of S. pneumoniae pathogenesis using a combination of mouse pneumonia and bacteremia selection models have been published (Table 1) [3,8,9]. An important finding in all three screens was the consistent detection of genes involved in basic metabolic processes, including complex carbohydrate catabolism. In the most comprehensive of the three S. pneumoniae STM studies, investigators identified dozens of genes putatively involved in carbohydrate utilization as contributing to pneumococcal virulence, thereby providing many novel pathogenesis investigative pathways [3]. Similarly, an STM study of GBS using a mouse bacteremia model found that inactivation of genes involved in complex carbohydrate metabolism, including a maltose-binding protein, a phosphotransferase (PTS) and a sucrose hydrolase, attenuated virulence [10].
Table 1. Examples of carbohydrate catabolism genes identified by genome-wide analyses of streptococcal host-pathogen interaction.
| Carbohydrate catabolism gene (putative function of encoded protein) | Organism | Type of study | Model | Refs |
|---|---|---|---|---|
|
nanA (hydrolysis of N-acetylneuraminic acid) msmK (sugar transport) |
S. pneumoniae | STM | Mouse bacteremia | [8] |
|
aga (α-galactosidase) fco (α-fucosidase) manL (mannose transporter) |
S. pneumoniae | STM | Mouse pneumonia and bacteremia |
[9] |
|
strH (β-N-acetylglucosaminidase) pulA (pullulanase) bgaA (β-galactosidase) glgB (glycogen biosynthesis) lacA (lactose catabolism) amy (α-amylase) aga (α-galactosidase) malX (maltodextrin-binding protein) msmK (sugar transport) |
S. pneumoniae | STM | Mouse pneumonia and bacteremia |
[3] |
|
malX (maltodextrin-binding protein) ptsK (carbohydrate transport) scrB (sucrose hydrolase) |
Group B Streptococcus | STM | Mouse bacteremia | [10] |
|
manN (mannose transport) lacA/lacB (lactose metabolism) malA (maltose metabolism) fba (fructose-bisphsophate aldolase) |
S. pneumoniae | Expression microarray |
Mouse bacteremia Rabbit meninigits Human pharyngeal cells |
[11] |
|
malE (maltodextrin-binding protein) manL (mannose transport) lacE (lactose transport) spy0252 (sialic-acid-binding protein) sptR/S (carbohydrate gene regulation) |
Group A Streptococcus | Expression microarray |
Nonhuman primate pharyngitis |
[14] |
|
malE (maltodextrin-binding protein) pulA (pullulanase) spy0252 (sialic-acid-binding protein) lacE (lactose transport) sptR/S (carbohydrate gene regulation) |
Group A Streptococcus | Expression microarray |
Human saliva | [15] |
|
manL (mannose transport) spy1986 (maltose transport) malE (maltodextrin-binding protein) amyA (α-amylase) |
Group A Streptococcus | Expression microarray |
Human blood | [13] |
| spy1592 (sugar transport) amyA (α-amylase) lacE (lactose transport) malE (maltodextrin-binding protein) |
Group A Streptococccus | Expression microarray |
Mouse soft tissue | [12] |
Expression microarray analysis also has been used as an effective method to identify genes whose products might contribute to streptococcal pathogenesis. Analysis of gene transcript levels of S. pneumoniae grown in mouse blood, rabbit cerebrospinal fluid and during interaction with human pharyngeal cells found upregulation of numerous genes encoding proteins with known or putative carbohydrate metabolism function [11]. Complex carbohydrate metabolism genes identified by the expression microarray approach overlapped with those found by STM, including genes involved in maltodextrin and lactose metabolism. Similarly, several studies have been published on the GAS transcriptome during host-pathogen interaction, including experimental pharyngitis in the nonhuman primate (Table 1) [12-15]. GAS transcript levels of genes involved in carbohydrate metabolism, including those involved in maltodextrin and mannose catabolism, were maximal during the initial primate colonization phase [14]. These data suggest that complex carbohydrate metabolism genes were either being induced or derepressed during the growth of the organism in vivo, suggesting a role for such genes in organism proliferation during the initial stages of pharyngeal infection [14]. In summary genome-wide studies have provided a foundation for more detailed investigation of the molecular mechanisms by which complex carbohydrate catabolism contributes to streptococcal pathogenesis.
