Abstract
Methylation of ribosomal RNA (rRNA) is required for optimal protein synthesis. Multiple 2'-O-ribose methylations are carried out by small (nucleolar) box C/D guide ribonucleoproteins (s(no)RNPs), which are ubiquitous in nature from archaea to eukaryotes. Each site of methylation is dictated by base pairing between the specific guide s(no)RNA component of the s(no)RNP and the target rRNA. Here we present the first structure of a reconstituted and catalytically active box C/D sRNP from the archaeon Methanocaldococcus jannaschii determined by single-particle electron microscopy. Our results reveal that archaeal box C/D sRNPs unexpectedly form a dimeric structure with a novel organization of their RNA and protein components, challenging the conventional view of box C/D s(no)RNP architecture. Mutational analysis demonstrates that the di-sRNP structure is relevant for the function of archaeal box C/D sRNPs as RNP enzymes.
In all three kingdoms of life, nucleotides in ribosomal RNA (rRNA) are post-transcriptionally modified, with as many as 200 nucleotide modifications in the human ribosome (1). The nucleotide modifications cluster in phylogenetically conserved regions and are important for ribosome structure, stability, and function (2). One of the most common nucleotide modifications is the 2'-O-methylation of ribose residues. In archaea and eukaryotes, most 2'-O-ribose methylations are performed by box C/D small ribonucleoproteins (sRNPs) and small nucleolar ribonucleoproteins (snoRNPs), respectively, and are guided by base pairing of the RNA component of the s(no)RNP with its target rRNA (3, 4). In addition, several box C/D snoRNPs are also essential for pre-rRNA processing (5). Understanding the mechanism of action of these s(no)RNPs demands information on their organization and architecture.
Methylation guide s(no)RNPs consist of a small box C/D s(no)RNA, which is characterized by the conserved box C, C', D, and D' sequence elements, and the core box C/D proteins (Fig. 1A) (reviewed in 5, 6). In archaea, the core proteins include L7Ae, Nop5 (also called Nop56/58), and fibrillarin, which are homologs of mammalian 15.5K, Nop56 and Nop58, and fibrillarin, respectively. While the core proteins are common to all box C/D s(no)RNPs, the s(no)RNAs differ in sequence, especially in the D and D' guide regions that base pair with the target RNA and thereby confer specificity to the methylation event (3, 4, 7, 8). Fibrillarin, the methyltransferase enzyme in box C/D s(no)RNPs, methylates the target nucleotide base paired to the fifth nucleotide upstream of box D or D' (3, 7, 9, 10).
Fig. 1.
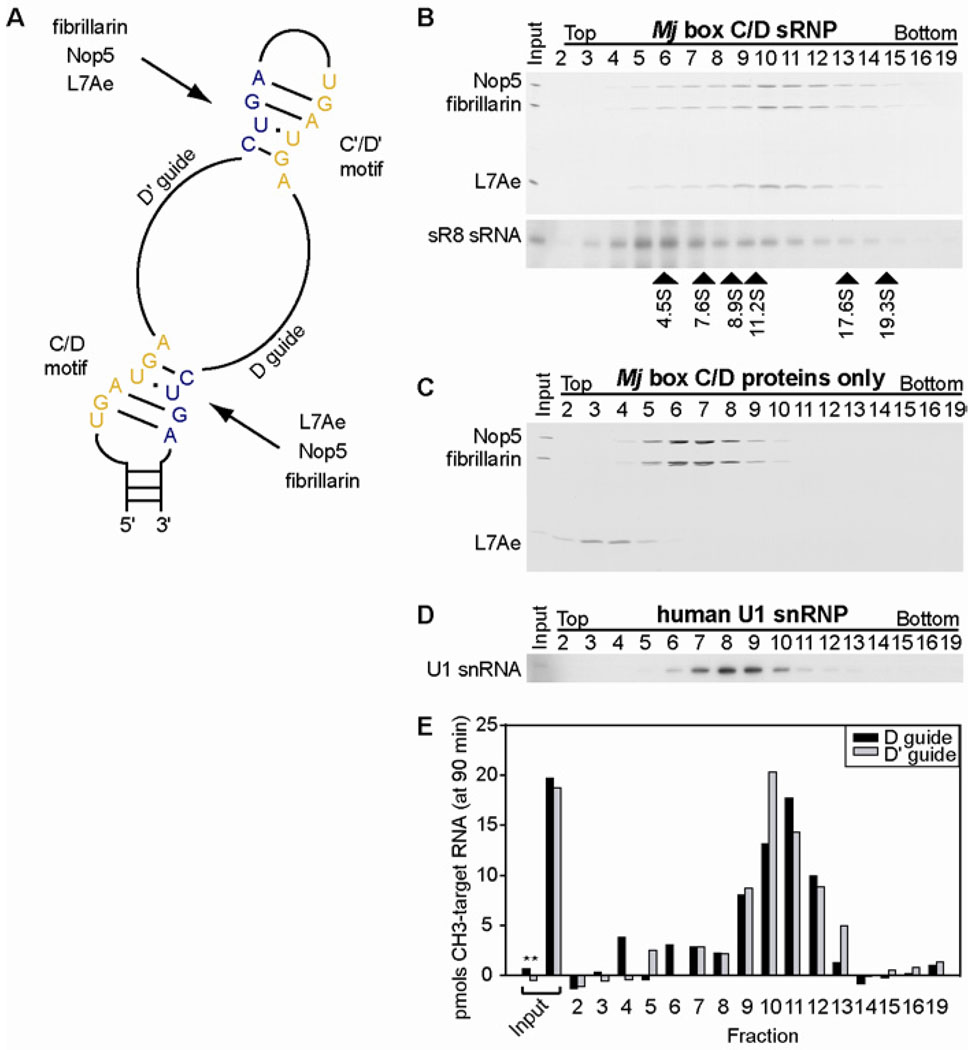
