Abstract
Genomic imprinting, where the effects of alleles depend on their parent-of-origin, can be an important component of the genetic architecture of complex traits. Although there has been a rapidly increasing number of studies of genetic architecture that have examined imprinting effects, none have examined whether imprinting effects depend on genetic background. Such effects are critical for the evolution of genomic imprinting because they allow the imprinting state of a locus to evolve as a function of genetic background. Here we develop a two-locus model of epistasis that includes epistatic interactions involving imprinting effects and apply this model to scan the mouse genome for loci that modulate the imprinting effects of quantitative trait loci (QTL). The inclusion of imprinting leads to nine orthogonal forms of epistasis, five of which do not appear in the usual two-locus decomposition of epistasis. Each form represents a change in the imprinting status of one locus across different classes of genotypes at the other locus. Our genome scan identified two different locus pairs that show complex patterns of epistasis, where the imprinting effect at one locus changes across genetic backgrounds at the other locus. Thus, our model provides a framework for the detection of genetic background-dependent imprinting effects that should provide insights into the background dependence and evolution of genomic imprinting. Our application of the model to a genome scan supports this assertion by identifying pairs of loci that show reciprocal changes in their imprinting status as the background provided by the other locus changes.
Introduction
Genomic imprinting refers to the epigenetic phenomenon of parent-of-origin-dependent expression of alleles, where alleles at a locus are expressed solely from the maternally or paternally inherited copy (Reik and Walter 2001). Because imprinting determines the pattern of allelic expression, it can play an important role in structuring the genotype–phenotype relationship and, consequently, can have important effects on patterns of phenotypic variation in populations. Studies of genetic architecture have exploited this phenotypic manifestation of imprinting to map putatively imprinted loci that underlie variation in complex traits (Wolf et al. 2008a). These studies have identified a number of quantitative trait loci (QTL) across several species that show a phenotypic signature of imprinting. As expected, many iQTL map to genomic regions known to contain imprinted genes (e.g., Nezer et al. 1999; Van Laere et al. 2003; Wolf et al. 2008b), but analyses of iQTL have also provided strong evidence for the presence of iQTL in genomic regions where there are currently no known imprinted genes (Cheverud et al. 2008; de Koning et al. 2000; Wolf et al. 2008b), although bioinformatics studies have predicted their presence in some of these locations (Luedi et al. 2005, 2007). Thus, studies aimed at mapping imprinted loci have led to important advances in our understanding of genomic imprinting.
Although there is rapidly growing interest in mapping iQTL underlying variation in complex traits, no previous mapping study has included the possibility that imprinting effects may themselves be dependent on the genetic background. Studies of QTL that include context-dependent (epistatic) genetic effects are becoming increasingly common and have led to great insights into the importance of gene interactions for patterns of phenotypic variation (Cheverud 2000). Epistatic interactions are important conceptually because they indicate that the effects of alleles are not an autonomous property of the gene or locus but rather depend upon the genetic context provided by other loci throughout the genome. Empirically, this implies that allelic effects cannot be characterized without specifying the genetic background being examined. For example, a number of studies have shown that mutations of major effect on one genetic background may have almost no influence on trait expression on other genetic backgrounds (Templeton 2000). From an evolutionary perspective, this context dependence of genetic effects means that genetic effects can evolve as the genetic background evolves in a population (Cheverud 2000; Templeton 2000). The evolutionary change in genetic effects plays a key role in a number of evolutionary processes, including population differentiation and speciation, adaptation, and the evolution of genetic architecture (Wolf et al. 2000). Therefore, understanding the genetic background dependency of genomic imprinting effects should provide insights into the degree to which imprinted expression is a property of a locus or is dependent on the background. Of course, interactions are reciprocal; therefore, it is also possible that the imprinting status of a gene has cascading effects at other loci through various types of interaction networks, where variation in the imprinting status of one gene determines or influences the effects of other loci.
Epistasis involving imprinting effects can arise from many underlying mechanisms, but perhaps most notable is the role of trans-acting regulatory effects such as those arising from noncoding RNA. A large proportion of the known imprinted transcriptional units in mammals (ca. 30%) are noncoding RNAs, including antisense transcripts, small nucleolar RNAs, and microRNAs, several of which appear to be involved in the regulation of imprinting of other protein-coding genes (Morison et al. 2005). This suggests that many imprinted transcriptional units may either regulate or be regulated by trans effects, making the possibility of epistasis involving imprinting effects highly likely. The first of these trans effects on imprinting was documented in maize (Kermicle 1978), where the maternal de-repression of the r1 (mdr1) locus influences the parent-of-origin-dependent pattern of expression of a second locus, r1 (Alleman and Doctor 2000). Similar patterns have been documented in other plant species (e.g., imprinting with maternal expression of MEA in Arabidopsis is under the control of other genes; Choi et al. 2002; Xiao et al. 2003), and, more generally, it appears that a number of plant genes show allele-specific imprinting that may arise from trans-acting effects of other loci (Grossniklaus 2005), which determine whether particular alleles are imprinted (e.g., dzr1 in maize; Chaudhuri and Messing 1994). Such effects could arise from large-scale alterations to genes that regulate methylation. For example, mutations in the decreasedDNAmethylation1 (DDM1) gene have been shown to reduce genome-wide levels of methylation by ca. 70% (Vongs et al. 1993).
Epistatic effects mediated by noncoding RNA that involve imprinting have also been documented at a “local” genomic level, such as the RNAi-mediated trans effects identified for the Dlk1-Gtl2 domain that harbors the callipyge locus in sheep. At the callipyge locus several RNAs from maternally expressed genes interfere with (i.e., degrade) the mRNA from a paternally expressed gene (Davis et al. 2005). The complex interactions between genes in the Dlk1–Gtl2 domain produce the phenomenon of polar overdominance, where a mutation of the callipyge gene (Freking et al. 2002) is manifested as hindquarter muscular hypertrophy only when the mutation is inherited from the father and is paired with the wild-type allele (Cockett et al. 1996). Although the interactions in this example are between closely linked loci, such effects, where one locus interferes with the phenotypic expression of another locus, could exist at a larger genomic scale and could lead to the appearance of epistatic effects involving imprinting.
Although there is a relative dearth of studies that have looked for epistatic interactions affecting complex traits that involve imprinting effects, studies by Reilly et al. (2004, 2006) and Walrath et al. (2009) provide a valuable example of the potential importance of such effects. Their work has demonstrated that cancer susceptibility associated with a major mutation present in a mouse model that develops neural tumors is modified by parent-of-origin-specific effects of loci on other chromosomes. Their results emphasize the complexity of interaction effects and the importance of considering allelic parent-of-origin when studying the effects of alleles. The identification of the loci that modify the effects of their focal locus was achieved by carefully controlled alteration of genetic background. However, these sorts of epistatic effects involving genomic imprinting are analogous to the types of epistasis that have long been considered in quantitative genetics and are becoming commonplace in analyses of QTL (Lynch and Walsh 1998). Therefore, it is logical that such effects could be identified in a genome scan using a mapping model that includes both parent-of-origin-dependent effects (which correspond to imprinting effects; see Wolf et al. 2008a) and epistatic interactions. In such an analysis, the epistatic interaction terms that include imprinting effects will, logically, reflect the genetic background dependence of imprinting effects. Therefore, the patterns of interaction effects on cancer susceptibility detected using genetic constructs in the studies by Reilly et al. (2004, 2006; Walrath et al. 2009) could presumably be detected in a mapping study in a population that had segregating variation at the interacting loci (such as an experimental intercross population). Thus, our goal here is to provide a framework in which to consider context-dependent imprinting effects.
Genetic background-dependent imprinting effects are important because they also are expected to play a key role in the evolution of genomic imprinting by providing a means for the imprinting status of a gene to evolve as genetic background evolves (Spencer and Williams 1997; Van Cleve and Feldman 2007; Wilkins and Haig 2002). Therefore, the presence of epistasis involving imprinting can be viewed as a source of genetic variation that allows imprinting itself to evolve. In general, evolutionary theories that predict imprinting evolution are consistent with either cis-effects on imprinting, where a locus determines its own imprinting status (Arnaud and Feil 2006), or trans effects on imprinting, where the imprinting status of one locus is determined by another locus (or loci). That is, essentially all models for the evolution of genomic imprinting assume that a locus starts in a nonimprinted state and evolves to a state of being either maternally or paternally expressed, whether that be via a cis- or a trans effect. Of course, the simple fact that imprinting status of genes is known to shift evolutionarily means that at some point in evolutionary time, there must have been variation in imprinting through either of these molecular pathways. For example, a recent review of imprinting in mammals (Morison et al. 2005) lists 83 known imprinted transcriptional units, only 29 of which are imprinted in both mice and humans which indicates a substantial rate of evolutionary divergence. Furthermore, some of the genes that are imprinted in both species show a different pattern of imprinting in the two species (Morison et al. 2005). Similar results have been found in a comparison of predicted imprinted genes (Luedi et al. 2005, 2007). Together these results highlight the substantial evolutionary differences that can be found between species with respect to the imprinting status of genes, reinforcing the evolutionary lability of imprinting.