Gene-focused studies examining the role of complex carbohydrate proteins in streptococcal pathogenesis
Studies of the role of complex carbohydrate metabolism in streptococcal pathogenesis have focused on three major mechanisms: acquisition of crucial nutrients, adherence to eukaryotic cells and interference with the function of host immunity proteins (Figure 1). The pathways are not mutually exclusive; a single complex carbohydrate metabolism enzyme might affect host-pathogen interaction by more than one mechanism. In addition to an examination of the role of a gene or gene product in pathogenesis, research on streptococcal complex carbohydrate catabolism also has been important in elucidating novel physiological characteristics of the protein and/or pathway being analyzed and have added to the understanding of streptococcal host-pathogen interactions.
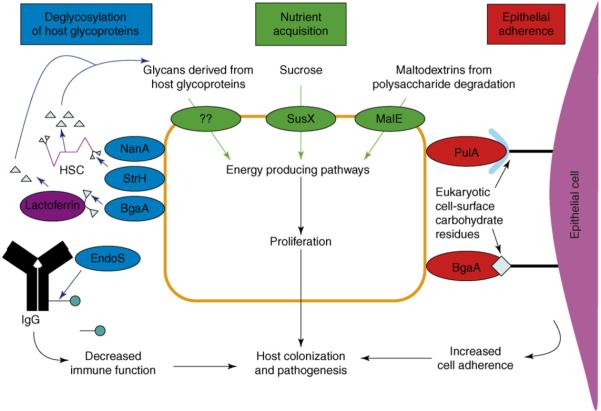
Figure 1.
Summary of mechanisms by which carbohydrate utilization proteins contribute to pathogenesis in S. pneumoniae and group A Streptococcus. Proteins involved in deglycosylation of host glycoproteins are shown in blue. NanA, StrH and BgaA are cell-surface exoglycosidases studied in S. pneumoniae. EndoS is a secreted endoglycosidase studied in group A Streptococcus. HSC, human secretory component. Pathways for nutrient acquisition are shown in green. SusX and MalE are cell-surface lipoprotein components of ATP-binding-cassette transport systems. Bacterial cell-wall-linked proteins involved in binding to eukaryotic cell surface carbohydrate residues are shown in red. PulA is a pullulanase mainly studied in group A Streptococcus. BgaA is a β-galactosidase studied in S. pneumoniae.
With respect to nutrient acquisition, a contribution to host-pathogen interaction has been demonstrated for streptococcal proteins involved in the creation and transport of maltodextrins, sucrose and glycans released from enzymatic degradation of host glycoproteins (Figure 1). S. pneumoniae has long been known to express neuraminidase activity or the ability to cleave terminal sialic acid residues from glycosides [16]. More recently, King et al. [17] demonstrated that S. pneumoniae has three cell-wall-linked exoglycosidases that cleave terminal carbohydrate moieties in addition to sialic acid. Although these exoglycosidases have been thought to contribute to pathogenesis through adhesion or immune evasion mechanisms (see below), Burnaugh et al. [18] recently discovered that these proteins also provide S. pneumoniae with the ability to use human glycoproteins as a nutrient source.
The role of sucrose utilization in S. pneumoniae infectivity recently was demonstrated by Iyer et al. [19]. They discovered that a sucrose ATP-binding-cassette (ABC) transport system contributed to the ability of S. pneumoniae to cause pneumonia, whereas a sucrose PTS was important for colonization of the nasopharynx. The findings are particularly noteworthy in that S. pneumoniae is the major streptococcal organism responsible for pneumonia in humans. Paralogs of the sucrose ABC transport system are absent in the genomes of streptococci such as GAS, GBS or S. mutans - organisms that rarely cause pneumonia.
