Abstract
The advent of high-throughput platforms for the interrogation of biological systems at the cellular and molecular level have allowed living cells to be observed and understood at a hitherto unprecedented level of detail and have enabled the construction of comprehensive, predictive in silico models. Here, we review the application of such high-throughput, systems-biological techniques to mycobacteria—specifically to the pernicious human pathogen Mycobacterium tuberculosis (MTb) and its ability to survive in human hosts. We discuss the development and application of transcriptomic, proteomic, regulomic, and metabolomic techniques for MTb as well as the development and application of genome-scale in silico models. Thus far, systems-biological approaches have largely focused on in vitro models of MTb growth; reliably extending these approaches to in vivo conditions relevant to infection is a significant challenge for the future that holds the ultimate promise of novel chemotherapeutic interventions.
Keywords: systems biology, transcriptomics, metabolomics, metabolic networks, proteomics, pathogen
Introduction
The pathogenesis of MTb infection is determined by a complex interplay of response of host cells to bacilli that invade and replicate in macrophages and to extracellular bacilli as well as response of the pathogen to the changing environments in the host. Global approaches to understanding the interaction between host and pathogen offer the prospect of a more integrated understanding of the dynamic behavior of the system. This requires integration of large-scale data that can be used to build a model of this interaction which in turn can answer disease relevant questions and generate testable hypotheses. To unravel the network of such a system, whether regulatory, structural or catalytic, requires heterogeneous datasets to be integrated with such datasets collected from high-throughput techniques, more recently referred to as the “omics” approaches. To date, most of the data has consisted of transcriptional data superimposed on reconstructed metabolic pathways based on genomic information of both the host and pathogen [1–6]. However, systems approaches benefit from a broad range of large scale data collection including proteins, metabolites, small stable RNAs, lipids, protein-protein interactions and protein-nucleic acid interactions. We will discuss the omics approaches that have been applied to MTb and finally how some of the data collected from these omics approaches have been applied to reconstruct metabolic models of MTb metabolism.
Transcriptomics
Early studies with knockout mutants indicated the importance of iron, nutrient, cation and carbon source (reviewed in [7]), but the underlying networks were unknown. DNA microarrays have to date proven the most sensitive tool for unraveling responses of pathogens to their host and of host response to pathogen (reviewed in [8]). Transcriptional profiling under in vivo relevant conditions has been used to understand the regulatory networks and potential metabolic pathways that may be active during host pathogenesis. Thus, microarray analysis under defined in vitro conditions such as exposure to lung surfactant, starvation, nitrosative stress, iron restriction, low oxygen and acidic pH [9–19] has indicated the importance of certain transcriptional pathways for adaptation or survival under these in vivo pertinent conditions. Transcriptional profiling of MTb during parasitism of macrophages has enabled the regulatory and catalytic pathways that are relevant during infection, growth and survival of these host cells to be explored and to be correlated to previous in vitro transcriptional analyses [20,21]. Thus, the importance of the dormancy response regulator DosR in orchestrating the metabolic slow-down that occurs when respiration on oxygen as terminal electron acceptor becomes limiting [22,23], of WhiB3 in maintaining redox homeostasis [24,25], of the Rv3574-encoded transcriptional regulator in controlling cholesterol metabolism [26], of the RshA-PknB-SigH regulatory pathway in adaptation to stress [27], of IdeR in regulating iron assimilation (reviewed in [28]), of RelA in coordinating the stringent response [11] and of PhoP in regulating expression of pathogenic lipids and other virulence factors [29] in vivo was elegantly demonstrated by a combination of in vitro transcriptional profiling, mutant knockout studies and analysis of gene expression in infected hosts. Transcriptional profiling along with analysis of stored lipids in the form of triacylglycerides of MTb expectorated in patient sputum has indicated that these bacilli are probably derived from an environment that allows slow replication based on the observation of an upregulated dormancy response and a high content of triacylglycerides which are associated with stress adaptation [30]. Microarray analyses of MTb derived from other in vivo sources such as chronically infected mice [16,17] and a variety of lesions from human TB patients [31] has corroborated the relevance of in vitro findings but also pointed to the environmental heterogeneity that MTb experiences in vivo. The multitudes of environmental cues that MTb experiences in vivo is apparent not only by analysis of different lesions [31] but also evident within one lesion [32] which makes simple extrapolation of data from pooled in vivo samples impossible. The transcriptional profiling under defined in vitro conditions has been and will be useful in finding indicator genes, proteins or metabolites of environmental cues that can be used as a sensor to explore the microenvironment of MTb during human pathogenesis [33].
