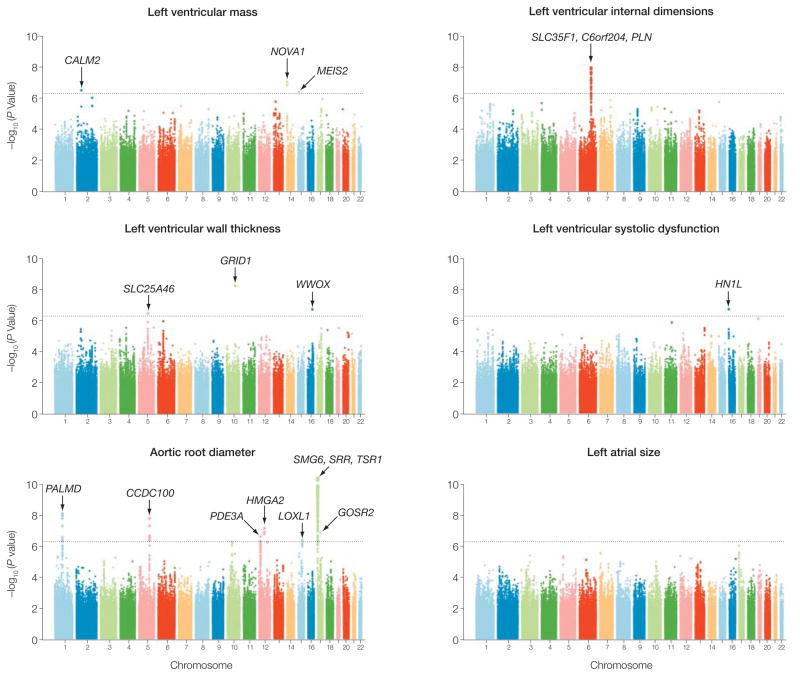
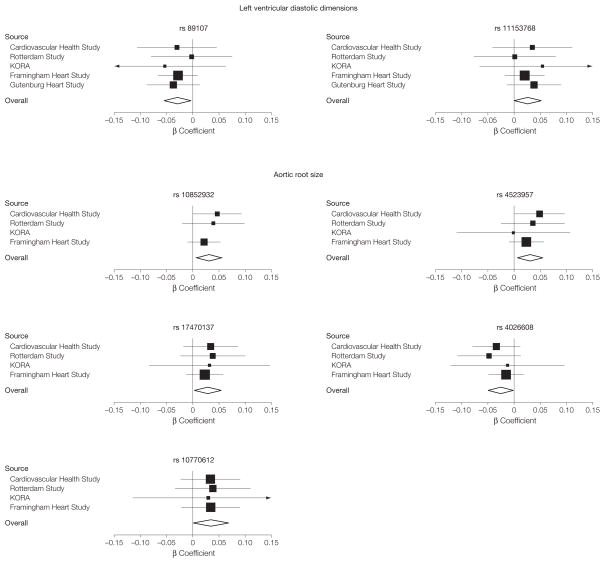
Ramachandran S Vasan
Dr Ramachandran S Vasan, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Nicole L Glazer
Dr Nicole L Glazer, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Janine F Felix
Dr Janine F Felix, MD, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Wolfgang Lieb
Dr Wolfgang Lieb, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Philipp S Wild
Dr Philipp S Wild, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Stephan B Felix
Dr Stephan B Felix, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Norbert Watzinger
Dr Norbert Watzinger, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Martin G Larson
Dr Martin G Larson, ScD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Nicholas L Smith
Dr Nicholas L Smith, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Abbas Dehghan
Dr Abbas Dehghan, MD, DSc
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Anika Großhennig
Dr Anika Großhennig, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Arne Schillert
Dr Arne Schillert, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Alexander Teumer
Mr Alexander Teumer, Dipl-Math
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Reinhold Schmidt
Dr Reinhold Schmidt, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Sekar Kathiresan
Dr Sekar Kathiresan, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Thomas Lumley
Dr Thomas Lumley, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Yurii S Aulchenko
Dr Yurii S Aulchenko, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Inke R König
Dr Inke R König, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Tanja Zeller
Dr Tanja Zeller, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Georg Homuth
Dr Georg Homuth, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Maksim Struchalin
Dr Maksim Struchalin
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Jayashri Aragam
Dr Jayashri Aragam, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Joshua C Bis
Dr Joshua C Bis, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Fernando Rivadeneira
Dr Fernando Rivadeneira, MD, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Jeanette Erdmann
Dr Jeanette Erdmann, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Renate B Schnabel
Dr Renate B Schnabel, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Marcus Dörr
Dr Marcus Dörr, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Robert Zweiker
Dr Robert Zweiker, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Lars Lind
Dr Lars Lind, MD, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Richard J Rodeheffer
Dr Richard J Rodeheffer, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Karin Halina Greiser
Dr Karin Halina Greiser, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Daniel Levy
Dr Daniel Levy, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Talin Haritunians
Dr Talin Haritunians, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Jaap W Deckers
Dr Jaap W Deckers, MD, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Jan Stritzke
Dr Jan Stritzke, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Karl J Lackner
Dr Karl J Lackner, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Uwe Völker
Dr Uwe Völker, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Erik Ingelsson
Dr Erik Ingelsson, MD, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Iftikhar Kullo
Dr Iftikhar Kullo, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Johannes Haerting
Dr Johannes Haerting, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Christopher J O’Donnell
Dr Christopher J O’Donnell, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Susan R Heckbert
Dr Susan R Heckbert, MD, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Bruno H Stricker
Dr Bruno H Stricker, MB, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Andreas Ziegler
Dr Andreas Ziegler, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Thorsten Reffelmann
Dr Thorsten Reffelmann, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Margaret M Redfield
Dr Margaret M Redfield, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Karl Werdan
Dr Karl Werdan, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Gary F Mitchell
Dr Gary F Mitchell, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Kenneth Rice
Dr Kenneth Rice, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Donna K Arnett
Dr Donna K Arnett, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Albert Hofman
Dr Albert Hofman, MD, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
John S Gottdiener
Dr John S Gottdiener, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Andre G Uitterlinden
Dr Andre G Uitterlinden, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Thomas Meitinger
Dr Thomas Meitinger, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Maria Blettner
Dr Maria Blettner, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Nele Friedrich