Assembly, purification, and enzymatic activity of the reconstituted Mj box C/D sR8 sRNP. (A) Schematic of an archaeal box C/D sRNA. The conserved sequence motifs, boxes C and C' (yellow) and boxes D and D' (blue), as well as the guide sequences are indicated. Each box C/D and box C'/D' motif is bound by the core box C/D proteins, L7Ae, Nop5, and fibrillarin. (B) Purification of reconstituted sR8 sRNP by glycerol gradient centrifugation. Unpurified sRNP (input) and harvested fractions, as indicated, were analyzed for the presence of the protein components by SDS-PAGE and silver staining (top) and of the sRNA component by Northern blotting (bottom). Arrowheads below the gel indicate peaks of protein markers of corresponding S values run in a parallel gradient. (C) Sedimentation of box C/D sRNP protein components in the absence of the box C/D sRNA in glycerol gradients as carried out in (B). (D) Sedimentation of the human U1 snRNP (MW 240 kDa) in a glycerol gradient run in parallel to (B) using the same experimental conditions. (E) Methylation activity of the Mj box C/D sR8 sRNP in the unpurified material (input) and in the gradient fractions using substrate RNAs complementary to the D guide (black bars) and D' guide (gray bars) sequences of the sR8 sRNA, respectively. As a control for non-specific methylation activity, pre-methylated RNAs were used as substrates (*).
In contrast to their eukaryotic counterparts, enzymatically active archaeal box C/D sRNPs can be reconstituted in vitro (9). L7Ae binds first to the k-turn or k-loop structures formed by the C/D and C'/D' motifs of the sRNA, nucleating the subsequent assembly of Nop5 and fibrillarin (9, 11). For efficient 2'-O-ribose methylation, both the box C/D and box C'/D' motifs in one sRNA are required to assemble symmetrically with all three core proteins into an RNP that is conventionally illustrated as containing one sRNA and two copies of each of the three core proteins, suggesting interdependence of both motifs (Fig. 1A) (12–14). In co-crystal structures, Nop5 and fibrillarin form a heterotetrameric protein complex in which Nop5 dimerizes via its coiled-coil domain (15, 16). These structures appear to be in good agreement with the symmetric assembly model. However, it has been proposed that the estimated length of the guide sequences of archaeal box C/D sRNAs when bound to the target RNA, which determines the distance between the C/D and C'/D' motifs, is too short to accommodate the dimerized Nop5 coiled-coil and to position the active site of fibrillarin in the proximity of the guide nucleotide (17). Thus, structural information on enzymatically active box C/D sRNPs containing the full length sRNA is vital for establishing architectural models of methylation guide sRNPs. Here we report the first 3D structure of a reconstituted and catalytically competent methylation guide sRNP from the hyperthermophilic euryarchaeon Methanocaldococcus jannaschii (Mj) determined by electron microscopy (EM) and single-particle analysis.