GAS must obtain nutrients from the host oropharynx to proliferate and cause pharyngitis. As noted above, genome-wide studies suggested that complex carbohydrates might be a key nutrient source for GAS [14,15]. Human salivary α-amylase is known to degrade ingested polysaccharides to yield maltose and other maltodextrins [20]. Although previously unstudied, the GAS protein MalE was predicted to be a maltodextrin-binding protein based on homology with Escherichia coli MalE. Study of purified GAS MalE demonstrated that it binds maltodextrins with high affinity and rapidly transports the maltodextrins that result from polysaccharide digestion by human salivary α-amylase [21]. Moreover, a GAS ΔmalE isogenic mutant strain colonized the mouse oropharynx at significantly lower levels compared to the wild-type strain [22], confirming the importance of MalE in GAS pathogenesis. Another recent study determined that regulation of MalE production by the maltodextrin repressor MalR also contributes to GAS colonization of the mouse oropharynx, thereby reinforcing the importance of the maltodextrin system in GAS-host interaction in the oropharynx [23]. Of note human salivary α-amylase levels are highly variable, suggesting that host α-amylase activity might be a predisposing factor for host susceptibility to GAS pharyngitis [24].
Given that carbohydrate residues are present on many eukaryotic cell surfaces, like other microbes, streptococci have evolved mechanisms to use these residues for adherence (Figure 1). Most species of pathogenic streptococci sequenced to date contain a large LPXTG-containing (i.e. cell-wall linked) pullulanase that is immunogenic in humans [25]. PulA is thought to contribute to the pathogenesis of GAS and S. pneumoniae by mediating adherence to eukaryotic carbohydrate residues [26-28]. Similarly, a cell-wall-linked exoglycosidase (BgaA) produced by S. pneumoniae contributes to nasopharyngeal colonization and facilitates adhesion to the host epithelium [17,29,30]. Paralogs of BgaA are not present in the sequenced GAS or GBS genomes. The fucose operon of S. pneumoniae also has been linked to virulence, perhaps by binding human cell-surface blood-group antigens [31,32].
Deglycosylation of host molecules also contributes to streptococcal pathogenesis through attenuation of the host immune response (Figure 1). The glycosylation of components of the innate and acquired immune response is important for their proper function, and pathogen deglycosylation of host molecules has evolved as a means of immune evasion [33]. All GAS strains encode a secreted endoglycosidase (EndoS) that hydrolyzes the conserved asparagine-linked glycan present on the heavy chain of human IgG, resulting in decreased functional IgG activity and an increased ability of the organism to proliferate in human blood [34,35]. The gene encoding EndoS, paralogs of which are not present in sequenced S. pneumoniae or GBS strains, is present in the middle of a putative sucrose utilization operon, which suggests a role in nutrient acquisition in addition to immune modulation. As previously noted, S. pneumoniae encodes three cell-surface exoglycosidases that might contribute to pathogenesis through nutrient acquisition or adherence activity. In addition these exoglycosidases also have been shown to deglycosylate proteins involved in innate immunity such as lactoferrin, human secretory component (HSC) and immunoglobulin A [36].The functional effect of the deglycosylation of these proteins, however, has yet to be experimentally determined. Given that many putative carbohydrate utilization proteins present on the cell surface of streptococci have yet to be investigated, it is probable that additional contributions of complex carbohydrate utilization proteins to pathogenesis will be elucidated.
Regulatory links between classical virulence factors and complex carbohydrate utilization in streptococci
Acquisition of energy sources is a prime imperative for all bacterial species, including streptococci. Compared to human blood, glucose levels at other common sites of streptococcal infection are generally quite low, meaning that alternative energy sources need to be pursued (Table 2). In one sense, then, streptococcal damage to the host might be thought of as a by-product of the organisms’ nutrient-questing behavior [37]. It has been known for over 35 years that production of the antiphagocytic M protein and the cysteine protease streptococcal pyrogenic exotoxin B (SpeB) are affected by environmental carbohydrate type and concentration [38,39]. We are now beginning to elucidate the underlying molecular mechanisms linking carbohydrate metabolism to virulence factor production.
Table 2. Typical glucose levels at various human body sites.
| Body site | Glucose level (mM)a | Comments |
|---|---|---|
| Blood | 3.57-6.06 | |
| Saliva [64] | 0.02-0.40 | Substantial interindividual variability |
| Nasal secretions [65] | <1.0 | Can be present in patients with hyperglycemia |
| Lower airway secretions [66] | <0.5 |
To convert to mg/dL multiply by 18.15.