The regulatory networks that control Corynebacterium glutamicum transcription have been deduced based on transcriptional profiling data and analyses of individual regulators [34]. Corynebacterium is taxonomically related to the mycobacteria and the transcriptional networks that control MTb gene expression have been deduced from conserved genes and binding sites upstream of these genes between these two organisms [35]. A large-scale transcriptional network of MTb was compiled based on gene regulatory interaction from the literature, conserved transcription regulatory pairs between Escherichia coli and MTb as well as the complete list of MTb operons with the regulation of all genes in an operon assumed to occur as a single transcriptional unit affected by a transcription factor [36]. The response of this transcriptional network during MTb adaptation to hypoxia or starvation was explored using published microarray data which showed the temporal responses of transcriptional subnetworks at various stages of the transition to non-replicating persistence [36]. This systems levels analysis is useful for probing the in vivo responses of MTb to similar environments.
The information that is gained from microarray analyses of gene expression is of course only as good as the functional annotation of the pathogen’s genome. Sequencing of genomes of clinical isolates has revealed some differences in open reading frames between strains both in terms of differences in annotation as well as presence of unique genes [37]. In addition, polymorphisms and deletions in genome sequence have pointed to the relevance or lack thereof of certain metabolic pathways for certain aspects of TB pathogenesis [38–40]. The functional significance of genetic diversity between strains has also been explored by transcriptome analyses [41]. In these analyses, transcriptional profiles during in vitro growth could be correlated to the clade that the isolates belonged to, with certain gene clusters being commonly up- or down-regulated in one or more of the 5 global phylogeographical lineages included in the study. The variation of expression observed in the basal transcriptomes of the clinical strains was unexpectedly high if the absolute genomic diversity of MTb strains relative to other bacteria is taken into consideration [42]. The variation in transcription profiles during in vitro growth is, however, an indicator of the functional consequences of the high-proportion of non-synonomous single nucleotide polymorphisms that have been observed in the genome sequences between various clinical strains [41,42]. Growth in macrophages of these clinical isolates was associated with clade-, genotype-and strain-specific fitness profiles and transcriptional profiles of these strains during parasitism of resting or activated phagocytes allowed correlations to be made between gene expression signatures and intracellular phenotypes [41]. A core transcriptome associated with intracellular growth of a panel of MTb strains was derived consisting of metabolic pathways such as hypoxia, iron storage, oxidative and nitrosative stress, cell wall remodeling, and fatty acid metabolism that were previously reported to be important for macrophage survival [20,21,41]. Although intra-macrophage transcriptomes were more divergent than in vitro transcriptomes, the correlation was still higher between strains of the same lineage than inter-genotypic expression profiles and corroborated genotype-specific differences in virulence gene expression that were known to play a role during survival in the host.
The microarrays that have been used to date to analyze transcription of MTb have not probed the regulatory elements associated with intergenic regions such as small stable RNAs. Tiling arrays have been used to analyze the Listeria transcriptome including RNAs derived from intergenic regions in response to in vitro, ex vivo and in vivo growth which highlighted the importance of small stable RNAs, anti-sense RNAs and 5’ and 3’ untranslated regions including riboswitches in its metabolism [43]. The role of small RNAs in controlling gene expression in MTb has only recently started to be explored [44].
Genome wide gene essentiality
Whole genome transposon mutagenesis has allowed the role of genes during growth or survival under in vivo relevant conditions to be explored. The essentiality of genes for symbiosis of Bacteroides in the gut [45], Salmonella typhi tolerance to bile [46], Haemophilus influenza in the mouse lung [47] and in vitro fitness of Streptococcus pneumonia [48] was analyzed by insertion sequencing, high-throughput insertion tracking by deep sequencing or high-throughput parallel se quencing of transposon mutants before and after exposure to the respective stresses. This has allowed every gene in the genomes of these pathogens to be explored for their role under the conditions of interest in a systems-wide approach. Initial saturating transposon mutagenesis studies of MTb assessed the contribution of genes to growth of MTb during growth in rich medium in vitro [49,50]. The importance of genes that were non6 essential for in vitro growth in rich medium for survival in infected mice [51,52] as well as growth on in vivo-relevant carbon sources [53] could also be assessed using genome-wide transposon mutagenesis libraries of MTb.