To obtain sufficiently homogeneous sRNP complexes suitable for structural studies, we reconstituted box C/D sRNPs in vitro using recombinant Mj core proteins and in vitro transcribed Mj box C/D sR8 sRNA and subsequently purified the assembled RNP on glycerol gradients. In the sedimentation profile, all sRNP components, L7Ae, Nop5, fibrillarin and the sR8 sRNA co-migrate in peak fractions 10 and 11 (Fig. 1B). Assembly requires the presence of the sRNA because, consistent with previous results (13), L7Ae (fractions 3 and 4 in Fig. 1C) does not associate with Nop5 or fibrillarin (fractions 6 and 7 in Fig. 1C) without the sRNA. Importantly, the purified sRNP is catalytically active for methylation of an RNA substrate using either guide sequence in the sR8 sRNA (Fig. 1E). Methylation is specific to the fifth nucleotide upstream of boxes D and D', as no activity is observed using a substrate with a pre-methylated target nucleotide (Fig. 1E). Comparison of the reconstituted sRNP to molecular weight markers of indicated S values (Fig. 1B) as well as to the human U1 snRNP (MW 240 kDa; Fig. 1D) shows that the catalytically active box C/D sRNP migrates at 12 S, faster than the human U1 snRNP, indicating a much larger complex than what would be expected based on the conventional model of box C/D sRNP architecture (predicted MW 183 kDa). In contrast, Nop5 and fibrillarin, which form a heterotetrameric complex (MW 136 kDa) that would account for 75% of the mass of the sRNP based on the conventional model (12, 15, 16), migrate at ∼6.5 S in glycerol gradients, predicting a sRNP based on the conventional model that would be significantly smaller than the catalytically active 12 S sRNP that we observe (Fig. 1C, fig. S1). Analysis of both the reconstituted sRNP and the Nop5-fibrillarin complex by gel filtration chromatography support these conclusions (fig. S2A–C). Furthermore, glycerol gradient sedimentation, gel filtration chromatography, as well as native gel electrophoresis (fig. S2E) also demonstrate that the reconstituted and catalytically active box C/D sRNPs are constituted by a single species, and are thus biochemically homogeneous and appropriate for structural studies.
EM analysis of negatively stained purified complexes from peak gradient fractions shows a monodisperse population of particles (Fig. 2A and fig. S2D). Three-dimensional reconstruction of the box C/D sRNP by single-particle analysis of EM images was successfully achieved ab initio using the random conical tilt method (RCT, 18) (figs. S3–S8). The EM structure was subsequently refined to a resolution of 27 Å (fig. S6B) (19). The experimental class averages and projections of the refined volume are in good agreement with each other (Fig. 2B).