Similar to other bacteria, pathogenic streptococci rely on two major types of transcriptional regulators: two-component gene regulatory systems (TCSs) and stand-alone regulators. TCSs consist of a cell-membrane-embedded sensor histidine kinase that influences the phosphorylation state of a cognate response regulator, which in turn either represses or activates gene expression via DNA binding (Figure 2). The best studied TCS in GAS is the ‘control of virulence’ (CovR/S) system that regulates transcription of several key GAS virulence factors and genes involved in carbohydrate catabolism, including those encoding the aforementioned pullulanase and maltodextrin-binding proteins [40,41]. A GBS ortholog of CovR/S also regulates the transcription of genes encoding proteins involved in carbohydrate metabolism and virulence [42]. Sitkiewicz et al. recently discovered that the GAS TCS M5005_spy0680/0681 controls expression of carbohydrate metabolism and virulence factor genes [43]. Subsequent experiments using an isogenic mutant strain showed that the TCS was necessary for GAS persistence in human saliva, leading to the name ‘SptR/S’ for saliva persistence regulator [15]. Analysis of transcript levels during experimental infection of nonhuman primates has suggested a central role for SptR/S in GAS pharyngitis, perhaps through its ability to couple complex carbohydrate metabolism with virulence factor production [14,15]. Similarly, the TCS CiaR/H in S. pneumoniae has been found to regulate transcription of maltose utilization genes and the virulence factor htra [44]. CiaR/H has been shown to contribute to the ability of S. pneumoniae to colonize the nasopharynx and lungs and to cause systemic infection in mice [45-47]. In summary data from GAS and S. pneumoniae demonstrate that streptococcal TCSs play a key role in connecting the expression of complex carbohydrate metabolism genes together with that of classical virulence factors, thereby contributing to pathogenesis.
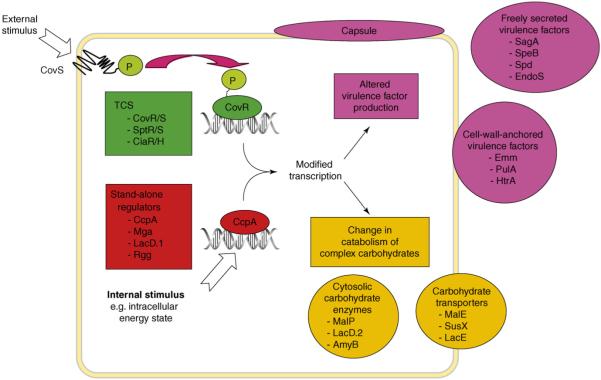
Figure 2.
Regulatory pathways in streptococci linking virulence factor production with complex carbohydrate metabolism. The sensor kinase component of a two-component gene regulatory system (TCS, in green) responds to external stimuli by phosphorylating its cognate response regulator, which in turn affects binding of the regulator to DNA. Regulator DNA binding leads to altered transcription of genes encoding virulence factors (in pink) and proteins involved in complex carbohydrate utilization (in orange). Virulence factors listed include SagA (cytotoxin), speB (cysteine protease), Spd (DNase), EndoS (immunoglobulin-cleaving ezyme), Emm (M protein), PulA (pulullanase) and HtrA (serine protease). Carbohydrate metabolic enzymes listed include MalP (maltose phosphorylase), LacD.2 (fructose bisphosphate aldolase), AmyB (neopullulanase), MalE (maltodextrin-binding protein), SusX (sucrose-binding protein) and LacE (lactose phosphotransferase enzyme).
Despite the importance of TCSs, the clearest data on links between complex carbohydrate utilization and classical virulence factor production pertains to stand-alone regulators (Figure 2). In the 1990s research on Bacillus species established that catabolite control protein A (CcpA) is a master regulator of genes involved in complex carbohydrate utilization [48]. In what is termed ‘catabolite repression’, CcpA represses the expression of genes involved in complex carbohydrate utilization in the presence of a readily metabolizable carbohydrate such as glucose [49]. Control of gene expression by CcpA occurs through CcpA binding to DNA at catabolite response elements (cre) sites for which consensus sequences have been determined in Bacillus species [49]. A CcpA ortholog was identified in the S. pneumoniae genome and inactivation of the gene (RegM) resulted in decreased virulence in a mouse model of bacteremia [50], providing the first evidence that CcpA-like regulation of gene expression contributes to streptococcal virulence. Polysaccharide capsule gene expression was decreased in the ΔregM strain, thereby providing a possible explanation for the effect of RegM inactivation on pathogenesis. A second study of a CcpA-inactivated strain in S. pneumoniae similarly showed a marked defect in virulence in mouse models of pneumonia and nasopharyngeal colonization [51]. Analysis of the cell-wall fractions of CcpA mutants showed altered cell-surface expression of several carbohydrate utilization proteins as well as enolase, which was previously shown to contribute to streptococcal virulence [52]. Thus, CcpA in S. pneumoniae clearly influences the colonization and infectivity of the organism in animal models, probably through a combination of effects on complex carbohydrate catabolism and virulence factor production.