Here we develop a quantitative genetic model that includes interactions involving imprinting effects on trait expression. This model is a simple extension of the traditional model for two-locus epistasis that has formed the foundation of QTL mapping approaches and a diversity of evolutionary genetic models (see Cheverud 2000). As with the forms of epistasis detected in existing mapping models, genetic background-dependent (epistatic) imprinting effect detected in our mapping model can be interpreted in different ways depending on the particular system being examined and can arise from different molecular or developmental mechanisms. Ultimately, because mapping models are used to explain phenotypic variation (i.e., trait expression), epistasis involving imprinting implies that the effect of imprinting on trait expression at one locus (i.e., the pattern of parent-of-origin-dependent effects) is dependent on the genotype present at another locus. These shifts in imprinting effects could correspond to simple changes in the magnitude of effect of a locus, where the imprinting pattern is stable but the parent-of-origin-dependent effect is larger on one genetic background than it is on another. Such a shift could be attributed to many mechanisms, including the possibility that a locus shows only imprinted expression on some genetic backgrounds while showing normal biallelic expression on others. It could also arise when the probability of imprinting is dependent on genetic background. For example, there might be a high probability of imprinted expression on one background and a low probability on another, resulting in a stronger average imprinting effect on the first background and a weaker effect on the latter. Alternatively, background-dependent imprinting effects might correspond to situations where there are complete shifts in the form of imprinting. For example, a locus shows maternal expression on one genetic background and paternal expression on another genetic background.
We start by presenting the two-locus model based on ordered genotypes with imprinting and then extend it to include interactions involving imprinting effects. We then apply this framework to scan the mouse genome for loci that modulate the imprinting status of a set of known QTL affecting early body weight in an experimental population.
A model of epistasis with imprinting
We build our model assuming that we have two loci, each with two alleles. This assumption is made not only for simplicity, but also because it follows the predominant framework used to decompose epistatic effects (Cheverud 2000; Kao and Zeng 2002). It also corresponds to the genetics of populations used in most experimental QTL mapping studies, such as inbred line intercrosses. However, the model can easily be expanded to include more than two loci, and we note how to extend the model to more loci in our presentation. We denote the first locus A, which has two alleles A1 and A2 and the second locus B, which has alleles B1 and B2. Because we wish to include genomic imprinting in our model, we keep track of the parent-of-origin of alleles, meaning that we distinguish between the two reciprocal heterozygotes. Thus, at each locus we have four “ordered” genotypes (i.e., alleles are ordered by parent-of-origin, with the paternally inherited allele listed first), e.g., at the A locus we have A1A1, A1A2, A2A1, and A2A2. The genotypic values, which are the average phenotypes of those having each of the genotypes, at the A locus are denoted as GAij and those for the B locus are GBij, where the subscript i indicates the identity of the paternally inherited allele (i.e., subscript i = 1 indicates that an individual inherited the “1” subscripted allele from their father) and the subscript j indicates the maternally inherited allele. The genotypic values for the A and B loci are arrayed in the vectors GA and GB, respectively, with genotypes ordered as G11, G12, G21, and G22 for both loci. With four genotypes at each of two loci, there are 16 two-locus ordered genotypes, which are described below when we present the two-locus model.
Single-locus model
In order to develop a two-locus model with imprinting and epistasis, we first introduce a single-locus model with imprinting. The single-locus model is analogous to the model presented by Mantey et al. (2005), but we build the model using the more general framework developed by Zeng et al. (2005) and expanded on by Álvarez-Castro and Carlborg (2007). This framework starts with an orthogonal single-locus model that is then expanded to include interactions between loci, although neither the Zeng et al. (2005) nor the Álvarez-Castro and Carlborg (2007) model includes imprinting effects, as both approaches are based on a model with unordered genotypes. We follow the nomenclature of Zeng et al. (2005), which is also used by Álvarez-Castro and Carlborg (2007). We focus on the structure of the model for locus A, noting that there is a directly analogous set of expressions for locus B.
The genotypic values are decomposed into a set of orthogonal genetic effects that describe the genotype–phenotype relationship (i.e., the structure of the genotype-to-phenotype map), where the genetic-effect design matrix for a locus (in this case locus A), denoted SA, gives the index values (or coefficients) of the genetic effects (i.e., defines the decomposition of genotypic values into the set of genetic effects, which can be viewed as the definition of the genotype-to-phenotype map) and EA gives the vector of genetic effects (again, for the A locus). The genetic-effects vector contains R, the reference point for the model (Álvarez-Castro and Carlborg 2007), aA, the additive effect, dA, the dominance effect, and iA, the imprinting effect (Mantey et al. 2005; Wolf et al. 2008b), i.e.,
Thus, in this model the SA matrix defines the relationship between the genetic effects for locus A (EA) and the genotypic values of locus A (GA):
| (1) |
The genetic-effect design matrix can be structured in various ways depending on the reference point of the system, but all produce analogous results (Álvarez-Castro and Carlborg 2007; Kao and Zeng 2002). Because our goal is to provide a first introduction to the two-locus model with imprinting, we simplify our presentation by building on the Cockerham (F2-metric) model, which is orthogonal for a population with a pair of alleles in equal frequency (Kao and Zeng 2002; Zeng et al. 2005). The Cockerham model has been championed by a number of researchers because it produces a simple set of orthogonal interaction terms that correspond to the definitions of the additive and dominance effects of the two loci (Kao and Zeng 2002). In Appendix 2 we present a more general model that can be applied to other sorts of populations in Hardy-Weinberg equilibrium.
Following Eq. 1, an arbitrary genotypic value can be defined by a regression equation:
| (2) |
where wA is the value of the additive effect design variable, vA is the dominance effect design variable, and uA is the imprinting effect design variable for genotype GAij (Mantey et al. 2005; Zeng et al. 2005). Under Cockerham’s model, extended here to include the imprinting effect, these variables are defined as
| (3) |
with the unweighted mean phenotype (i.e., unweighted mean of the genotypic values) as the reference point (R), the genetic-effect design matrix (SA) for locus A is defined as
| (4) |
making the corresponding vector of genetic effects:
| (5) |
The additive and dominance effects (genotypic values) in Eq. 5 correspond to the standard definitions for a model with unordered genotypes (Falconer and Mackay 1996), but with the two heterozygote classes separated and averaged. The additive genotypic value (aA) for a locus corresponds to half the difference between the average phenotype of the two homozygotes at that locus. The additive genotypic value measures the effect of alleles independent of the allele with which they are paired. The dominance genotypic value (dA) for a locus corresponds to the deviation of the average phenotype of the heterozygote from the midpoint between (i.e., average of) the two homozygotes at the locus. The dominance genotypic value is a measure of the interaction between alleles at a locus (where the effect of an allele depends on the allele with which it is paired). The imprinting effect (iA) of a locus corresponds to half the difference between the average phenotypes of the alternative heterozygotes at that locus (Knott et al. 1998; Mantey et al. 2005) and, therefore, is a measure if the parent-of-origin-dependent effect of alleles at the locus.
The two-locus model
The two-locus model is built directly from the combination of the single-locus models defined for the A locus and the B locus, as defined in Eqs. 1–5. Following Álvarez-Castro and Carlborg (2007), we build the order of the vector of two-locus genotypic values (GAB) by using the Kronecker product of the two single-locus genotypic value vectors, where GAB = GB ⊗ GA. Here, the Kronecker product is used to give the order of the genotypic values and does not imply that the single-locus genotypic values are themselves multiplied together. Rather, the products of the single-locus genotypic values are replaced by the corresponding multilocus genotypic values (cf. Appendix 1 in Álvarez-Castro and Carlborg 2007). The resulting vector of two-locus genotypic values is labeled Gijkl, where the first two subscripts refer to the paternally (subscript i) and maternally (subscript j) inherited A locus alleles and the last two refer to the maternally (subscript k) and paternally (subscript l) inherited B locus alleles. The construction of the two-locus genotypic values matrix GAB is shown in Appendix 1. Additional loci can easily be added to the vector of genotypic values, with the ordering of multilocus genotypic values again given use of the Kronecker product, where additional loci are computed to the left of the previous ones (Álvarez-Castro and Carlborg 2007).
Like the matrix of two-locus genotypic values, the structure of the vector of two-locus genetic effects (EAB), which now includes interactions between locus A and locus B, is defined by the Kronecker product of the two single-locus vectors (EA and EB), EAB = EB ⊗ EA, but with the reference point in each vector first replaced with a 1 (Álvarez-Castro and Carlborg 2007). The products of single-locus effects in the resulting vector are replaced with corresponding epistasis terms (e.g., aAaB would be replaced with aa, which corresponds to the interaction between additive effects of the two loci, see below), resulting in a vector that contains a total of 16 terms, 9 of which correspond to epistasis between the two loci. The construction of the two-locus genetic-effects vector is shown in Appendix 1. The 16 terms in the two-locus genetic-effects vector are described in detail below.
The two-locus genetic-effect design matrix SAB is defined as the Kronecker product of the single-locus genetic-effect design matrices SA (Eq. 4) and SB (which is analogous to Eq. 4 but written for locus B): SAB = SB ⊗ SA. Therefore, the two-locus genotypic values (contained in the vector GAB) can be decomposed into the 16 effects in the two-locus genetic-effect vector EAB as GAB = SAB • EAB. The two-locus genetic-effect design matrix, which therefore defines the relationship between two-locus genotypic values and genetic effects, is shown in tabular form in Table 1, with the columns of the matrix labeled to indicate the genetic effect and the rows labeled to indicate the genotypic values (note that the columns have been resorted in Table 1 to clarify the presentation). The vector of two-locus genetic effects is defined as
| (6) |
Equation 6 gives the definitions for the 15 genetic effects for the two-locus model, plus the reference point, in terms of the genotypic values. Most importantly, it is the inverse of the two-locus genetic-effect design matrix, that provides the definition of each of the 15 genetic effects in terms of the genotypic values. Therefore, to understand what the effect represents in the two-locus model with ordered genotypes, we present and discuss these 16 terms and present the inverse of the two-locus genetic-effect design matrix in Table 2.