Fig. 2.
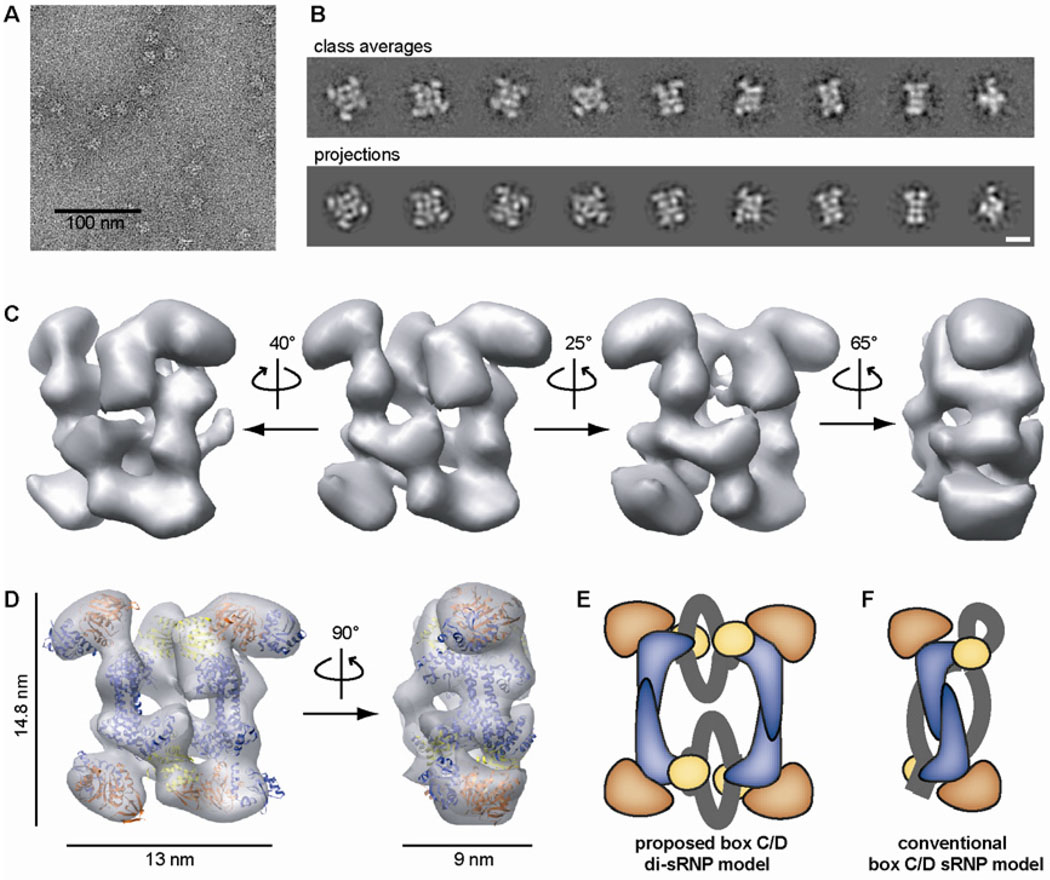
Electron microscopy and 3D reconstruction of the archaeal box C/D sR8 sRNP. (A) Electron micrograph of negatively stained sR8 sRNPs from peak fractions after glycerol gradient centrifugation. (B) Experimental class averages and corresponding 2D projections of the reconstructed 3D volume. Scale bar is 10 nm. (see also figs. S3 and S4) (C) Isodensity map of the reconstructed 3D volume of the archaeal box C/D sRNP rotated around the y-axis as indicated. (D) Docking of the crystal structures of Pyrococcus furiosus Nop5-fibrillarin (PDB 2nnw, 16) and Mj L7Ae (1xbi, 31) in the isodensity map. Nop5 - blue, fibrillarin - orange, L7Ae - yellow. The volumes are thresholded to 118% of the MW of the di-sRNP. (E) Proposed di-sRNP model contrasted with (F) the conventional model of archaeal box C/D sRNP architecture. Note that the Nop5-fibrillarin heterotetramer is seen along its 2-fold symmetry axis in the conventional model, whereas the Nop5-fibrillarin heterotetramer in the di-sRNP model is rotated 90° clockwise with respect to the view shown in the conventional model. Colors are as in (D) and the RNA is shown in grey. The orientation of the sRNA ends was purposefully left ambiguous.
The refined sRNP volume measures 14.8 × 13 × 9 nm and exhibits 2-fold pseudosymmetry (Fig. 2C). Consistent with our biochemical analyses (Fig. 1, figs. S1 and S2), the volume is significantly larger than was anticipated based on existing atomic resolution structures of box C/D core proteins and the conventional box C/D sRNP model (15, 16). Indeed, the dimensions of the heterotetrameric complex of Nop5 and fibrillarin were only about 13.8 × 7.5 × 5.4 nm in the X-ray structure, significantly smaller in two dimensions than what we observe. Surprisingly, docking of the crystal structures into our EM volume revealed that it can accommodate not one but two Nop5-fibrillarin heterotetramers, one along each long side (14.8 nm) of the volume, placing fibrillarin in the corners of the complex (Fig. 2D). Consistent with a relative stochiometry of L7Ae:Nop5:fibrillarin of 1:1:1, which we have determined by quantitative amino acid analysis (19), four L7Ae molecules can also be fit into the remaining density in the central part of the volume, placing L7Ae in proximity to the C-terminal domain of Nop5 (Fig. 2D). Thus, our results indicate that our sRNP EM volume contains four copies of each core protein.