Proteomics
Proteomics has lagged behind transcriptomics due to instrumental and sensitivity problems although recent technical advances especially in the field of tandem mass spectrometry have made it possible to study the pathogenesis of organisms by analyzing the spectrum of secreted proteins, the proteome of virulence mutants in regulatory proteins or small RNAs, the proteome of the pathogen in response to the host or to in vivo relevant environmental cues (reviewed in [54]). Differential proteomics where proteins synthesized under an environmental condition of interest are fluorescently or isotopically labeled or tagged to compare to a basal condition, have allowed subtle variations in protein expression as well as temporal changes in expression to be unraveled [54]. The proteome of MTb in response to deletion of the pyruvate kinase gene, a gene deletion observed in certain members of the MTb complex, was determined and used to define the metabolic pathways that were altered and the associated metabolic consequences deduced from the altered proteome [55]. These studies indicated that loss of pyruvate kinase was associated with a shift in metabolism to growth on lipids for energy generation as seen by increased expression of isocitrate dehydrogenase and beta-oxidation enzymes with concomitant decreased expression of isocitrate lyase and gluconeogenic proteins. The proteome of cell membrane associated proteins has been determined which has given further insight into the localization and associated role of a variety of plasma membrane associated proteins [56]. Proteome analysis of mycobacterial proteins likely targeted for proteasomal degradation by conjugation to a ubiquitin-like protein (PUP) has revealed that there were 103 candidate PUPylation targets of which 52 were confirmed [57]. An analysis of the pathways containing PUPylated proteins indicated a high preponderance of targets on metabolic pathways associated with rapid growth as well as an over-representation of proteins from the MTb S-nitroso proteome [57,58] suggesting the importance of proteasome degradation for proteasome remodeling during metabolic growth arrest as well as stress. Proteome studies of non-replicating as opposed to replicating MTb has indicated that there are very few differences in presence of components of the proteome, despite large-scale transcriptional differences. However, the discrepancy between transcriptional and proteomic studies can be addressed by studies of protein turnover which has revealed the subtle dynamics of the response to stresses such as iron starvation [59,60].
Proteomic analyses have been used to develop models of protein-protein interaction networks which can be based on interaction of proteins to form a functional enzymatically active complex or interaction through their role in metabolic or signaling pathways. Wet-lab experiments to experimentally identify protein-protein interactions are plagued by high numbers of false-negatives but especially false-positives. Improvements in methods to identify protein interactions by for example using dual affinity tagged bait proteins that are used for in vivo interaction experiments followed by in vivo cross-linking with formaldehyde or using dual affinity purification under fully denaturing conditions followed by liquid chromatography-tandem mass spectrometric identification have allowed networks of proteins that interact with virulence factors in Salmonella to be revealed [61]. Protein interaction networks that govern mycobacterial metabolism have been constructed and have led to novel insights of the function of proteins with previously unknown function as well as the presence of a potential signaling pathway [62]. The protein-protein interaction network of MTb was a critical component in the creation of a database for MTb drug target identification which also takes into account flux balance analysis, network analysis, information on protein essentiality, comparative genomics with the host to assess uniqueness of the target to the pathogen as opposed to its host, as well as structural analysis of proposed ligand binding sites [63].
Regulomics
High-throughput chromatin immunoprecipitation (ChIP) techniques for regulomic analysis of mycobacteria have been developed and applied. Rodrigue et al. [64] have applied ChIP-chip, where DNA fragments bound by immunoprecipitated proteins are identified using microarrays, for various σ factors in M. bovis BCG. More recently, ChIP-seq, where DNA fragments are instead identified using next-generation sequencing, has been applied to MTb [65]. ChIP-seq offers a number of advantages over ChIP-chip including lower cost, higher resolution, and a lower requirement for input material [66]. Because of the very small genomes of mycobacteria compared to the mammalian genomes for which ChIP-seq was first developed, it is easy to obtain very high sequencing coverage and hence very high resolution of the location and degree of interaction at protein-DNA interaction sites. For the DosR transcription factor of MTb, ChIP-seq followed by bioinformatics analysis with the computational method CSDeconv [65] allows accurate identification of individual binding sites separated by as few as 40 base pairs.