Table 1.
The two-locus genetic-effect design matrix (SAB)
| Genotypic values | Coefficients |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | aA | dA | iA | aB | dB | iB | aa | ad | ai | da | dd | di | ia | id | ii | |
| G1111 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |||||
| G1211 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||||
| G2111 | 1 | 0 | −1 | 1 | 0 | 0 | −1 | 0 | 0 | 0 | 0 | |||||
| G2211 | 1 | −1 | 0 | 1 | 0 | −1 | 0 | 0 | 0 | 0 | 0 | |||||
| G1112 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | |||||
| G1212 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |||||
| G2112 | 1 | 0 | −1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | −1 | |||||
| G2212 | 1 | −1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | −1 | 0 | |||||
| G1121 | 1 | 1 | 0 | 0 | −1 | 0 | 0 | 0 | 0 | −1 | 0 | |||||
| G1221 | 1 | 0 | 1 | 0 | −1 | 0 | 0 | 0 | 0 | 0 | −1 | |||||
| G2121 | 1 | 0 | −1 | 0 | −1 | 0 | 0 | 0 | 0 | 0 | 1 | |||||
| G2221 | 1 | −1 | 0 | 0 | −1 | 0 | 0 | 0 | 0 | 1 | 0 | |||||
| G1122 | 1 | 1 | 0 | −1 | 0 | −1 | 0 | 0 | 0 | 0 | 0 | |||||
| G1222 | 1 | 0 | 1 | −1 | 0 | 0 | −1 | 0 | 0 | 0 | 0 | |||||
| G2122 | 1 | 0 | −1 | −1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||||
| G2222 | 1 | −1 | 0 | −1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |||||
Because the genetic-effect design matrix gives the relationship between the vector of genotypic values (GAB) and the vector of genetic effects (EAB), i.e., GAB = SABEAB, we label the columns and rows to reflect this relationship. Thus, the rows are labeled to reflect the corresponding genotypic value (Gijkl, where ij are the paternally and maternally inherited alleles at the A locus, while kl are the paternally and maternally inherited alleles at the B locus) and the columns are labeled to reflect the 16 effects in the two-locus genetic-effect vector EAB. The first column is reference point for the model (R). The other 15 columns correspond to the single-locus (additive, dominance, and imprinting) effects of the A locus (aA, dA, and iA), the B locus (aB, dB, and iB), and the interaction between these single-locus effects (e.g., id is the interaction between the imprinting effect of the A locus and the dominance effect of the B locus and therefore is referred to as imprinting-by-dominance epistasis). The order of the columns has been rearranged to conform to a more natural order (e.g., as in Kao and Zeng 2002; Zeng et al. 2005), but the natural order of the columns can be seen in Appendix 1. These index scores are used in a regression model to estimate each of the 16 parameters and are given as X(y)ijkl, where y indicates the parameter (e.g., aA, aB, etc.) and ijkl indicates the particular genotypic value. For example, X(aA)1111 would be the aA index score for the G1111 genotypic value and would have the value 1
Table 2.
The inverse of the two-locus genetic-effect design matrix (i.e.,
| Genetic effects | Genotypic values |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1111 | G1211 | G2111 | G2211 | G1112 | G1212 | G2112 | G2212 | G1121 | G1221 | G2121 | G2221 | G1122 | G1222 | G2122 | G2222 | |
| R | ||||||||||||||||
| aA | 0 | 0 | 0 | 0 | ||||||||||||
| dA | ||||||||||||||||
| iA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||
| aB | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||
| dB | ||||||||||||||||
| iB | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||
| aa | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| ad | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||
| ai | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| da | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||
| dd | ||||||||||||||||
| di | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||
| ia | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| id | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||
| ii | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
The inverse of the genetic-effect design matrix in Table 1 gives the unique solution to the two-locus vector of genetic effects, EAB, i.e., Therefore, the resulting matrix gives the definition of the 16 terms in the two-locus vector of genetic effects. Thus, the columns of the matrix are labeled with the corresponding genotypic value and the rows are labeled with the corresponding genetic effect
Definitions of genetic effects
As in the Cockerham model of epistasis for unordered genotypes (Álvarez-Castro and Carlborg 2007; Kao and Zeng 2002; Zeng et al. 2005), the reference point (R) for the model is the unweighted (or equally weighted) mean of the 16 two-locus genotypic values (Table 2).
Single-locus effects
The single-locus effects at one locus are averaged (unweighted) over genotypes at the other locus (Álvarez-Castro and Carlborg 2007; Cheverud 2000; Kao and Zeng 2002; Zeng et al. 2005). For the two-locus model with ordered genotypes, the additive genotypic values for the A (aA) and B (aB) loci are defined as (Table 2)
| (7) |
For the two-locus model, the dominance genotypic values for the A (dA) and B (dB) loci are defined as
| (8) |
For the two-locus model, the imprinting genotypic values for the A (iA) and B (iB) loci are defined as
| (9) |
As in the additive and dominance effects, the imprinting effect of each locus is defined as an unweighted average over the genotypes at the other locus. Under these definitions of effects, when there is paternal expression of a locus the additive and imprinting genotypic values are expected to be the same (de Koning et al. 2002; Wolf et al. 2008b). This is because genotypes group by their paternally inherited allele such that the A1A1 and A1A2 genotypes should have the same genotypic value while the A2A1 and A2A2 genotypes have the same value. Likewise, when there is maternal expression the absolute values of the additive and imprinting genotypic values are expected to be the same, but they are expected to be of opposite sign.
Epistatic effects
As in the typical epistasis model based on the genotypic values of unordered genotypes at a pair of loci, the two-locus model based on ordered genotypes includes interaction terms corresponding to interactions between the additive and dominance effects at both loci (Cheverud 2000). These are the familiar four orthogonal forms of epistasis usually considered in an analysis of interactions between loci: additive-by-additive (aa), additive-by-dominance (ad), dominance-by-additive (da), and dominance-by-dominance (dd). These four forms of epistasis have simple interpretations. For example, additive-by-dominance indicates that the additive effect of the first locus depends on (i.e., changes as a function of) the genotype present at the second locus, while the dominance effect of the second locus depends on the genotype present at the first locus (Cheverud 2000). The interpretations of the other three forms follow this same logic. For the two-locus model with ordered genotypes, the four orthogonal forms of epistasis involving additive and dominance effects are defined as (Table 2)
| (10) |
which, as expected, matches the definitions of these effects in the two-locus model with unordered genotypes (Cheverud 2000; Kao and Zeng 2002) but with the separate genotypic values of the alternate heterozygotes included.
By including the imprinting effect in the single-locus model based on ordered genotypes, the two-locus epistasis model includes an additional five forms of epistatic effects that have not previously been included in a model of epistasis. These epistatic effects are the additive-by-imprinting (ai), imprinting-by-additive (ia), dominance-by-imprinting (di), imprinting-by-dominance (id) and the imprinting-by-imprinting (ii) interaction effects. Note that as with additive-by-dominance and dominance-by-additive interaction effects, the additive-by-imprinting and imprinting-by-additive effects are fundamentally the same, but with the roles of the two loci reversed. The same is the case for dominance-by-imprinting and imprinting-by-dominance interactions. These five orthogonal forms of epistasis involving imprinting effects are defined as (Table 2)
| (11) |
Table 3 illustrates patterns of the 16 genotypic values associated with the forms of epistasis involving imprinting effects.
Table 3.
Examples of the three orthogonal forms of epistasis involving imprinting
| Genotype | B1B1 | B1B2 | B2B1 | B2B2 |
|---|---|---|---|---|
| (A) Additive-by-imprinting | ||||
| A1A1 | 0 | 1 | −1 | 0 |
| A1A2 | 0 | 0 | 0 | 0 |
| A2A1 | 0 | 0 | 0 | 0 |
| A2A2 | 0 | −1 | 1 | 0 |
| (B) Dominance-by-imprinting | ||||
| A1A1 | 0 | 0 | ||
| A1A2 | 0 | 0 | ||
| A2A1 | 0 | 0 | ||
| A2A2 | 0 | 0 | ||
| (C) Imprinting-by-imprinting | ||||
| A1A1 | 0 | 0 | 0 | 0 |
| A1A2 | 0 | 1 | −1 | 0 |
| A2A1 | 0 | −1 | 1 | 0 |
| A2A2 | 0 | 0 | 0 | 0 |
(A) In the case of an additive-by-imprinting interaction, the sign of the imprinting effect of locus B changes as a function of the type of homozygote at locus A. On the A1A1 background, locus B shows a positive imprinting effect (i = 1), while on the A2A2 background, locus B shows a negative imprinting effect (i = −1). (B) In the case of dominance-by-imprinting, the sign of the imprinting effect of locus B changes as a function of whether individuals are homozygous or heterozygous at locus A. For genotypes that are homozygous at locus A, the B locus has a negative imprinting effect (i = −½), and for genotypes that are heterozygous at locus A, locus B has a positive imprinting effect (i = +½). (C) In the case of imprinting-by-imprinting epistasis, the imprinting effect of the B locus depends on which of the reciprocal heterozygote genotypes are present in the A locus and vice versa. Not shown are the reciprocal cases of imprinting-by-additive and imprinting-by-dominance epistasis. These are identical to the cases shown in A and B, respectively, but with the roles of the two loci reversed
Analysis of quantitative trait loci
The 16 effects in the full two-locus model with imprinting and epistasis (contained in the EAB vector) can be fitted empirically by regression using the orthogonal genotypic index values (coefficients) shown in Table 1 as the independent variables and individual phenotypic values (i.e., the trait values measured for individuals, not genotype classes) as the dependent variables (Cheverud 2000; Kao and Zeng 2002). That is, although we define the effects of the locus in terms of genotypic values for the 16 two-locus genotypes (shown in Eq. 6), the effects can be estimated empirically using individual phenotypes rather than the average values of each of the 16 genotype classes. The regression model that yields estimates of the 15 genetic effects is given as
| (12) |
where Py(ijkl) is the phenotypic value of individual y with two-locus ordered genotype ijkl, X(z)ijkl is the genotypic index value of parameter z (where z is one of the 15 genetic effects being estimated) for genotype ijkl, and ry is the residual from the model for individual y. The genotypic index values X(z)ijkl for the Cockerham model are given in Table 1. Supplementary Table 1 gives the more general index values for populations in Hardy–Weinberg equilibrium (which are developed in Appendix 2) but where allele frequencies are not ½ at each locus.