The presence of four sets of core proteins (L7Ae:Nop5:fibrillarin) in the sRNP volume strongly suggests that the box C/D sRNP contains two sRNA molecules. Two types of evidence support this conclusion; first, extensive biochemical evidence indicates that one box C/D sRNA molecule is bound by two sets of core proteins, one set binding to each of the C/D and C'/D' motifs (12, 13); second, Mj L7Ae does not interact with Nop5 or fibrillarin in the absence of the sRNA (Fig. 1C) (13, 17) and, since one sRNA has binding sites for two L7Ae proteins, two sRNAs are needed to recruit four copies of L7Ae into the sRNP. The box C/D k-turns and box C'/D' k-loops are most likely positioned in the remaining density in proximity to the sRNA binding protein L7Ae (20) and the C-terminal domain of Nop5, which can also bind the sRNA in the context of the sRNP but only if L7Ae is also bound (15). This topology is consistent with the recent crystal structure of hPrp31, a Nop5 homologous protein of the Nop domain family, complexed with 15.5K (the mammalian L7Ae homolog) and the k-turn containing fragment of the U4 snRNA (fig. S9) (21). In this structure the U4 snRNA is sandwiched between 15.5K and the RNA binding Nop domain of hPrp31.
While the resolution of our EM structure is too low to allow the sRNA molecules to be unambiguously localized, especially their single stranded guide sequences, the distribution of the remaining density in our EM volume that is not occupied by the core box C/D proteins implies that the two sRNAs connect the two Nop5-fibrillarin heterotetramers along the short sides (13 nm in Fig. 2D) of the sRNP volume and follow a different directionality than the Nop5 coiled-coil domains. Consistent with this interpretation, RNase treatment of the reconstituted sRNP yields significantly smaller complexes, the majority of which measure about 14 × 6 nm in projection, and are thus about half the size of untreated sRNPs (fig. S10). Taken together, these results are not compatible with the conventional model of box C/D sRNP architecture (Fig. 2F), but can be explained by a new model (Fig. 2E). Thus, we propose that archaeal box C/D sRNPs are di-sRNPs and that, in contrast to previous representations, one box C/D sRNP contains four copies of each core protein and two sRNAs.
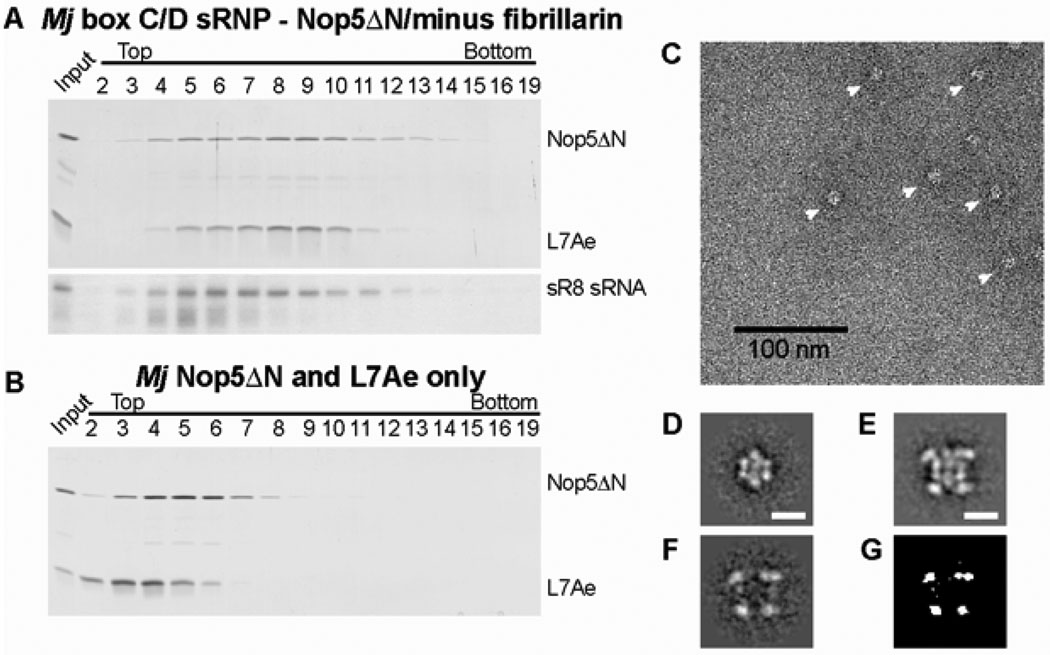
To verify the new model, we analyzed a complex lacking sRNP components by EM. This approach has been successfully used to map proteins into the EM structure of the U1 snRNP (22). Docking of the crystal structures into the EM volume (Fig. 2D) predicts that both fibrillarin and the N-terminal, fibrillarin-interacting domain of Nop5 occupy the density in the four corners of the sRNP volume. Therefore, we predict that a sRNP lacking both fibrillarin and the N-terminus of Nop5 (Nop5ΔN/minus fibrillarin) should lack those four regions of density. The Nop5ΔN/minus fibrillarin sRNP was assembled and purified on glycerol gradients. Consistent with a smaller size of these particles as compared to the fully assembled sRNP, the peak gradient fractions are shifted towards the top of the gradient (Figs. 3A and B), and the particles from peak gradient fractions are smaller when analyzed by EM, measuring only 10 × 10 nm (Fig. 3C and figs. S11 and S12). Class averages of the Nop5ΔN/minus fibrillarin sRNP indeed lack the strong density in the four corners that is characteristic of the fully assembled sRNP, whereas the central regions of both particles are very similar (Fig. 3D–G). These results provide further evidence that the box C/D sRNP contains four copies of fibrillarin, and likewise four copies of L7Ae and Nop5, and consequently support the di-sRNP model (Fig. 2E).