The relationship between CcpA and virulence has been further investigated in two recent GAS studies [53,54]. Analysis of the GAS genome identified a putative cre site in the promoter region of the gene encoding the multigene activator (Mga) protein, which activates transcription of M protein [53]. Purified GAS CcpA bound to the mga cre site, and genetic inactivation of ccpA led to decreased mga gene expression and Mga protein production. These results indicate that CcpA potentially regulates (indirectly) transcription of several key GAS virulence factors such as M protein, C5a peptidase and the collagen-binding protein SclA [55,56]. In another study, comparative transcriptome analysis of wild-type GAS versus an isogenic ΔccpA strain demonstrated that CcpA influenced many virulence factor and carbohydrate utilization genes during growth in a standard laboratory medium [54]. Moreover, when grown in the nutrient-limited medium of human saliva, CcpA influenced the transcript levels of additional virulence factors not affected during growth in standard laboratory medium, demonstrating that the particular growth medium affects how CcpA influences GAS virulence gene expression [54]. Purified GAS CcpA bound to the promoter region of streptolysin S, a key GAS cytotoxin, demonstrating that in addition to indirect regulation via Mga, CcpA also can directly regulate virulence factor gene expression in GAS. Analysis of the genomes of S. pneumoniae and GBS identified putative cre sites present in the promoter region of genes encoding known virulence factors, suggesting that CcpA regulation of virulence factor production could be occurring in other Gram-positive pathogens (S. Shelburne, unpublished).
Research into the regulation of the cysteine protease virulence factor SpeB in GAS has led to the discovery of additional stand-alone regulators that control expression of both carbohydrate utilization genes and virulence factor production (Figure 2). Transposon mutagenesis identified LacD.1, which is annotated as a tagatose-1,6-bisphosphate aldolase, as being a regulator of SpeB activity [57]. Loughman et al. confirmed the aldolase activity of LacD.1 and showed that it negatively influences transcription of genes encoding mannose and sucrose PTS and SpeB [57]. Interestingly, transcriptome analysis indicated that CcpA and Mga negatively influence lacD.1 transcript levels, further demonstrating the complex relationship between virulence factor production and complex carbohydrate utilization in GAS [54,55]. Similar to LacD.1, Rgg was initially identified via transposon mutagenesis as regulating SpeB activity, although unlike LacD.1, Rgg positively modifies speB transcription through direct binding to the speB promoter [58,59]. Transcriptome analysis of a Δrgg isogenic mutant strain demonstrated altered transcript levels of numerous genes involved in complex carbohydrate utilization [60]. In accordance with these data, the Rgg-deficient strain showed significant defects in catabolizing non-glucose carbon sources such as fructose, mannose and sucrose [60]. RovS, a paralog of Rgg in GBS, has been shown to influence virulence factor production, but the role of RovS in complex carbohydrate utilization has not been investigated [61]. In summary there are multiple levels of regulatory links between virulence factor production and complex carbohydrate catabolism in S. pneumoniae, GAS and GBS, demonstrating the close relationship between carbohydrate utilization and streptococcal pathogenesis (Figure 2).
Concluding remarks and future directions
Genome sequencing has allowed for the full assessment of gene content of pathogenic organisms and provided the platform for the design and execution of genome-wide screens of bacterial pathogenesis. In turn results from genome-wide investigations have broadened the approach to understanding bacterial infectivity to include aspects of bacterial physiology not traditionally associated with virulence. The increasing appreciation of the role of complex carbohydrate catabolism in the infectivities of S. pneumoniae, GAS and GBS is a prime example of how genome sequencing has paved the way for novel insights into host-pathogen interactions (Figure 1). Similarly, transcriptome analysis of streptococcal regulators has uncovered heretofore unappreciated links between regulation of virulence factors and complex carbohydrate genes (Figure 2). Taken together, recent investigations clearly have shown that streptococcal virulence and complex carbohydrate catabolism are closely entwined.