To illustrate the application of the two-locus model shown in Eq. 12, we provide an empirical example from an experimental population of mice that we have used to identify imprinted QTL that affect a number of growth- and size-related traits (Cheverud et al. 2008; Hager et al. 2009; Leamy et al. 2008; Wolf et al. 2008b).
Materials and methods
Empirical analysis of epistasis with imprinting
Study population
Our experimental population is the F3 generation of an intercross between the Large (LG/J) and Small (SM/J) inbred strains of mice [see Kramer et al. (1998) for details of this population]. These strains were originally independently derived as selection lines for large and small adult body weight (i.e., weight at 60 days of age) and have been inbred for more than 60 years (Chai 1956; Goodale 1938; MacArthur 1944). With more than 120 generations of inbreeding, the strains are devoid of within-strain allelic variation, other than in a very narrow genomic region around the agouti locus in the SM/J strain (Hrbek et al. 2006).
The experimental population was initiated by mating ten SM/J males to ten LG/J females. The resulting F1 population consisted of 52 individuals that were randomly mated to produce 510 F2 animals. These F2 animals were randomly mated to produce an F3 generation of 200 full-sibling families with a total of 1632 individuals. Half litters of most families (158/200) were reciprocally cross-fostered at random between pairs of females that gave birth on the same day. Pups were weaned at 21 days of age and randomly housed with three or four other same-sex individuals. Further details of the husbandry are given in Cheverud et al. (1996) and Vaughn et al. (1999).
Phenotypes
All pups were weighed weekly (to 0.1 g) into adulthood. To illustrate the analysis of epistasis we focus on the first period of growth represented by weight gain from week 1 to week 2 after birth, which we refer to as “early growth.” These age-specific weights were corrected for sex and litter size.
Genotypes
Individuals were genotyped at 353 autosomal single nucleotide polymorphism (SNP) markers that show allelic differences between the SM/J and LG/J strains. Details of the markers are given in Wolf et al. (2008b). The average map distance between markers in the F2 generation is ca. 4 cM, with markers evenly placed throughout the genome based on the physical position of the markers [see Wolf et al. (2008b) for details], except for regions in which LG/J and SM/J have been found to be monomorphic (Hrbek et al. 2006). A complete list of the markers along with their physical and map positions are given in the supplementary information in Wolf et al. (2008b).
These SNP genotypes from the F2 parents and their F3 offspring were used to reconstruct haplotypes for all animals using the integer linear programming algorithm (ILP) in the program PedPhase (Li and Jiang 2003a, b). The haplotype reconstruction yielded a set of unordered haplotypes for the F2 animals and a set of ordered haplotypes (i.e., ordered by parent-of-origin of alleles) for the F3 animals. The ordered genotypes of the F3 allowed us to distinguish between the four possible genotypes at a given locus.
Analysis of single-locus effects
We started by scanning the genome for QTL that have significant main effects on early growth. This was done using an analog of the model shown in Eq. 12 but with only the single-locus model terms corresponding to Eq. 5. Model fitting was done following the procedure described in Wolf et al. (2008b) so we only briefly describe the process here.
The regression model was fitted as a mixed model using Restricted Maximum Likelihood (REML) as implemented in the Mixed Procedure of SAS (SAS version 9.1; SAS Institute, Cary, NC, USA). The significance of the individual genetic effects was determined using a mixed model with the a, d, and i index values as fixed regression effects and family as a random effect. To generate a test for the overall significance of a locus, a model with ordered genotype class as a fixed effect (rather than the 3 orthogonal regression effects) and family as a random effect was fitted using REML. Family was included as a random effect in these models to account for variation among families not attributable to the effects of the locus in question, which significantly increases power (Wolf et al. 2008b).
The mixed model was used to scan the genome for QTL, generating for each locus the probability associated with the overall effect of the locus and the individual additive, dominance, and imprinting effects. These probability values were transformed to a logarithmic probability ratio (LPR) in order to make them comparable to the LOD scores typically seen in QTL analyses (LPR = −log10[probability]). Significance thresholds were determined using a Bonferroni correction, which was calculated using the effective number of markers method (Li and Ji 2005). The thresholds for the whole genome and for individual chromosomes are given in Supplementary Table 3. Once a QTL was identified, we used post hoc tests, with a LPR significance threshold of 1.3 (i.e., p < 0.05) to determine the effects at a locus. Confidence intervals for the positions of iQTL were determined using a one LOD drop (using LPR values) following Lander and Botstein (1989). Because our goal in this study was to identify epistatic interactions between QTL and not the QTL per se, we used an approach that maximized the number of loci that were considered QTL. In the significance tests, all nonsignificant terms were dropped for a locus when generating the LPR value for a locus. This approach is logical in that there is not necessarily an a priori expectation that a locus showing one form of effect (additive, dominance, or imprinting) will necessarily show other forms of effect, and so the overall test of a locus based on the full three degrees of freedom is conservative.
When multiple peaks were identified on a chromosome we tested for the presence of multiple QTL using a likelihood ratio test. A model was first fitted as described above, but by maximum likelihood, with a single locus at the “consensus” position on the chromosome that corresponded to the location of the highest LPR (this is the “single-QTL model”). A second model was then fitted with either some effects moved to alternate positions (e.g., a model could be fitted where the additive effect was moved to a different location) or additional terms included to account for the effects of other loci (e.g., the model could be fitted with two additive effects corresponding to two different positions; this is the “multiple-QTL model”). The difference in the −2 log likelihood values of the two models (single-QTL model minus the multiple-QTL model) was calculated; these values are approximately χ2 distributed with the number of degrees of freedom corresponding to the sum of the number of terms that changed position and the number of additional terms. A chromosome was determined to have multiple QTL when the multiple-QTL model had a significantly better fit than the single-QTL model.
Because apparent additive and parent-of-origin effects at a locus can also be caused by a maternal effect of that locus rather than by genomic imprinting, we tested all loci to confirm that the appearance of main effects could not be attributed to maternal effects following the procedure outlined in Hager et al. (2008).
Analysis of epistasis
We started by testing whether the QTL identified in the single-locus analysis interacted to affect early growth. We took this approach for two main reasons: First, a blind scan using all pairs of loci in the genome has the major disadvantage of including a very large number of tests, which greatly inflates significance thresholds and reduces power. Although the complete two-way scan of the genome has the clear benefit of including all possible interactions, interactions would have to be exceptionally strong to stand out from the background and, as a result, it would be very difficult to identify true epistatic interactions in the large set of tests. The second reason is that we might expect, a priori, that loci with main effects on trait expression are likely to interact to affect trait expression, perhaps because they are involved in some shared molecular or developmental process. Because there is uncertainty in the location of QTL, we scanned the genome for interactions that mapped within the confidence interval for the QTL rather than looking just at the location of the QTL. We also did a blind scan of the genome, where we tested for interactions between all pairs of loci in the genome. As stated above, this scan lacks power because of the large number of tests being done, but it allows us to identify any locations that show strong interactions, even if they do not show significant main effects.
As in the single-locus model, the overall significance test was based on an ANOVA model (Lynch and Walsh 1998), with the four ordered genotypes at each of the two loci coded as a pair of class variables (with values 1, 2, 3, and 4 for the genotypes ordered as in Eq. 1). The epistatic interaction was included in the model as an interaction between these two class variables. This produces a test for overall epistasis that is nearly identical to an overall test of the nine orthogonal forms of epistasis. The ANOVA model, with the ordered genotypes at the two loci and their interaction included as fixed-effect class variables and family included as a random-effect class variable, was fitted using Minimum Variance Quadratic Unbiased Estimation (MIVQUE0) as implemented in the Mixed Procedure of SAS (SAS version 9.1; SAS Institute, Cary, NC, USA). MIVQUE0 was used because it is noniterative, allowing the two-way genome scan to run significantly faster while providing similar results to REML or ML. Family was again included as a random effect to account for variation among families not attributable to the effects of the pair of loci being tested.
Once epistatic pairs were detected, we used the full regression model shown in Table 1 and Eq. 12 to test the nine orthogonal forms of epistasis. This model was again fitted using the repeated-measures model in the Mixed Procedure of SAS, including the six main effects of the two loci and nine interaction tests as fixed regression effects and family as a random-effect class variable. Because the identification of epistatic interactions was based on an overall test of interaction between loci, our approach was not biased toward finding any particular form of interaction. Interactions between loci on the same chromosome were not included because of strong multicolinearity between linked markers.