Metabolomics
Metabolomic analyses of pathogens are extremely challenging. For MTb in particular, quenching of metabolism in such a way as to preserve the relative metabolite concentrations at a specific point in time without causing leakage of metabolites from cells is extremely challenging. The half-life of cellular metabolites is generally in the order of milliseconds or less thus immediate quenching requires addition of a vast excess of sub-zero temperature solvent and confirmation that the quenching method does not lead to cell leakage. In MTb metabolite analyses have been done on monolayers of cells grown on nitrocellulose filters to overcome the excessive dilution and cell leakage issues of quenching cell suspension [67,68]. These metabolite analyses have highlighted the role of gluconeogenesis in MTb during growth on beta-oxidation substrates and the importance of this pathway during macrophage infection as well as the unique flow of metabolites through the two halves of the tricarboxylic acid cycle. Substrate availability influences intermediary metabolite concentrations and this in turn affects the production of end-products such as cell wall lipids. Jain and co-workers [69] demonstrated that growth on propionate and other uneven chain length fatty acids which are expected to increase intracellular pools of methylmalonyl-CoA led to affected the production as well as length of methyl-branched lipids such as sulfolipid-1 and phthiocerol dimycocerosate. The chain-length of phthiocerol dimycocerosate was then used as an indicator of carbon substrate pools in MTb infected mice which suggested that MTb may oxidize odd-chain length fatty acids in vivo as evidenced by the increased chain length of this cell wall lipid [69].
In silico metabolic modeling
In silico metabolic modeling of living organisms offers the lucrative prospect of integrating data from multiple omics platforms to allow fast, inexpensive in silico experiments and fundamental understanding of function and survival. Such modeling falls into two major classes: kinetic modeling, where chemical reaction dynamics are modeled using coupled differential equations, and constraint-based modeling, where only the stoichiometric constraints of metabolic reactions are imposed and predictions of metabolic behavior are made by assuming that organisms regulate metabolic fluxes so as to achieve an objective such as growth maximization. Kinetic modeling is more detailed but suffers from the requirement of knowing many parameters, such as reaction rate constants, which cannot currently be measured on a large scale, and its application is therefore restricted to specific, well-studied pathways. For MTb, kinetic modeling has been applied to describe the d ynamics of the tricarboxylic acid (TCA) cycle and glyoxylate bypass [70].
Constraint-based modeling, on the other hand, can be achieved by simply knowing reaction stoichiometries and can therefore be applied on a genome-wide scale. Indeed, genome-scale constraint-based modeling has proven to be powerful tool for understanding and accurately predicting metabolic behavior in a large variety of organisms [71]. An example of such a model is iAF1260 [72], which models the metabolism of E. coli and accounts for the action of 1260 enzyme-coding ORFs and for the presence of 2077 reactions and 1039 metabolites.
Using the method of flux-balance analysis (FBA) [73,74], constraint-based models can be used to give predictions of metabolic behavior, such as growth rates and metabolite consumption and production rates. To make predictions using FBA, the assumption needs to be made that organisms regulate metabolic fluxes to achieve particular objectives, such as growth maximization or maximization of ATP production. Such an assumption, though seemingly artificial, nevertheless produces high predictive capability. For example, FBA can be used to predict whether or not an organism grows under genetic modifications such as knockouts and in the presence or absence of particular nutrients and, using iAF1260, FBA predicts E. coli’s ability to grow under glucose aerobic conditions with any single knockout of the 1260 modeled ORFs with 92% accuracy and its ability to grow on over 300 different sources of carbon, nitrogen, phosphorous, and sulfur with approximately 76% accuracy [72]. FBA also predicts specific growth rates and metabolite consumption and production rates for E. coli with high accuracy over a variety of nutrient sources and over time [75,76].
For MTb, three constraint-based models have been constructed: the mycolic-acid pathway (MAP) model, and two genome-scale models—GSMN-TB and iNJ661 (see Table 1). The MAP model is a relatively small model focusing only on the mycolic acid synthesis pathway of MTb. This pathway is significant because mycolic acids are known to be important for the growth, survival, and pathogenicity of MTb, and InhA—a key enzyme in the pathway—is the target of the front-line anti-tubercular drugs isoniazid and ethionamide. Using this model, it is possible to identify other potential targets for anti-tubercular drugs inhibiting mycolic acid production [77].
Table 1.
Constraint-based models of MTb.