Despite the advances reviewed herein, key questions remain unanswered regarding the role of carbohydrate catabolism in streptococcal pathogenesis. For example, what are the key nutrients needed for streptococcal proliferation in the host and does host diet and/or genetic makeup contribute to susceptibility to streptococcal infections? Similarly, it is well known that streptococcal virulence is dependent on coordinate interaction between multiple regulatory networks [62]. Yet, we have limited information regarding how the CovR/S, SptR/S, CcpA, Mga, LacD.1 and Rgg systems interact to coordinate virulence factor production in response to environmental carbohydrate conditions [63]. Increased understanding of these regulatory pathways is needed as a prerequisite for rational interruption strategies. Given that many of the carbohydrate catabolism genes and regulators discussed herein are highly conserved among Gram-positive pathogens, insights into the role of carbohydrate catabolism in streptococcal host-pathogen interactions might have implications for a wide range of microbes.
Acknowledgements
We thank K. Stockbauer for suggestions to improve the manuscript. This work was supported by American Heart Association grants 0565133Y and 0765115Y (S.A.S.) and National Institute Allergy and Infectious Diseases K08 Career Development Award AI-064564 (S.A.S.). We apologize to all authors whose work could not be included due to space limitations.
Glossary
- ATP-binding cassette
transport system consisting of cell-surface lipoprotein and permease utilizing ATP as energy source.
- Cellobiose
disaccharide found in plants that is composed of two glucose monomers linked by a β-(1,4) bond.
- Endoglycosidase
enzyme that cleaves carbohydrate residues at internal sites.
- Exoglycosidase
enzyme that cleaves terminal carbohydrate residues.
- Glycan
generic term for oligosaccharide or polysaccharide.
- Glycoprotein
conjugated molecule consisting of a protein plus a carbohydrate.
- Glycoside
a sugar that is bonded to a non-sugar.
- Maltodextrin
carbohydrate composed of repeating glucose monomers. Maltose is the smallest maltodextrin comprising two-linked glucose monomers.
- Phosphotransferase
transport system consisting of cell membrane protein utilizing phosphate transfer to create transport gradient.
- Pullulan
polysaccharide composed of repeating maltotriose units.
- Pullulanase
cell-surface enzyme that degrades pullulan.
- Sialic acid
generic term for derivatives of the nine-carbon monosaccharide neuraminic acid. Also refers to N-acetylneuraminic acid, a common component of animal and bacterial glycoproteins.
- Sucrose
disaccharide composed of glucose and fructose.
References
- 1.Wassenaar TM, Gaastra W. Bacterial virulence: can we draw the line? FEMS Microbiol. Lett. 2001;201:1–7. doi: 10.1111/j.1574-6968.2001.tb10724.x. [DOI] [PubMed] [Google Scholar]
- 2.Smith H. Studies on bacterial pathogenicity since 1950 and their future. In: Brogden K, editor. Virulence Mechanisms of Bacterial Pathogens. ASM Press; 2007. p. 329. [Google Scholar]
- 3.Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 4.Tchawa Yimga M, et al. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 2006;74:1130–1140. doi: 10.1128/IAI.74.2.1130-1140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moyrand F, et al. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol. Microbiol. 2007;64:771–781. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- 6.Abranches J, et al. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J. Bacteriol. 2008;190:2340–2349. doi: 10.1128/JB.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polissi A, et al. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau GW, et al. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 2001;40:555–571. doi: 10.1046/j.1365-2958.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- 10.Jones AL, et al. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 2000;37:1444–1455. doi: 10.1046/j.1365-2958.2000.02099.x. [DOI] [PubMed] [Google Scholar]
- 11.Orihuela CJ, et al. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 2004;72:5582–5596. doi: 10.1128/IAI.72.10.5582-5596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham MR, et al. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am. J. Pathol. 2006;169:927–942. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham MR, et al. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am. J. Pathol. 2005;166:455–465. doi: 10.1016/S0002-9440(10)62268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Virtaneva K, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shelburne SA, et al. Central role of a two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly RT, et al. Neuraminidase activities of clinical isolates of Diplococcus pneumoniae. J. Bacteriol. 