Significance thresholds were determined using a Bonferroni correction. In the case of the test for interactions between QTL, we used a threshold corresponding to the number of QTL pairs (55) to generate a LPR threshold of 3.03. For the genome scan we used an approach that implements the effective number of markers method (Li and Ji 2005) adapted for an interaction test (i.e., adapted for the effective number of marker pairs; see Cheverud 2000). Due to correlations between linked markers, our interaction test included approximately 9200 effective tests. This results in a Bonferroni threshold LPR at the 5% level (i.e., α = 0.05) of 5.25. Once an epistatic pair of QTL was identified, we used post-hoc tests, with a LPR significance threshold of 1.3 (i.e., p < 0.05), to determine the form of epistasis.
Because interactions between maternal genotypes and their offspring’s genotypes can lead to the appearance of a pattern of epistasis in the offspring, even though loci do not interact within individuals (Wolf 2001), we did post-hoc tests where we restricted the analysis to offspring of heterozygous mothers at both loci. By doing so, we removed genetic variation among mothers and thereby removed the possibility that maternal-zygotic interactions could explain the patterns of within-genome epistasis that we detected.
To determine the relative proportion of variance explained by the interaction between loci overall and by interactions involving genomic imprinting effects, we calculated the approximate variance (Vg) using the expectation:
| (13) |
which is the genetic variance of a two-locus system in a population where the two alleles at each locus are approximately at equal frequency and in Hardy–Weinberg equilibrium (cf. Eq. 12 in Kao and Zeng 2002). The analytical expectation was used because REML does not compute sums of squares and the corresponding R2. The proportion of phenotypic variance explained by the two-locus system, or particular effects, is the variance component (or total Vg) divided by the phenotypic variance (Vp).
Results and discussion
Extension of the two-locus model using ordered genotypes allows one to test for the genetic background dependence of imprinting effects. The presence of epistasis involving imprinting effects can most simply be interpreted as resulting from a scenario where the imprinting status of a locus changes as a function of alleles present at a second locus (i.e., imprinting status is background-dependent). That is, although the interaction between loci is reciprocal, it seems most logical to view epistatic effects involving imprinting as cases where the genotype at one locus modulates the imprinting state at a second locus. In the two-locus model with ordered genotypes, there are five orthogonal forms of epistasis that involve imprinting effects (in addition to the usual four forms that do not). These correspond to three basic patterns (since there are two pairs of forms that are identical, but with the roles of the two loci reversed): additive-by-imprinting (with imprinting-by-additive being the same pattern but with the roles of the loci reversed), dominance-by-imprinting (or imprinting-by-dominance), and imprinting-by-imprinting. These three types of interaction effects are illustrated in Table 3. In the example of additive-by-imprinting interaction (Table 3A), the B locus shows a positive imprinting effect on the A1A1 background (i.e., B1B2 has the phenotypic value +1, while B2B1 has the value −1, making the imprinting effect i = 1), no imprinting effect on either heterozygous A locus background, and a negative imprinting effect (i = −1) on the A2A2 background. Dominance-by-imprinting is analogous, with the imprinting effect changing depending on whether individuals are homozygotes or heterozygotes at the A locus. In the example of dominance-by-imprinting shown in Table 3B, the imprinting effect i for the B locus is −½ on the two homozygous A locus backgrounds and +½ on the two heterozygous A locus backgrounds. Imprinting-by-imprinting epistasis occurs when the imprinting effect at one locus depends on which type of heterozygote (i.e., which of the two reciprocal heterozygotes) is present at the other locus and vice versa. In the example shown in Table 3C, both loci switch from having an imprinting effect of +1 on one heterozygous background (the 12 background) at the other locus to −1 on the alternative heterozygous background (the 21 background).
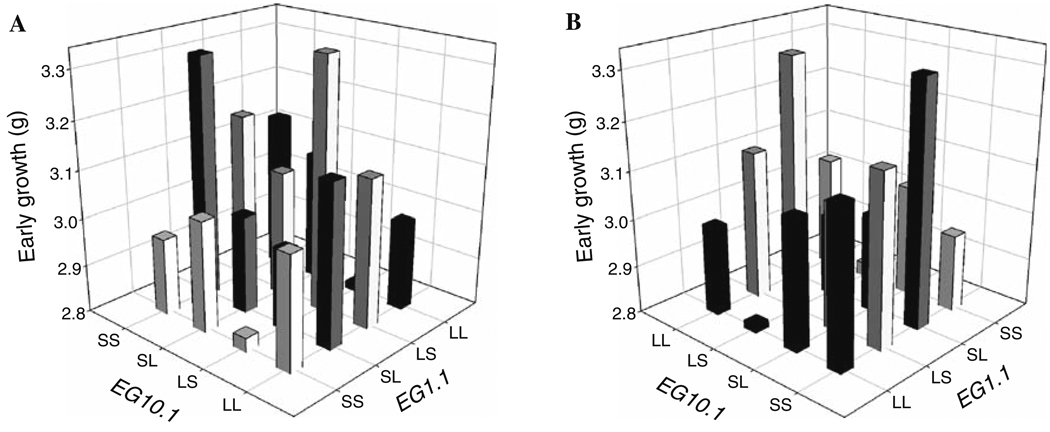
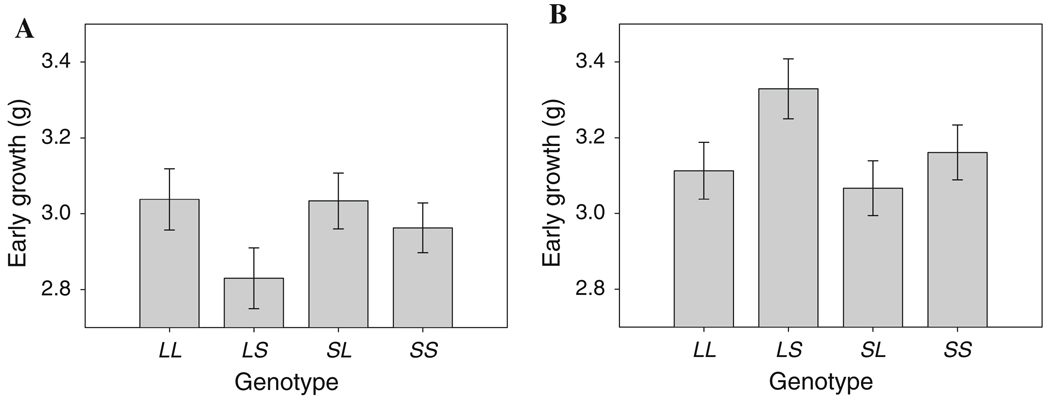
Our genome scan identified 11 QTL on nine chromosomes, three of which showed a significant parent-of-origin (imprinting) effect. These QTL are shown in Table 4 and have been named EGX.Y, where EG indicates an early growth locus, X is the chromosome, and Y is the number of that QTL on the chromosome. The application of our model to test for interactions between these loci identified an interaction between EG1.1 and EG10.1. Both of these loci show a single-locus dominance effect and they show a reciprocal interaction associated with dominance-by-imprinting, imprinting-by-dominance, and (relatively weak) imprinting-by-imprinting interactions (Table 5; illustrated in Fig. 1). The overall pattern is complex and is made more complex by the single-locus dominance effects at the two loci. The dominance-by-imprinting and the reciprocal imprinting-by-dominance effects indicate that each locus shows an imprinting effect that is significantly different between the homozygous and heterozygous backgrounds at the other locus (as in Table 3B). To illustrate this example we first denote the two alleles at each locus as the L allele, for the allele derived from the LG/J strain, and the S allele, for the allele derived from the SM/J strain. An example of this shift is illustrated in Fig. 2, which shows that EG10.1 has a polar underdominance effect on the SS background at EG1.1, where the LS heterozygote grows less than the other three genotypes and the imprinting effect i is negative, but has a polar over-dominance effect on the LS background at EG1.1, where the LS heterozygote grows significantly more than the other three genotypes and the imprinting effect is positive (see Wolf et al. 2008b for a discussion of the various forms of imprinting effects).
Table 4.
QTL affecting growth (gain in body mass) from 1 to 2 weeks of age
| Chr | QTL | Marker | Map position (cM) |
Physical position (Mb) |
LPR | Effect(s) |
|---|---|---|---|---|---|---|
| 1 | EG1.1 | rs3692731 | 44.27 | 105.9 | 4.40 | d = 0.20 |
| 3 | EG3.1 | rs3684333 | 54.17 | 107.3 | 2.43 | a = −0.12 |
| i = −0.10 | ||||||
| 5 | EG5.1 | mCV25284008 | 0.90 | 12.5 | 2.26 | d = −0.14 |
| 8 | EG8.1 | rs6410533 | 4.22 | 14.3 | 8.82 | a = 0.20 |
| d = 0.20 | ||||||
| 10 | EG10.1 | rs13480652 | 30.94 | 74.1 | 2.58 | d = −0.16 |
| 13 | EG13.1 | rs6215262 | 0.00 | 3.7 | 2.43 | i = 0.12 |
| 13 | EG13.2 | rs6271232 | 15.71 | 41.9 | 2.41 | a = −0.12 |
| 13 | EG13.3 | rs3718727 | 28.46 | 66.2 | 2.88 | d = 0.16 |
| 14 | EG14.1 | rs3718262 | 39.17 | 92.4 | 3.70 | a = 0.16 |
| 15 | EG15.1 | rs4230908 | 38.13 | 83.2 | 3.71 | a = 0.12 |
| d = 0.14 | ||||||
| 19 | EG19.1 | rs3711945 | 53.97 | 58.6 | 2.44 | i = −0.12 |
Shown for each locus are the chromosome, QTL name, marker name, map position (in F2 centimorgan [cM] units), physical position (in megabases [Mb], according to mouse genome build 37 from www.ensembl.org), LPR for the significance test, and the effects of the locus in standard deviation units. Only significant effects are shown (i.e., effects where the LPR for the given effect is > 1.3, which corresponds to p = 0.05)
Table 5.