Fig. 3.
Localization of fibrillarin and the N-terminal domain of Nop5 in the EM structure. Sedimentation profile of (A) the Nop5ΔN/minus fibrillarin sRNP and (B) L7Ae and Nop5ΔN in the absence of the sR8 sRNA analyzed by glycerol gradient centrifugation. Glycerol gradient centrifugation was performed and analyzed as described in Fig. 1B. (C) Electron micrograph and (D) representative class average of negatively stained Nop5ΔN/minus fibrillarin sRNP particles from peak glycerol gradient fractions. Particles are marked with white arrowheads in (C). See also fig. S11 for full-sized images and additional class averages. (E) Class average of the reconstituted box C/D sRNP particles containing fibrillarin and the N-terminus of Nop5. Additional class averages are provided in fig. S3A. Scale bars are 10 nm in (D) and (E). (F) Difference map between the class averages of the two different RNPs as shown in (D) and (E). (G) Statistically significant region with a p-value of p≤0.001.
The di-sRNP model predicts a molecular mass of 366 kDa for the Mj box C/D sRNP. This size is consistent with the results obtained both by glycerol gradient centrifugation (Fig. 1B) and by gel filtration chromatography (fig. S2A). Furthermore, the S value calculated from the EM volume is 11.7 S (19), very close to the observed S value of 12 S (Fig. 1B). Di-sRNP formation was also apparent if we used a different EM staining technique, the GraFix method of glycerol gradient centrifugation (23), or different constructs for protein expression (figs. S13–S16).
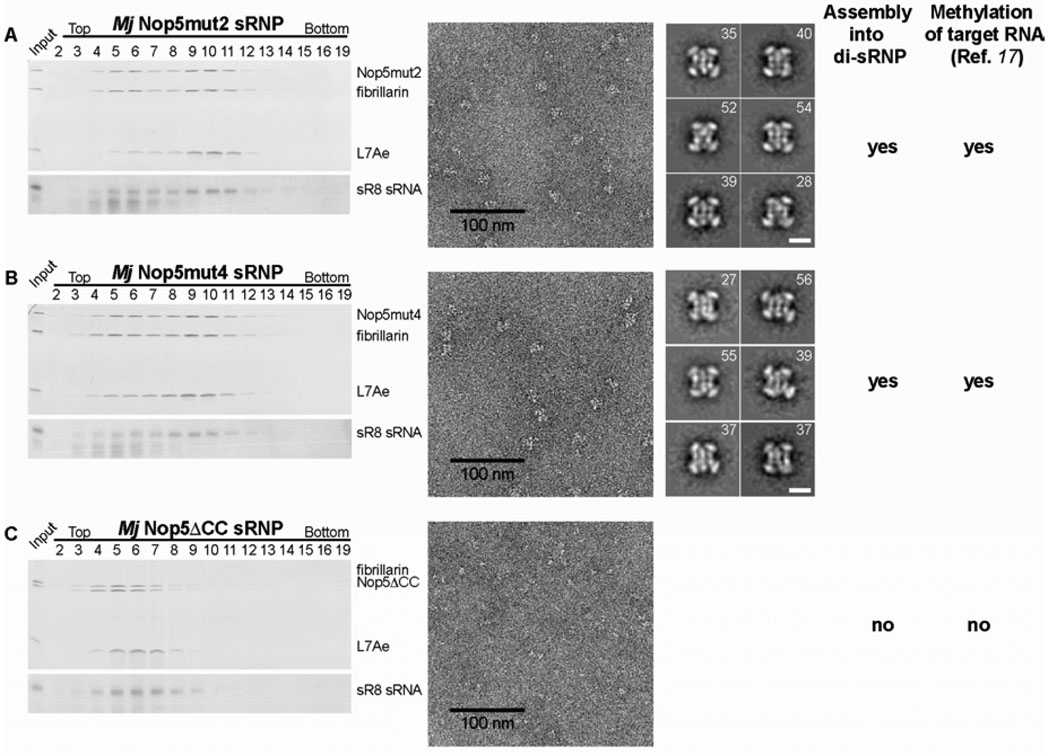
According to our proposed model of a dimeric box C/D sRNP, the Nop5 proteins are critical for maintaining the di-sRNP conformation, as they orchestrate the positioning of two different sRNAs in the complex (Figs. 2D and 2E). Previous biochemical experiments showed that some mutations in the coiled-coil domain of Nop5 (Nop5 CC) abrogated the methylation activity of the sRNP, while others were tolerated (12, 17). We asked whether the catalytic activity of the Nop5 CC mutant sRNPs correlates with their ability to form the di-sRNP structure. We tested three different Nop5 CC mutants: the point mutants Nop5mut2 (L143K, V217E) and Nop5mut4 (D135A, L143K, V217E, E224A), as well as the deletion mutant Nop5ΔCC (Δ123–241). Both Nop5mut2 and Nop5mut4 sRNPs have been previously shown to methylate substrate RNAs, whereas the Nop5ΔCC sRNP is methylation deficient (17). Glycerol gradient and EM analyses of these mutant complexes demonstrate that Nop5mut2 and Nop5mut4 assemble into sRNPs of similar size and dimensions as sRNPs containing the wild type Nop5, whereas Nop5ΔCC assembles into complexes that are significantly smaller (Figs. 4A to C, fig. S17). As predicted based on our interpretation of the box C/D sRNP EM structure, the Nop5ΔCC mutant still assembles into complexes containing all box C/D sRNP components, including the sRNA and L7, but they are not di-sRNPs (fig. S17C and S18). Collectively, our results indicate that the formation of the di-sRNP structure correlates with efficient methylation activity of the box C/D sRNP.