Dr Nele Friedrich, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Thomas J Wang
Dr Thomas J Wang, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Bruce M Psaty
Dr Bruce M Psaty, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Cornelia M van Duijn
Dr Cornelia M van Duijn, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
H-Erich Wichmann
Dr H-Erich Wichmann, MD, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Thomas F Munzel
Dr Thomas F Munzel, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Heyo K Kroemer
Dr Heyo K Kroemer, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Emelia J Benjamin
Dr Emelia J Benjamin, MD, ScM
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Jerome I Rotter
Dr Jerome I Rotter, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Jacqueline C Witteman
Dr Jacqueline C Witteman, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Heribert Schunkert
Dr Heribert Schunkert, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Helena Schmidt
Dr Helena Schmidt, MD, PhD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Henry Völzke
Dr Henry Völzke, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1,
Stefan Blankenberg
Dr Stefan Blankenberg, MD
1Framingham Heart Study: National Heart, Lung, and Blood Institute’s Framingham Heart Study, Framingham (Drs Vasan, Larson, Kathiresan, Aragam, Levy, O’Donnell, Mitchell, Wang, and Benjamin); Departments of Medicine, Preventive Medicine and Cardiology Sections, Boston University School of Medicine (Drs Vasan and Benjamin) and Department of Mathematics and Statistics, Boston University (Dr Larson), Boston, Massachusetts; and National Heart, Lung, and Blood Institute (Drs O’Donnell and Levy), Bethesda, Maryland. The Cardiovascular Health Study: Cardiovascular Health Research Unit and Department of Medicine (Drs Glazer, Bis, and Psaty), Departments of Biostatistics (Drs Lumley and Rice), Epidemiology (Drs Heckbert, Smith, and Psaty), and Health Services (Dr Psaty), University of Washington, Seattle; Seattle Epidemiologic Research and Information Center of the Department of Veterans Affairs Office of Research and Development (Dr Smith) and Center for Health Studies, Group Health (Dr Psaty), Seattle; Department of Epidemiology, University of Alabama at Birmingham (Dr Arnett); Division of Cardiology, University of Maryland Hospital, Baltimore (Dr Gottdiener); and Medical Genetics Institute, Cedars-Sinai Medical Center, West Los Angeles, California (Drs Haritunians and Rotter). Rotterdam Study: Departments of Epidemiology (Drs J. Felix, Dehghan, Aulchenko, Struchalin, Stricker, Hofman, van Duijn, and Witteman), Internal Medicine (Drs Rivadeneira and Uitterlinden), Cardiology (Dr Deckers), Erasmus MC Rotterdam, the Netherlands; Member of the Netherlands Consortium on Healthy Aging (Drs J. Felix and Witteman). MONICA/KORA: Medical Clinic 2 (Drs Lieb, Großhennig, Erdmann, Stritzke, and Schunkert) and Institute of Medical Biometry and Statistics (Drs Großhennig, König), University of Lübeck, Lübeck; Institutes of Epidemiology (Dr Wichmann) and Human Genetics (Dr Meitinger), Helmholtz Zentrum München, München; German Research Center for Environmental Health, Neuherberg and Ludwig Maximilians University (Dr Wichmann) and German Research Center for Environmental Health, Neuherberg, Technische Universität München (Dr Meitinger), Munich, Germany. Gutenberg Heart Study: Departments of Medicine II (Drs Wild, Zeller, Schnabel, Münzel, and Blankenberg), Clinical Chemistry and Laboratory Medicine (Dr Lackner), Institute of Medical Biometry, Epidemiology, and Informatics (Dr Blettner), Johannes Gutenberg-University, Mainz, and Institute for Medical Biometry and Statistics (Drs Schillert and Ziegler), University Lübeck, Germany. Study of Health in Pomerania: Department of Internal Medicine B (Drs S. Felix, Dörr, and Reffelmann), Interfaculty Institute for Genetics and Functional Genomics (Drs Teumer, Homuth, and Völker), Institute of Pharmacology (Dr Kroemer), and Institute for Community Medicine (Drs Friedrich and Völzke), Ernst-Moritz-Arndt-Universität, Greifswald, Germany (Drs S. Felix, Homuth, Dörr, Völker, Reffelmann, Friedrich, Kroemer, and Völzke and Mr Teumer). Austrian Stroke Prevention Study: Department of Internal Medicine, Division of Cardiology (Drs Watzinger and Zweiker), Department of Neurology (Dr R. Schmidt), and Institute for Molecular Biology and Biochemistry (Dr H. Schmidt), Medical University Graz, Graz, Austria. PIVUS Study: Department of Medical Sciences, Uppsala University, Uppsala (Dr Lind) and Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm (Dr Ingelsson). Division of Cardiovascular Diseases, Mayo Clinic, Rochester, Minnesota (Drs Rodeheffer, Kullo, and Redfield). CARLA Study: Institute of Medical Epidemiology, Biostatistics and Informatics (Drs Greiser and Haerting) and Martin-Luther-University, Halle-Wittenberg, Halle (Saale), Germany; and Center for Population Studies, National Heart, Lung, and Blood Institute, Bethesda, Maryland (Dr Levy)
1