Regression analysis of the interaction between the QTL EG1.1 and EG10.1 affecting growth (weight gain) from week 1 to week 2
| Effect | Estimate | SE | LPR | % |
|---|---|---|---|---|
| aA | 0.018 | 0.028 | 0.30 | – |
| dA | 0.153 | 0.034 | 4.97 | 1.23 |
| iA | 0.031 | 0.028 | 0.57 | – |
| aB | −0.036 | 0.029 | 0.67 | – |
| dB | −0.090 | 0.035 | 1.95 | 0.43 |
| iB | −0.029 | 0.029 | 0.50 | – |
| aa | −0.054 | 0.035 | 0.92 | – |
| ad | −0.024 | 0.047 | 0.21 | – |
| ai | −0.013 | 0.035 | 0.15 | – |
| da | −0.038 | 0.047 | 0.38 | – |
| dd | 0.005 | 0.064 | 0.02 | – |
| di | 0.172 | 0.046 | 3.68 | 0.78 |
| ia | 0.031 | 0.035 | 0.42 | – |
| id | 0.148 | 0.047 | 2.78 | 0.58 |
| ii | 0.074 | 0.035 | 1.48 | 0.29 |
The 15 genetic-effect estimates were fitted using the regression model shown in Eq. 12 and the genotypic index scores shown in Table 1. The single-locus effects with the subscript A correspond to the effects of EG1.1 while those with the subscript B correspond to the locus EG10.1. Shown are the parameter estimates (in grams), the standard error of the estimate (SE), the LPR for the test of that term (where LPR = −log10[probability]), and the percent of phenotypic variance (%) explained by that term (given only for significant effects). Together, the significance test of the epistatic interaction has an LPR of 3.29. The combined effects of the locus explain 3.9% of the phenotypic variance (3.3% for significant effects). An LPR value of 1.3 corresponds to a p value of 0.05. Significant effects are highlighted in boldface
Fig. 1.
The genotypic values (average phenotypes) for early weight of the 16 possible two-locus genotypes involved in the interaction between loci EG1.1 and EG10.1. The four ordered genotypes at each locus (with the paternal allele listed first) are given as LL, LS, SL, and SS, with the L allele coming from the LG/J strain and the S allele coming from the SM/J strain. Panels a and b show two different views of the same pattern because some bars are obstructed in each view
Fig. 2.
An example of the change in the pattern of effect of EG10.1 across genetic backgrounds at EG1.1. In all cases, genotypes at both loci are labeled LL, LS, SL, and SS, as in Fig. 1. Bars correspond to the genotypic values (i.e., mean phenotypes) derived from the mixed model for early weight of each genotype, with error bars being standard errors of these means. a The pattern of expression at EG10.1 on the SS background at EG1.1, which is consistent with polar underdominance. b The pattern of expression of EG10.1 on the LS background at EG1.1, which is consistent with a pattern of polar overdominance
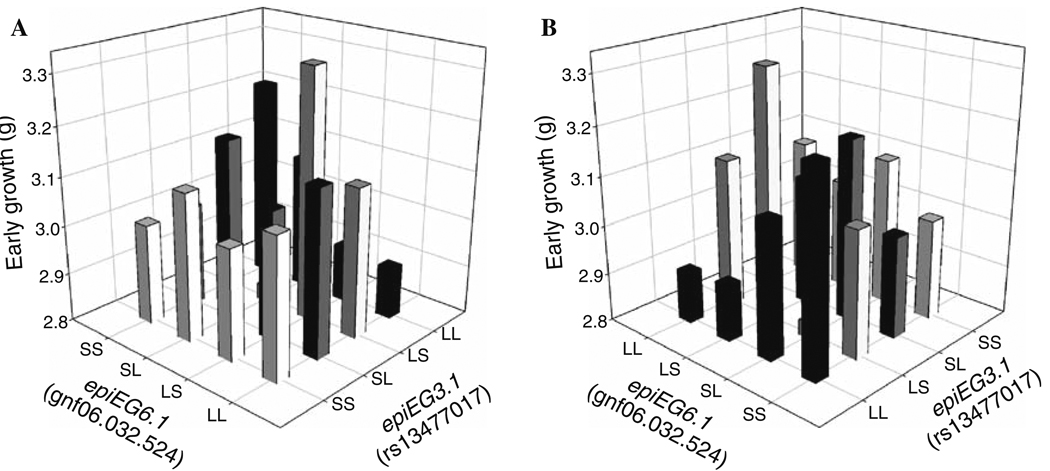
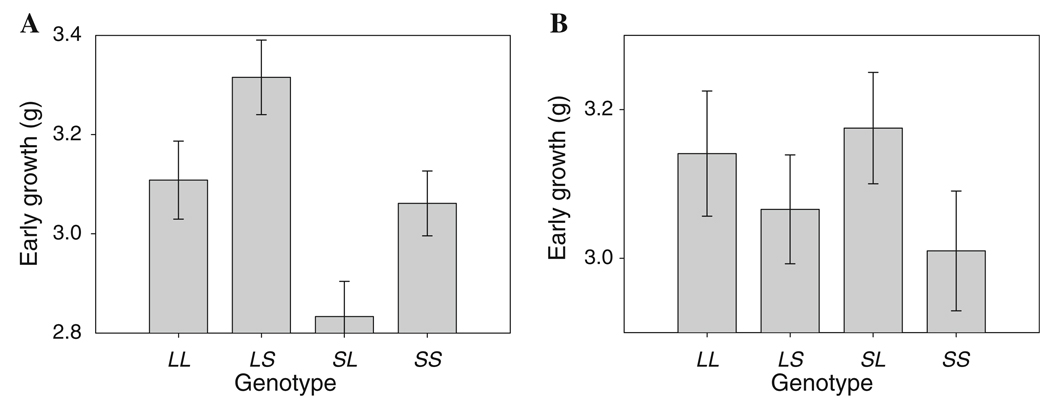
The full two-way scan, where we blindly scanned the genome for all two-locus interactions, identified a single pair of loci that showed a significant interaction effect after Bonferroni correction (Fig. 3, Table 6). The first of these loci is located at 7.9 cM on chromosome 3 (corresponding to marker rs13477017) and has been named epiEG3.1 (for an epistatic locus effecting early growth) and the second is at 12.4 cM on chromosome 6 (corresponding to marker gnf06.032.524) and has been named epiEG6.1. Neither locus in this pair showed a significant main (single-locus) effect of any sort, but they showed a complex pattern of epistasis that included additive-by-additive, dominance-by-additive, dominance-by-imprinting, and imprinting-by-imprinting interactions. The strongest of these interactions is the imprinting-by-imprinting interaction in which both loci change their imprinting effect across the two reciprocal heterozygote backgrounds at the other locus. This results in a case similar to that shown in Fig. 2 in which epiEG3.1 shows polar overdominance, where the LS heterozygote grows more than the other three genotypes (making i positive), on the LS background at epiEG6.1, and polar underdominance, where the LS heterozygote grows significantly less than the other three genotypes (making i negative), on the SL background at epiEG6.1. Because interactions are necessarily reciprocal, we find that epiEG6.1 also shows a shift in the imprinting effect (Fig. 4), going from positive on the LS background at epiEG3.1, associated with a bipolar pattern of imprinting (Wolf et al. 2008b), to negative on the SL background at epiEG3.1. In the latter case, the pattern is consistent with maternal expression, but the differences between the genotype classes are more subtle.
Fig. 3.
The genotypic values (average phenotypes) for early weight of the 16 possible two-locus genotypes involved in the interaction between loci epiEG3.1 and epiEG6.1. The four ordered genotypes at each locus (with the paternal allele listed first) are given as LL, LS, SL, and SS, with the L allele coming from the LG/J strain and the S allele coming from the SM/J strain. Panels a and b show two different views of the same pattern because some bars are obstructed in each view
Table 6.
Regression analysis of the interaction between a chromosome 3 locus (epiEG3.1, corresponding to marker rs13477017) and a chromosome 6 locus (epiEG6.1, corresponding to marker gnf06.032.524) affecting growth (weight gain) from week 1 to week 2
| Effect | Estimate | SE | LPR | % |
|---|---|---|---|---|
| aA | −0.013 | 0.027 | 0.20 | – |
| dA | 0.046 | 0.034 | 0.76 | – |
| iA | −0.009 | 0.027 | 0.13 | – |
| aB | −0.006 | 0.029 | 0.08 | – |
| dB | −0.002 | 0.036 | 0.02 | – |
| iB | 0.017 | 0.029 | 0.25 | – |
| aa | −0.097 | 0.034 | 2.35 | 0.50 |
| ad | −0.040 | 0.047 | 0.41 | – |
| ai | −0.022 | 0.035 | 0.27 | – |
| da | 0.101 | 0.047 | 1.52 | 0.27 |
| dd | 0.039 | 0.064 | 0.27 | – |
| di | 0.153 | 0.046 | 3.10 | 0.62 |
| ia | −0.021 | 0.036 | 0.26 | – |
| id | −0.028 | 0.048 | 0.25 | – |
| ii | 0.148 | 0.034 | 4.81 | 1.16 |
The 15 genetic-effect estimates were fitted using the regression model shown in Eq. 12 and the genotypic index scores shown in Table 1. The single-locus effects with the subscript A correspond to the effects of epiEG3.1 while those with the subscript B correspond to the locus epiEG6.1. Together, the significance test of the epistatic interaction has an LPR of 5.54. The combined effects of the locus explain 2.8% of the phenotypic variance (2.5% for significant effects). An LPR value of 1.3 corresponds to a p value of 0.05. Significant effects are highlighted in boldface. Other details are given in Table 5
Fig. 4.