GSMN-TB and iNJ661, on the other hand, are genome-scale models in that they both model the effect of a significant fraction of metabolic genes in the MTb genome. Both models give good agreement with experimentally observed growth rates and metabolite consumption rates. Moreover, both models were validated by comparing experimental measurements of essentiality using transposon site hybridization (TraSH) [50] with in silico predictions and achieved high levels of accuracy (78% and 65% correct predictions, respectively [78,79]).
Given genome-scale metabolic models, it becomes possible to interrogate these models to simulate and understand MTb. In particular, it becomes possible to predict, in silico, gene or gene combinations that are necessary for the survival of MTb in defined growth media or in an environment reflecting that of host infection. targetTb [63], an in silico pipeline for tuberculosis drug target identification, uses predictions obtained from FBA to identify potential targets.
It becomes possible also to understand mycobacterial metabolic capability and how particular perturbations, such as the application of a drug, affect this capability and the organism’s ability to survive. In a novel method for coupling FBA with measurements of gene expression called E-flux, Colijn et al. [80] use the GSMN-TB and MAP models coupled with over 400 genome-wide expression measurements [19,81] to identify drugs, drug combinations, and nutrient conditions that inhibit mycolic acid production and the specificity of this inhibition. E-flux correctly predicts seven of the eight known fatty acid inhibitors in the tested expression compendium, including isoniazid and ethionamide, and makes accurate predictions regarding the specificity of the action of these compounds. Further, E-flux predicts a number of additional potential modulators of mycolic acid biosynthesis in MTb. Thus, E-flux demonstrates the capability of in silico modeling to characterize the mechanism of drugs with unknown mechanism.
In addition to employing either kinetic modeling or constraint-based modeling, it is possible to simultaneously employ both approaches, using kinetic modeling for detailed modeling of particular pathways of interest and using constraint-based modeling to extend beyond these pathways to the genome scale. Fang et al. [82] have employed such a combined approach to quantitatively model the response of MTb to the growth inhibitors 3-nitropropinate and 5’-O-(N-salicylsulfamoyl) adenosine under particular nutrient conditions, and achieved good correspondence with experimentally measured dose-response curves.
Conclusion
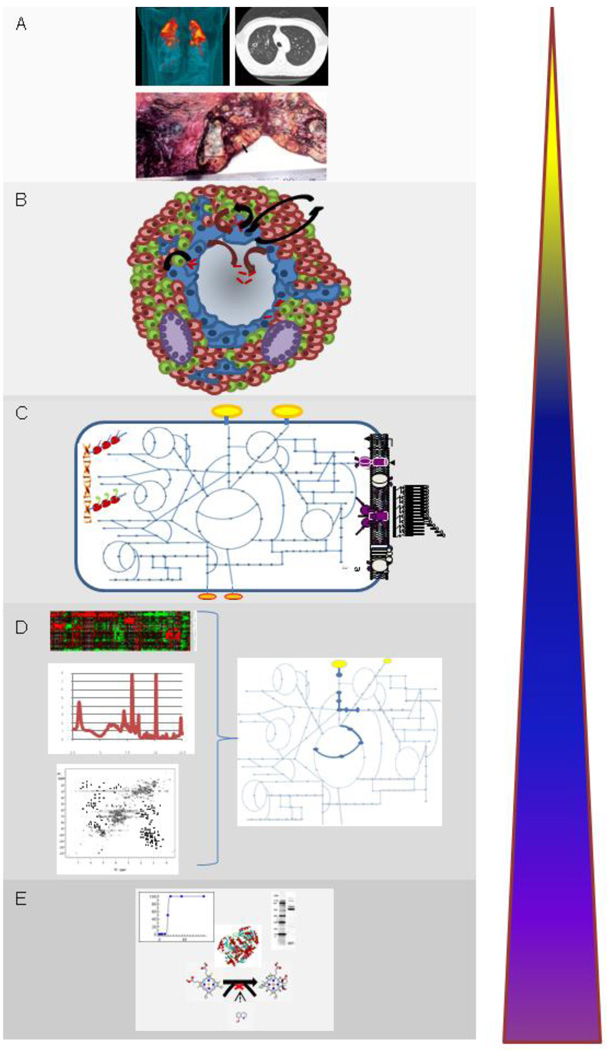
The ultimate goal of systems biology of pathogens such as MTb is to understand the metabolism of the organism in the context of infection (Fig. 1). In the last few years much progress has been made in the various omics approaches aimed at understanding MTb metabolism. Clearly, the biggest challenge will be applying these high-throughput methods to in vivo models of this disease where the vast excess of host proteins, DNA, RNA and metabolites makes identification of the bacterial derived products extremely challenging. Some transcriptional data is available for mouse and even human tissues infected with MTb [16,17,31] but converting this data to a form that allows modeling of metabolism has proven difficult. Thus, the most robust data is currently only available for various in vitro models of MTb growth. An understanding of the environment in which MTb resides in the host is, however, required to reconstruct the metabolic models that would be most pertinent to in vivo pathogenesis. Thus, determination of extracellular metabolites as well as oxygen concentration in the various types of granulomas would allow flux through the metabolic pathways to be modeled. A recent study measured the oxygen concentration in MTb- and M. bovis- infected rabbit granulomas to be measured which demonstrated the extreme hypoxic environment in which MTb can reside [83]. However, these studies do not address the spectrum of oxygen concentrations and associated substrate concentrations that can be expected in the various heterogeneous granulomas that are observed in human TB patients. Metabolomic analyses of granulomas would be key to developing systems biology approaches to MTb in the context of human pathogenesis. This would allow us to fully exploit metabolic models for the identification of metabolic chokepoints in vivo that could be targeted for chemotherapeutic intervention.
Figure 1.