Fig. 4.
Formation of the box C/D di-sRNP structure correlates with efficient enzymatic activity. Box C/D sRNPs assembled with Nop5 containing point mutations in the coiled coil domain [mut2 in (A) and mut4 in (B)] or a deletion of the coiled-coil domain [ΔCC in (C)] were analyzed by glycerol gradient centrifugation (as in Fig. 1B) and complexes in peak fractions were visualized by electron microscopy after negative staining. Representative class averages of sRNP particles assembled with Nop5mut2 and Nop5mut4, respectively, are shown. The number of images in each class is indicated in the right upper corner of each class average. The sRNP assembled with Nop5ΔCC was too small for accurate image processing and calculation of meaningful class averages.
Our work provides the first 3D structure of a catalytically active box C/D sRNP, revealing an unexpected di-sRNP topology. Multimeric RNPs composed of individual RNPs are not unprecedented. Examples, among others, include the U4/U6 di-snRNP, the U4/U6.U5 tri-snRNP, the U11/U12 di-snRNP, and telomerase (24–29). In many cases these complexes are composed of different individual RNPs of different RNA and protein composition, e.g. the U4 and U6 snRNPs in the U4/U6 di-snRNP. In contrast, telomerase appears to be a homodimeric RNP. However, no structure of the telomerase RNP is available yet and no consensus has been reached as to how dimerization occurs.
Since the first discovery of a box C/D s(no)RNA almost 30 years ago (30), box C/D s(no)RNPs have been assumed to consist of one s(no)RNA molecule and one or two sets of each core protein. In contrast to this original assumption, the structure of archaeal box C/D sRNPs presented here argues that these complexes form an unanticipated di-sRNP structure with an organization of the RNA and protein components that is novel. Our results further demonstrate that this structure is functionally relevant: 1) the di-sRNPs are catalytically active, 2) no RNPs representing the conventional model of box C/D sRNP architecture are detectable by biochemical and EM methods, 3) the catalytic activity peaks with the di-sRNP and not with the S value of box C/D sRNPs predicted based on the conventional model, and 4) mutations in conserved domains that impede di-sRNP formation also interfere with sRNP function. Therefore, the di-sRNP structure is relevant to its function as an RNP enzyme. Considering the conserved nature of the box C/D sRNAs beyond the guide sequences, it is possible that box C/D di-sRNPs composed of two different box C/D sRNAs but the same box C/D core proteins could form as well. This di-sRNP architecture may provide an efficient means by which these RNP chaperones can participate in folding of the long pre-ribosomal RNA.