An example of the change in the pattern of effect of epiEG6.1 across genetic backgrounds at epiEG3.1. In all cases, genotypes at both loci are labeled LL, LS, SL, and SS, as in Fig. 3. Bars correspond to the genotypic values (i.e., mean phenotypes) derived from the mixed model for early weight of each genotype, with error bars being standard errors of these means. a The pattern of expression at epiEG6.1 on the LS background at epiEG3.1, which is consistent with bipolar dominance. b The pattern of expression of epiEG6.1 on the SL background at epiEG3.1, which is consistent with weak maternal expression
From a genetic perspective, these pairs of loci point out the potential importance of trans effects in controlling the pattern of imprinting. They also highlight the importance of considering multiple genetic backgrounds when defining the imprinting status of a gene. If one were to consider any one of these four loci on a single genetic background, one would get a misleading picture of the imprinting pattern. In each case, because the patterns of epistasis are symmetrical across genetic backgrounds, none of these loci shows a significant imprinting effect overall, despite the fact that they all appear to show imprinted expression on some backgrounds. These results suggest that there may be significant flexibility in imprinted expression and causes one to question whether it makes sense to assign a particular pattern of expression to a locus if the pattern is dependent on trans effects and is, therefore, not consistent across genetic backgrounds. These results also imply that imprinting effects may be variable across mouse strains, and, as a result, characterizing a locus as being maternally or paternally expressed (or showing more complex patterns) may be strain (i.e., background) dependent.
The presence of epistasis involving imprinting effects could play an important role in the evolution of imprinting effects. For example, consider a case where there is an epistatic interaction between loci A and B corresponding to an additive-by-imprinting interaction (as in Table 3A), while the B locus also shows an additive main effect. In this scenario (illustrated in Table 7), the B locus shows paternal expression (where genotypes group phenotypically by the paternally inherited allele) on the A1A1 background but maternal expression (where genotypes group phenotypically by the maternally inherited allele) on the A2A2 background. Such a pattern would allow for an evolutionary switch from paternal to maternal expression (or vice versa depending on the allelic effects), as the population evolved from being near fixation for the A1 allele to near fixation of the A2 allele. The patterns we identified show more complex reciprocal shifts in imprinting state, most of which are associated with changes in the heterozygote state at the other locus, and so would be associated with more complex shifts in imprinting effects as allele frequencies shift. Whether such shifts in imprinting effects across genetic backgrounds are widespread and correspond to patterns predicted to allow for imprinting effects to evolve is currently unknown, but the framework presented here should allow for the genetic dissection of such effects.
Table 7.
Example of a pattern that allows for the evolution of imprinting
| Genotype | B1B1 | B1B2 | B2B1 | B2B2 |
|---|---|---|---|---|
| A1A1 | 1 | 1 | −1 | −1 |
| A1A2 | 1 | 0 | 0 | −1 |
| A2A1 | 1 | 0 | 0 | −1 |
| A2A2 | 1 | −1 | 1 | −1 |
In this scenario, the A locus shows an additive-by-imprinting interaction as illustrated in Table 3A, while the B locus also shows an additive main effect in which the B1 allele increases the phenotypic value of an individual. The B locus shows paternal expression on the A1A1 background but maternal expression on the A2A2 background
Supplementary Material
Acknowledgments
We thank Larry Leamy and Charles Roseman for insightful discussions during the development of this work and Reinmar Hager and Will Pitchers for help with the haplotype reconstruction. This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (UK) to JBW and by NIH grant DK055736 to JMC.
Appendix 1: model details
The order of the vector of two-locus genotypic values (GAB) is constructed using the Kronecker product of the two single-locus genotypic value vectors, where GAB = GB ⊗ GA (cf. Appendix 1 in Álvarez-Castro and Carlborg 2007). The Kronecker product gives the order of the genotypic values, not the actual products (i.e., multiplication) of values. Since each single-locus genotypic value has two subscripts corresponding to the maternally and paternally inherited alleles at the locus, the two-locus genotypic values simply contain the combination of subscripts taken from the two single-locus genotypic values, with the A locus subscripts appearing first. Thus, in the resulting vector of two-locus genotypic values, Gijkl, the first two subscripts refer to the two A locus alleles (i = paternally inherited allele, j = maternally inherited allele) and the last two refer to the B locus alleles (k = maternally inherited allele, l = paternally inherited allele). This can be visualized as
Like the matrix of two-locus genotypic values, the structure of the vector of two-locus genetic effects (EAB) is defined by the Kronecker product of the two single-locus vectors (EA and EB), EAB = EB ⊗ EA, but with the reference point in each vector first replaced with a 1 (Álvarez-Castro and Carlborg 2007). The products of single-locus effects in the resulting vector are replaced with corresponding epistasis terms (e.g., aAaB would be replaced with aa, which corresponds to the interaction between additive effects of the two loci, see below), resulting in a vector that contains a total of 16 terms, 9 of which correspond to epistasis between the two loci:
Appendix 2: extension of the model to arbitrary allele frequencies
In this appendix we extend the model to populations with arbitrary allele frequencies that are in Hardy–Weinberg equilibrium. This approach follows the general two-allele model presented in Zeng et al. (2005), which has been called the G2A model by Álvarez-Castro and Carlborg (2007). It can be extended to any population using the framework in Álvarez-Castro and Carlborg (2007). We do not present a full description of the G2A model since a full description and justification is presented by Zeng et al. (2005). The G2A model differs from the model presented in the text above in that the values of the genotypic index variables (coefficients) depend on allele frequencies at the loci (to maintain orthogonality across allele frequencies). When the two alleles are at equal frequency, the G2A model reduces to the Cockerham model presented in the text.
Because the genotypic index variables depend on allele frequency, we denote the frequency of the A1 allele as p1 and the frequency of the A2 allele as p2. Under the G2A model the additive and dominance effect design variables, wA and vA, respectively are defined as (extended to consider the two heterozygotes separately)
| (14) |
The inclusion of the imprinting genotypic index variable in the G2A model is straightforward because the two heterozygotes are always at equal frequency at Hardy-Weinberg equilibrium. As a result, the value of the imprinting genotypic index variable uA does not depend on allele frequencies and therefore is orthogonal from the additive and the dominance genotypic index variables at all frequencies. The value of the genotypic index variable for the G2A model matches that given in Eq. 3. This makes the genetic-effect design matrix for the G2A model:
| (15) |
and the corresponding vector of genetic effects:
| (16) |
The reference point for the model remains the mean of the genotypic values but is now weighted by their frequency. In the G2A model, the measure of the additive effect aA is frequency-dependent and now represents the average effect of an allele substitution rather than the typical frequency-independent additive effect or additive genotypic value. The dominance and imprinting effects, however, are independent of allele frequencies for the single-locus model (but not for multiple loci Zeng et al. 2005).
The two-locus extension of the G2A model for ordered genotypes follows the extension of the single-locus Cockerham model to the two-locus case. The structure of the two-locus vectors of genotypic values (GAB) and genetic effects (EAB) match those in Appendix 1. To extend the model to two loci, one-first needs to define the allele frequencies at the B locus; therefore, we denote the frequency of the B1 allele as q1 and the frequency of the B2 allele as q2 = 1 − q1. Using these allele frequencies, a genetic-effect design matrix for locus B can be constructed following the structure in Eq. 15:
| (17) |
As in the Cockerham model presented in the text, the two single-locus genetic-effect design matrices (Eqs. 15 and 17) can be used to derive the genotypic index values for the two-locus model as SAB = SB ⊗ SA. The resulting matrix, which is given in Supplementary Table 2, is analogous to that presented in Table 1 of Zeng et al. (2005), but with the additional dimensions added for the ordered genotypes. As in the model of unordered genotypes from Zeng et al. (2005), the values of all coefficients involving additive and dominance effects (including all epistatic terms involving interactions between these effects) depend on allele frequencies. However, coefficients for the imprinting effects of both loci (iA and iB) and their interaction (ii) do not depend on allele frequencies. The inverse of the two-locus genetic-effect design matrix defines the two-locus genetic effects in terms of two-locus genotypic values. The inverse of the two-locus genetic-effect design matrix, , which gives the definitions of the two-locus genetic effects in terms of the genotypic values, is given in Supplementary Table 3.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00335-009-9209-2) contains supplementary material, which is available to authorized users.