Development of systems approaches for understanding MTb infection requires integrating information from all levels of understanding of disease. At the highest level (A), information is gathered from tuberculosis patients ranging from understanding the pathology of disease from clinical data and imaging studies and the epidemiology of human genetic variation in disease susceptibility and progression. Modeling of the dynamics of infection at the level of the granuloma (B) builds on information acquired from host immunology as well as pathogen biology. Genomic information of MTb as well as clinical strains of MTb has allowed metabolic networks (C) to be constructed that mirror the metabolic capabilities of this organism as well as the production of metabolites that play a role in growth, survival and virulence. Data gathered from “omics” studies such as transcriptomics, metabolomics and proteomics have allowed reconstruction of dynamic metabolic networks that capture information of metabolic fluxes that occur under various environmental conditions (D). The majority of work to date has been performed at the base level which includes focused studies of proteins, their function and structure and their interaction with small molecules (E).
Acknowledgements
This work was supported in part by the Intramural Research Program of the NIH, NIAID. We thank Clifton Barry 3rd for helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rizzetto L, Cavalieri D. A systems biology approach to the mutual interaction between yeast and the immune system. Immunobiology. doi: 10.1016/j.imbio.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Napolitani G, et al. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat Immunol. 2005;6(8):769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirschner DE, et al. Tuberculosis: global approaches to a global disease. Curr Opin Biotechnol. doi: 10.1016/j.copbio.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan DE, et al. Data integration for dynamic and sustainable systems biology resources: challenges and lessons learned. Chem Biodivers. 7(5):1124–1141. doi: 10.1002/cbdv.200900317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang A, et al. A systemic network for Chlamydia pneumoniae entry into human cells. J Bacteriol. 192(11):2809–2815. doi: 10.1128/JB.01462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang YC, et al. Global screening of potential Candida albicans biofilm-related transcription factors via network comparison. BMC Bioinformatics. 11:53. doi: 10.1186/1471-2105-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hingley-Wilson SM, et al. Survival perspectives from the world's most successful pathogen, Mycobacterium tuberculosis. Nat Immunol. 2003;4(10):949–955. doi: 10.1038/ni981. [DOI] [PubMed] [Google Scholar]
- 8.Sturdevant DE, et al. Host-microbe interaction systems biology: lifecycle transcriptomics and comparative genomics. Future Microbiol. 5(2):205–219. doi: 10.2217/fmb.09.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacon J, et al. Lipid composition and transcriptional response of Mycobacterium tuberculosis grown under iron-limitation in continuous culture: identification of a novel wax ester. Microbiology. 2007;153(Pt 5):1435–1444. doi: 10.1099/mic.0.2006/004317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betts JC, et al. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43(3):717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 11.Dahl JL, et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A. 2003;100(17):10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deb C, et al. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One. 2009;4(6):e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher MA, et al. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol. 2002;184(14):4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rustad TR, et al. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One. 2008;3(1):e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwab U, et al. Transcriptional responses of Mycobacterium tuberculosis to lung surfactant. Microb Pathog. 2009;46(4):185–193. doi: 10.1016/j.micpath.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talaat AM, et al. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci U S A. 2004;101(13):4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talaat AM, et al. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J Bacteriol. 2007;189(11):4265–4274. doi: 10.1128/JB.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198(5):705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boshoff HIM, et al. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism. Journal of Biological Chemistry. 2004;279(38):40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 20.Rohde KH, et al. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2(5):352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Schnappinger D, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198(5):693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leistikow RL, et al. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J Bacteriol. 192(6):1662–1670. doi: 10.1128/JB.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park HD, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48(3):833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh A, et al. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5(8):e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banaiee N, et al. Regulation of Mycobacterium tuberculosis whiB3 in the mouse lung and macrophages. Infect Immun. 2006;74(11):6449–6457. doi: 10.1128/IAI.00190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kendall SL, et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol Microbiol. 