Supplementary Material
Footnotes
Summary
Archaeal methylation guide box C/D sRNPs form a di-sRNP structure, and the ability to assemble into this structure correlates with efficient enzymatic activity.
References and Notes
- 1.Decatur WA, Fournier MJ. Trends Biochem Sci. 2002;27:344. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 2.Liang XH, Liu Q, Fournier MJ. Mol Cell. 2007;28:965. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 4.Omer AD, et al. Science. 2000;288:517. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 5.Matera AG, Terns RM, Terns MP. Nat Rev Mol Cell Biol. 2007;8:209. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 6.Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G. Nucleic Acids Res. 2007;35:1452. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiss-Laszlo Z, Henry Y, Kiss T. EMBO J. 1998;17:797. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaspin C, Cavaille J, Erauso G, Bachellerie JP. J Mol Biol. 2000;297:895. doi: 10.1006/jmbi.2000.3593. [DOI] [PubMed] [Google Scholar]
- 9.Omer AD, Ziesche S, Ebhardt H, Dennis PP. Proc Natl Acad Sci U S A. 2002;99:5289. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Boisvert D, Kim KK, Kim R, Kim SH. EMBO J. 2000;19:317. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein DJ, Schmeing TM, Moore PB, Steitz TA. EMBO J. 2001;20:4214. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rashid R, et al. J Mol Biol. 2003;333:295. doi: 10.1016/j.jmb.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Tran EJ, Zhang X, Maxwell ES. EMBO J. 2003;22:3930. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omer AD, Zago M, Chang A, Dennis PP. RNA. 2006;12:1708. doi: 10.1261/rna.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aittaleb M, et al. Nat Struct Biol. 2003;10:256. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- 16.Oruganti S, et al. J Mol Biol. 2007;371:1141. doi: 10.1016/j.jmb.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, et al. RNA. 2006;12:1092. doi: 10.1261/rna.2230106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radermacher M, Wagenknecht T, Verschoor A, Frank J. J Microsc. 1987;146:113. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 19.Materials and methods are available as supporting material on Science Online.
- 20.Kuhn JF, Tran EJ, Maxwell ES. Nucleic Acids Res. 2002;30:931. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, et al. Science. 2007;316:115. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 22.Stark H, Dube P, Luhrmann R, Kastner B. Nature. 2001;409:539. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- 23.Kastner B, et al. Nat Methods. 2008;5:53. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 24.Stark H, Luhrmann R. Annu Rev Biophys Biomol Struct. 2006;35:435. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- 25.Beattie TL, Zhou W, Robinson MO, Harrington L. Mol Cell Biol. 2001;21:6151. doi: 10.1128/MCB.21.18.6151-6160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prescott J, Blackburn EH. Genes Dev. 1997;11:2790. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fouche N, Moon IK, Keppler BR, Griffith JD, Jarstfer MB. Biochemistry. 2006;45:9624. doi: 10.1021/bi060313s. [DOI] [PubMed] [Google Scholar]
- 28.Cohen SB, et al. Science. 2007;315:1850. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 29.Wenz C, et al. EMBO J. 2001;20:3526. doi: 10.1093/emboj/20.13.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy R, Henning D, Busch H. J Biol Chem. 1979;254:11097. [PubMed] [Google Scholar]
- 31.Suryadi J, Tran EJ, Maxwell ES, Brown BA., 2nd Biochemistry. 2005;44:9657. doi: 10.1021/bi050568q. [DOI] [PubMed] [Google Scholar]
- 32.We thank Monika Golas, Peter Moore, Karin Reinisch, Joan Steitz, Tom Steitz, Hongwei Wang, and Sandra Wolin for helpful discussion and/or critical reading of the manuscript. In addition, we thank the Yale University Biomedical High Performance Computing Center (supported by NIH grant RR19895). This work was supported by the Boehringer Ingelheim Fonds PhD scholarship to F.B., NIH grants R01GM69699 to B.A.B.II and R01GM52581 to S.J.B, and NSF grant MCB0543741 to E.S.M. The EM map will be deposited upon acceptance and released upon publication of this manuscript (entry code XXX).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.