Contributor Information
Jason B. Wolf, Faculty of Life Sciences, University of Manchester, Manchester M13 9PT, UK, jason@evolutionarygenetics.org
James M. Cheverud, Department of Anatomy and Neurobiology, Washington University School of Medicine, St Louis, MO 63110, USA cheverud@pcg.wustl.edu
References
- Alleman M, Doctor J. Genomic imprinting in plants: observations and evolutionary implications. Plant Mol Biol. 2000;43:147–161. doi: 10.1023/a:1006419025155. [DOI] [PubMed] [Google Scholar]
- Álvarez-Castro JM, Carlborg O. A unified model for functional and statistical epistasis and its application to quantitative trait loci analysis. Genetics. 2007;176:1151–1167. doi: 10.1534/genetics.106.067348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud P, Feil R. MEDEA takes control of its own imprinting. Cell. 2006;124:468–470. doi: 10.1016/j.cell.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Chai CK. Analysis of quantitative inheritance of body size in mice. I. Hybridization and maternal influence. Genetics. 1956;41:167–178. doi: 10.1093/genetics/41.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S, Messing J. Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation. Proc Natl Acad Sci USA. 1994;91:4867–4871. doi: 10.1073/pnas.91.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM. Detecting epistasis among quantitative trait loci. In: Wolf JB, Brodie ED III, Wade MJ, editors. Epistasis and the evolutionary process. New York: Oxford University Press; 2000. pp. 58–81. [Google Scholar]
- Cheverud JM, Routman EJ, Duarte FAM, Swinderen BV, Cothran K, et al. Quantitative trait loci for murine growth. Genetics. 1996;142:1305–1319. doi: 10.1093/genetics/142.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Hager R, Roseman C, Fawcett G, Wang B, et al. Genomic imprinting effects on adult body composition in mice. Proc Natl Acad Sci USA. 2008;105:4253–4258. doi: 10.1073/pnas.0706562105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- Cockett NE, Jackson SP, Shay TL, Farnir F, Berghmans S, Snowder GD. Polar overdominance at the ovine callipyge locus. Science. 1996;273:236–238. doi: 10.1126/science.273.5272.236. [DOI] [PubMed] [Google Scholar]
- Davis E, Caiment F, Tordoir X, Cavaillé J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- de Koning D-J, Rattink AP, Harlizius B, van Arendonk JAM, Brascamp EW, Groenen MAM. Genome-wide scan for body composition in pigs reveals important role of imprinting. Proc Natl Acad Sci USA. 2000;97:7947–7950. doi: 10.1073/pnas.140216397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning D-J, Bovenhuis H, Van Arendonk JAM. On the detection of imprinted quantitative trait loci in experimental crosses of outbred species. Genetics. 2002;161:931–938. doi: 10.1093/genetics/161.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to quantitative genetics. 4th edn. Essex: Longman; 1996. [Google Scholar]
- Freking BA, Murphy SK, Wylie AA, Rhodes SJ, Keele JW, Leymaster KA, Jirtle RL, Smith TPL. Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res. 2002;12:1496–1506. doi: 10.1101/gr.571002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodale HD. A study of the inheritance of body weight in the albino mouse by selection. J Hered. 1938;29:101–112. [Google Scholar]
- Grossniklaus U. Genomic imprinting in plants: a predominantly maternal affair. In: Meyer P, editor. Annual plant reviews: plant epigenetics. Sheffield: Blackwell; 2005. pp. 174–200. [Google Scholar]
- Hager R, Cheverud JM, Wolf JB. Maternal effects as the cause of parent-of-origin effects that mimic genomic imprinting. Genetics. 2008;178:1755–1762. doi: 10.1534/genetics.107.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager R, Cheverud JM, Wolf JB. Relative contribution of additive, dominance and imprinting effects to phenotypic variation in body size and growth between divergent selection lines of mice. Evolution Int J org Evolution. 2009;63:1118–1128. doi: 10.1111/j.1558-5646.2009.00638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrbek T, Alves de Brito R, Wang B, Pletscher LS, Cheverud JM. Genetic characterization of a new set of recombinant inbred lines (LGXSM) formed from the intercross of SM/J and LG/J inbred mouse strains. Mamm Genome. 2006;17:417–429. doi: 10.1007/s00335-005-0038-7. [DOI] [PubMed] [Google Scholar]
- Kao C-H, Zeng Z-B. Modeling epistasis of quantitative trait loci using Cockerham’s model. Genetics. 2002;160:1243–1261. doi: 10.1093/genetics/160.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle JL. Imprinting of gene action in maize endosperm. In: Walden DB, editor. Maize breeding and genetics. New York: Wiley; 1978. pp. 357–371. [Google Scholar]
- Knott SA, Marklund L, Haley CS, Andersson K, Davies W, Ellegren H, Fredholm M, Hansson I, Hoyheim B, Lundstrom K, Moller M, Andersson L. Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics. 1998;149:1069–1080. doi: 10.1093/genetics/149.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MG, Vaughn TT, Pletscher LS, King-Ellison K, Adams E, Cheverud JM. Genetic variation for body weight gain and composition in the intercross of Large (LG/J) and Small (SM/J) inbred strains of mice. Genet Mol Biol. 1998;21:211–218. [Google Scholar]
- Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–189. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy LJ, Klingenberg CP, Sherratt E, Wolf JB, Cheverud JM. A search for quantitative trait loci exhibiting imprinting effects on mouse mandible size and shape. Heredity. 2008;101:518–526. doi: 10.1038/hdy.2008.79. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analysis using the eigenvalues of a correlaton matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Li J, Jiang T. Efficient inference of haplotypes from genotypes on a pedigree. J Bioinform Comput Biol. 2003a;1:41–69. doi: 10.1142/s0219720003000204. [DOI] [PubMed] [Google Scholar]
- Li J, Jiang T. Efficient rule-based haplotyping algorithms for pedigree data; Proceedings of the 7th annual international conference on research computational molecular biology; 2003b. pp. 197–206. [Google Scholar]
- Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sunderland: Sinauer Associates; 1998. [Google Scholar]
- MacArthur J. Genetics of body size and related characters. I. Selection of small and large races of the laboratory mouse. Am Naturalist. 1944;78:142–157. [Google Scholar]
- Mantey C, Brockmann GA, Kalm E, Reinsch N. Mapping and exclusion mapping of genomic imprinting effects in mouse F2 families. J Hered. 2005;96:329–338. doi: 10.1093/jhered/esi044. [DOI] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Nezer C, Moreau L, Brouwers B, Coppieters W, Detilleaux J, Hanset R, Karim L, Kvasz A, Leroy P, Georges M. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet. 1999;21:155–156. doi: 10.1038/5935. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Reilly KM, Tuskan RG, Christy E, Loisel DA, Ledger J, Bronson RT, Smith CD, Tsang S, Munroe DJ, Jacks T. Susceptibility to astrocytoma in mice mutant for Nf1 and Trp53 is linked to chromosome 11 and subject to epigenetic effects. Proc Natl Acad Sci USA. 2004;35:13008–13013. doi: 10.1073/pnas.0401236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly KM, Broman KW, Bronson RT, Tsang S, Loisel DA, Christy E, Sun J, Diehl J, Munroe DJ, Tuskan RG. An imprinted locus epistatically influences Nstr1 and Nstr2 to control resistance to nerve sheath tumors in a neuorofibromatosis type 1 mouse model. Cancer Res. 2006;66:62–68. doi: 10.1158/0008-5472.CAN-05-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HG, Williams JM. The evolution of genomic imprinting: two modifier-locus models. Theor Popul Biol. 1997;51:23–35. doi: 10.1006/tpbi.1997.1293. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Epistasis and complex traits. In: Wolf JB, Brodie ED III, Wade MJ, editors. Epistasis and the evolutionary process. New York: Oxford University Press; 2000. pp. 41–57. [Google Scholar]
- Van Cleve J, Feldman MW. Sex-specific viability, sex linkage and dominance in genomic imprinting. Genetics. 2007;176:1101–1118. doi: 10.1534/genetics.107.071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laere A-S, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, Archibald AL, Haley CS, Buys N, Tally M, Andersson G, Georges M, Andersson L. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425:832–836. doi: 10.1038/nature02064. [DOI] [PubMed] [Google Scholar]
- Vaughn TT, Pletscher LS, Peripato A, King-Ellison K, Adams E, Erikson C, Cheverud JM. Mapping quantitative trait loci for murine growth—a closer look at genetic architecture. Genet Res. 1999;74:313–322. doi: 10.1017/s0016672399004103. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabid-opsisthaliana DNA methylation deficient mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Walrath JC, Fox K, Truffer E, Alvord WG, Quiñones OA, Reilly KM. Chr 19A/J modifies tumor resistance in a sex- and parent-or-origin-specific manner. Mamm Genome. 2009;20:214–223. doi: 10.1007/s00335-009-9179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. Parental modifiers, antisense transcripts and loss of imprinting. Proc R Soc B Biol Sci. 2002;269:1841–1846. doi: 10.1098/rspb.2002.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB. Gene interactions from maternal effects. Evolution Int J org Evolution. 2001;54:1882–1898. doi: 10.1111/j.0014-3820.2000.tb01235.x. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Brodie ED, III, Wade MJ. Epistasis and the evolutionary process. Oxford: Oxford University Press; 2000. [Google Scholar]
- Wolf JB, Hager R, Cheverud JM. Genomic imprinting effects on complex traits: a phenotype-based perspective. Epigenetics. 2008a;3:295–299. doi: 10.4161/epi.3.6.7257. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Cheverud JM, Roseman C, Hager R. Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet. 2008b;4:e1000091. doi: 10.1371/journal.pgen.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL. Imprinting of the MEA polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell. 2003;5:891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
- Zeng Z-B, Wang T, Zou W. Modeling quantitative trait loci and interpretation of models. Genetics. 2005;169:1711–1725. doi: 10.1534/genetics.104.035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.