2007;65(3):684–699. doi: 10.1111/j.1365-2958.2007.05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park ST, et al. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2008;105(35):13105–13110. doi: 10.1073/pnas.0801143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez GM. Control of iron metabolism in Mycobacterium tuberculosis. Trends Microbiol. 2006;14(7):320–327. doi: 10.1016/j.tim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalo-Asensio J, et al. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One. 2008;3(10):e3496. doi: 10.1371/journal.pone.0003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garton NJ, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 2008;5(4):e75. doi: 10.1371/journal.pmed.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rachman H, et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun. 2006;74(2):1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan GJ, et al. Multiple M. tuberculosis phenotypes in mouse and guinea pig lung tissue revealed by a dual-staining approach. PLoS One. 5(6):e11108. doi: 10.1371/journal.pone.0011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timm J, et al. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc Natl Acad Sci U S A. 2003;100(24):14321–14326. doi: 10.1073/pnas.2436197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baumbach J. CoryneRegNet 4.0 - A reference database for corynebacterial gene regulatory networks. BMC Bioinformatics. 2007;8:429. doi: 10.1186/1471-2105-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krawczyk J, et al. From Corynebacterium glutamicum to Mycobacterium tuberculosis--towards transfers of gene regulatory networks and integrated data analyses with MycoRegNet. Nucleic Acids Res. 2009;37(14):e97. doi: 10.1093/nar/gkp453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balazsi G, et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol Syst Biol. 2008;4:225. doi: 10.1038/msb.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy TB, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37:D499–D508. doi: 10.1093/nar/gkn652. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caws M, et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4(3):e1000034. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsolaki AG, et al. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J Clin Microbiol. 2005;43(7):3185–3191. doi: 10.1128/JCM.43.7.3185-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsolaki AG, et al. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci U S A. 2004;101(14):4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Homolka S, et al. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 6(7):e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hershberg R, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6(12):e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459(7249):950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 44.Arnvig KB, Young DB. Identification of small RNAs in Mycobacterium tuberculosis. Mol Microbiol. 2009;73(3):397–408. doi: 10.1111/j.1365-2958.2009.06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6(3):279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langridge GC, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19(12):2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gawronski JD, et al. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A. 2009;106(38):16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Opijnen T, et al. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6(10):767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamichhane G, et al. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2003;100(12):7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sassetti CM, et al. Genes required for mycobacterial growth defined by high density mutagenesis. Molecular Microbiology. 2003;48(1):77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 51.Lamichhane G, et al. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect Immun. 2005;73(4):2533–2540. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100(22):12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A. 2008;105(11):4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhavsar AP, et al. Proteomics as a probe of microbial pathogenesis and its molecular boundaries. Future Microbiol. 5(2):253–265. doi: 10.2217/fmb.09.114. [DOI] [PubMed] [Google Scholar]