1967;94:272–273. doi: 10.1128/jb.94.1.272-273.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King SJ, et al. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 2006;59:961–974. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- 18.Burnaugh AM, et al. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 2008;190:221–230. doi: 10.1128/JB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer R, Camilli A. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol. Microbiol. 2007;66:1–13. doi: 10.1111/j.1365-2958.2007.05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaczmarek MJ, Rosenmund H. The action of human pancreatic and salivary isoamylases on starch and glycogen. Clin. Chim. Acta. 1977;79:69–73. doi: 10.1016/0009-8981(77)90462-4. [DOI] [PubMed] [Google Scholar]
- 21.Shelburne SA, 3rd, et al. MalE of group A Streptococcus participates in the rapid transport of maltotriose and longer maltodextrins. J. Bacteriol. 2007;189:2610–2617. doi: 10.1128/JB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shelburne SA, 3rd, et al. Maltodextrin utilization plays a key role in the ability of group A Streptococcus to colonize the oropharynx. Infect. Immun. 2006;74:4605–4614. doi: 10.1128/IAI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shelburne SA, 3rd, et al. Regulation of polysaccharide utilization contributes to the persistence of group A Streptococcus in the oropharynx. Infect. Immun. 2007;75:2981–2990. doi: 10.1128/IAI.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellavia SL, et al. alpha-Amylase activity of human neonate and adult saliva. Arch. Oral Biol. 1979;24:117–121. doi: 10.1016/0003-9969(79)90059-1. [DOI] [PubMed] [Google Scholar]
- 25.Reid SD, et al. Postgenomic analysis of four novel antigens of group A Streptococcus: growth phase-dependent gene transcription and human serologic response. J. Bacteriol. 2002;184:6316–6324. doi: 10.1128/JB.184.22.6316-6324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Bueren AL, et al. Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nat. Struct. Mol. Biol. 2007;14:76–84. doi: 10.1038/nsmb1187. [DOI] [PubMed] [Google Scholar]
- 27.Hytönen J, et al. Use of flow cytometry for the adhesion analysis of Streptococcus pyogenes mutant strains to epithelial cells: investigation of the possible role of surface pullulanase and cysteine protease, and the transcriptional regulator Rgg. BMC Microbiol. 2006;6:18. doi: 10.1186/1471-2180-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hytonen J, et al. Streptococcus pyogenes glycoprotein-binding strepadhesin activity is mediated by a surface-associated carbohydrate-degrading enzyme, pullulanase. Infect. Immun. 2003;71:784–793. doi: 10.1128/IAI.71.2.784-793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zahner D, Hakenbeck R. The Streptococcus pneumoniae beta-galactosidase is a surface protein. J. Bacteriol. 2000;182:5919–5921. doi: 10.1128/jb.182.20.5919-5921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufman GE, Yother J. CcpA-dependent and -independent control of beta-galactosidase expression in Streptococcus pneumoniae occurs via regulation of an upstream phosphotransferase system-encoding operon. J. Bacteriol. 2007;189:5183–5192. doi: 10.1128/JB.00449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boraston AB, et al. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J. Biol. Chem. 2006;281:35263–35271. doi: 10.1074/jbc.M607620200. [DOI] [PubMed] [Google Scholar]
- 32.Embry A, et al. Regions of Diversity 8, 9 and 13 contribute to Streptococcus pneumoniae virulence. BMC Microbiol. 2007;7:80. doi: 10.1186/1471-2180-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnold JN, et al. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 34.Collin M, Olsen A. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 2001;20:3046–3055. doi: 10.1093/emboj/20.12.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collin M, et al. EndoS and SpeB from Streptococcus pyogenes inhibit immunoglobulin-mediated opsonophagocytosis. Infect. Immun. 2002;70:6646–6651. doi: 10.1128/IAI.70.12.6646-6651.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King SJ, et al. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol. Microbiol. 2004;54:159–171. doi: 10.1111/j.1365-2958.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- 37.Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 38.Pine L, Reeves MW. Correlation of M protein production with those factors found to influence growth and substrate utilization of Streptococcus pyogenes. Infect. Immun. 1972;5:668–680. doi: 10.1128/iai.5.5.668-680.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen JO. Effect of culture medium composition and pH on the production of M protein and proteinase by group A streptococci. J. Bacteriol. 1969;99:737–744. doi: 10.1128/jb.99.3.737-744.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Churchward G. The two faces of Janus: virulence gene regulation by CovR/S in group A streptococci. Mol. Microbiol. 2007;64:34–41. doi: 10.1111/j.1365-2958.2007.05649.x. [DOI] [PubMed] [Google Scholar]