- 55.Chavadi S, et al. Global effects of inactivation of the pyruvate kinase gene in the Mycobacterium tuberculosis complex. J Bacteriol. 2009;191(24):7545–7553. doi: 10.1128/JB.00619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha S, et al. Proteome analysis of the plasma membrane of Mycobacterium tuberculosis. Comp Funct Genomics. 2002;3(6):470–483. doi: 10.1002/cfg.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watrous J, et al. Expansion of the mycobacterial "PUPylome". Mol Biosyst. 6(2):376–385. doi: 10.1039/b916104j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhee KY, et al. S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci U S A. 2005;102(2):467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao PK, Li Q. Principal Component Analysis of Proteome Dynamics in Iron-starved Mycobacterium Tuberculosis. J Proteomics Bioinform. 2009;2(1):19–31. doi: 10.4172/jpb.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao PK, et al. Protein dynamics in iron-starved Mycobacterium tuberculosis revealed by turnover and abundance measurement using hybrid-linear ion trap-Fourier transform mass spectrometry. Anal Chem. 2008;80(18):6860–6869. doi: 10.1021/ac800288t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chowdhury SM, et al. A method for investigating protein-protein interactions related to salmonella typhimurium pathogenesis. J Proteome Res. 2009;8(3):1504–1514. doi: 10.1021/pr800865d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cui T, et al. Uncovering new signaling proteins and potential drug targets through the interactome analysis of Mycobacterium tuberculosis. BMC Genomics. 2009;10:118. doi: 10.1186/1471-2164-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raman K, et al. targetTB: A target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome, and genome-scale structural analysis. BMC Systems Biology. 2008;2:109. doi: 10.1186/1752-0509-2-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodrigue S, et al. Identification of mycobacterial σ binding sites by chromatin immunoprecipitation assays. Journal of Bacteriology. 2007;189(5):1505–1513. doi: 10.1128/JB.01371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lun DS, et al. A blind deconvolution approach to high-resolution mapping of transcription factor binding sites from ChIP-seq data. Genome Biology. 2009;10(12):R142. doi: 10.1186/gb-2009-10-12-r142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mardis ER. ChIP-seq: welcome to the new frontier. Nature Methods. 2007;4(8):613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 67.de Carvalho LP, et al. Activity-based metabolomic profiling of enzymatic function: identification of Rv1248c as a mycobacterial 2-hydroxy-3-oxoadipate synthase. Chem Biol. 17(4):323–332. doi: 10.1016/j.chembiol.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marrero J, et al. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci U S A. 107(21):9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain M, et al. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci U S A. 2007;104(12):5133–5138. doi: 10.1073/pnas.0610634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh VK, Ghosh I. Kinetic modeling of tricarboxylic acid cycle and glyoxylate bypass in Mycobacterium tuberculosis, and its application to assessment of drug targets. Theoretical Biology and Medical Modelling. 2006;3:27. doi: 10.1186/1742-4682-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milne CB, et al. Accomplishments in genome-scale in silico modeling for industrial and medical biotechnology. Biotechnology Journal. 2009;4(12):1653–1670. doi: 10.1002/biot.200900234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feist AM, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Molecular Systems Biology. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orth JD, et al. What is flux balance analysis? Nature Biotechnology. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raman K, Chandra N. Flux balance analysis of biological systems: applications and challenges. Briefings in Bioinformatics. 2009;10(4):435–449. doi: 10.1093/bib/bbp011. [DOI] [PubMed] [Google Scholar]
- 75.Mahadevan R, et al. Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys J. 2002;83(3):1331–1340. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edwards JS, et al. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nature Biotechnology. 2001;19(2):125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- 77.Raman K, et al. Flux balance analysis of mycolic acid pathway: Targets for anti-tubercular drugs. PLoS Computational Biology. 2005;1(5):e46. doi: 10.1371/journal.pcbi.0010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beste DJ, et al. GSMN-TB: a web-based genome-scale network model of Mycobacterium tuberculosis metabolism. Genome Biology. 2007;8(5):R89. doi: 10.1186/gb-2007-8-5-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jamshidi N, Palsson BO. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targetsc. BMC Systems Biology. 2007;1:26. doi: 10.1186/1752-0509-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colijn C, et al. Interpreting expression data with metabolic flux models: Predicting Mycobacterium tuberculosis mycolic acid production. PLoS Computational Biology. 2009;5(8):e1000489. doi: 10.1371/journal.pcbi.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karakousis PC, et al. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. Journal of Antimicrobial Chemotherapy. 2008;61:323–331. doi: 10.1093/jac/dkm485. [DOI] [PubMed] [Google Scholar]
- 82.Fang X, et al. A systems biology framework for modeling metabolic enzyme inhibition of Mycobacterium tuberculosis. BMC Systems Biology. 2009;3:92. doi: 10.1186/1752-0509-3-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Via LE, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76(6):2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]