- 41.Dalton TL, et al. RscA, a member of the MDR1 family of transporters, is repressed by CovR and required for growth of Streptococcus pyogenes under heat stress. J. Bacteriol. 2006;188:77–85. doi: 10.1128/JB.188.1.77-85.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamy MC, et al. CovS/CovR of group B Streptococcus: a two-component global regulatory system involved in virulence. Mol. Microbiol. 2004;54:1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x. [DOI] [PubMed] [Google Scholar]
- 43.Sitkiewicz I, Musser JM. Expression microarray and mouse virulence analysis of four conserved two-component gene regulatory systems in group A Streptococcus. Infect. Immun. 2006;74:1339–1351. doi: 10.1128/IAI.74.2.1339-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mascher T, et al. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 2003;185:60–70. doi: 10.1128/JB.185.1.60-70.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Throup JP, et al. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 2000;35:566–576. doi: 10.1046/j.1365-2958.2000.01725.x. [DOI] [PubMed] [Google Scholar]
- 46.Sebert ME, et al. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 2002;70:4059–4067. doi: 10.1128/IAI.70.8.4059-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marra A, et al. Differential fluorescence induction analysis of Streptococcus pneumoniae identifies genes involved in pathogenesis. Infect. Immun. 2002;70:1422–1433. doi: 10.1128/IAI.70.3.1422-1433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hueck CJ, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol. Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 49.Warner JB, Lolkema JS. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 2003;67:475–490. doi: 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giammarinaro P, Paton JC. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect. Immun. 2002;70:5454–5461. doi: 10.1128/IAI.70.10.5454-5461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer R, et al. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergmann S, et al. alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 2001;40:1273–1287. doi: 10.1046/j.1365-2958.2001.02448.x. [DOI] [PubMed] [Google Scholar]
- 53.Almengor AC, et al. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator mga in the group A Streptococcus. J. Bacteriol. 2007;189:8405–8416. doi: 10.1128/JB.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shelburne SA, 3rd, et al. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ribardo DA, McIver KS. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A Streptococcus. Mol. Microbiol. 2006;62:491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 56.Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 2007;66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- 57.Loughman JA, Caparon MG. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 2006;25:5414–5422. doi: 10.1038/sj.emboj.7601393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lyon WR, et al. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neely MN, et al. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 2003;185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dmitriev AV, et al. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J. Bacteriol. 2006;188:7230–7241. doi: 10.1128/JB.00877-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samen UM, et al. The transcriptional regulator RovS controls the attachment of Streptococcus agalactiae to human epithelial cells and the expression of virulence genes. Infect. Immun. 2006;74:5625–5635. doi: 10.1128/IAI.00667-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kreikemeyer B, et al. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 63.Roberts SA, Scott JR. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Mol. Microbiol. 2007;66:1506–1522. doi: 10.1111/j.1365-2958.2007.06015.x. [DOI] [PubMed] [Google Scholar]
- 64.Gough H, et al. Human salivary glucose analysis by high-performance ion-exchange chromatography and pulsed amperometric detection. Arch. Oral Biol. 1996;41:141–145. doi: 10.1016/0003-9969(95)00121-2. [DOI] [PubMed] [Google Scholar]
- 65.Wood DM, et al. Effect of hyperglycaemia on glucose concentration of human nasal secretions. Clin. Sci. (Lond.) 2004;106:527–533. doi: 10.1042/CS20030333. [DOI] [PubMed] [Google Scholar]
- 66.de Prost N, Saumon G. Glucose transport in the lung and its role in liquid movement. Respir. Physiol. Neurobiol. 2007;159:331–337. doi: 10.1016/j.resp.2007.02.014. [DOI] [PubMed] [Google Scholar]