Abstract
Expressed sequence tags (ESTs) are randomly sequenced cDNA clones. Currently, nearly 3 million human and 2 million mouse ESTs provide valuable resources that enable researchers to investigate the products of gene expression. The EST databases have proven to be useful tools for detecting homologous genes, for exon mapping, revealing differential splicing, etc. With the increasing availability of large amounts of poorly characterised eukaryotic (notably human) genomic sequence, ESTs have now become a vital tool for gene identification, sometimes yielding the only unambiguous evidence for the existence of a gene expression product. However, BLAST-based Web servers available to the general user have not kept pace with these developments and do not provide appropriate tools for querying EST databases with large highly spliced genes, often spanning 50 000–100 000 bases or more. Here we describe Gene2EST (http://woody.embl-heidelberg.de/gene2est/), a server that brings together a set of tools enabling efficient retrieval of ESTs matching large DNA queries and their subsequent analysis. RepeatMasker is used to mask dispersed repetitive sequences (such as Alu elements) in the query, BLAST2 for searching EST databases and Artemis for graphical display of the findings. Gene2EST combines these components into a Web resource targeted at the researcher who wishes to study one or a few genes to a high level of detail.
INTRODUCTION
Ten years ago, it was proposed that databases of randomly-selected cDNA sequences would have applications in the discovery of new human genes, mapping of the human genome and identification of coding regions in genomic sequences (1). The fast throughput (allowed by only a minimum of characterisation) has allowed the expressed sequence tag (EST) databases to grow rapidly. While many uses for ESTs have indeed been found, there are two features of mammalian genomes for which the establishment of these databases has proved particularly helpful: (i) the numerous large families of homologous genes, many of which are functionally redundant (2,3), and (ii) the high degree of RNA splicing, with genes split into tens or even hundreds of exons, often assembled into several alternatively spliced mRNA variants (4). Alternative splicing may greatly increase the complexity of functional gene products encoded by a genome (for a recent review, see 5). To a greater or lesser degree, most eukaryotic genomes possess these characteristics. Thus, ESTs can help to provide a more dynamic view of DNA coding content than arises from sequencing just a single cDNA product and the gene itself, as was most often done. Seen in this way, ESTs provide a most useful counterpart to large scale genome sequencing projects. Enormous effort has gone into the public human and mouse EST projects with >4.4 million ESTs currently available for these two species. Their genome sequencing projects benefit directly in terms of genes revealed, exons predicted and differentially spliced products mapped. Hence, due to the limitations of current de novo gene prediction algorithms (6,7), EST matches are intrinsic to the gene prediction protocol in the ‘real time’ human genome annotation project ENSEMBL (http://www.ensembl.org). In this light, it seems regrettable that the two completed animal genomes (8,9), Caenorhabditis elegans and Drosophila melanogaster, have not been accompanied by similar large scale EST projects. The 80 000 Drosophila ESTs are only informative for highly expressed genes (10).
Despite the utility of ESTs for gene expression analysis, the many researchers who are primarily dependent on publicly available servers may have found it difficult to use EST databases in a sensitive and efficient manner. They are likely to be faced with a variety of problems, amongst which are the following. (i) Providers of popular online BLAST servers need to set restrictions on the query size or CPU time per job to prevent resource hogging: many vertebrate genes are too large to be submitted in a single query. (ii) Eukaryotic genomes are full of dispersed repetitive sequences such as the Alu and LINE-1 retroelements in the human genome (11–13): many of these are transcribed by RNA polymerase III and/or are often present in RNA polymerase II-transcribed 3′ non-coding exons and can fill up the top scores in the BLAST output, hindering detection of the true spliced exons. (iii) ESTs from closely related genes (e.g. recently duplicated paralogous genes) may be present in the output and could outnumber the true ESTs. (iv) Analysing the results from BLAST output alone is time consuming and inefficient, while other commonly used tools may be inappropriate: for example, the Clustal W multiple sequence alignment program was not designed for aligning (and cannot align) exons from multiple ESTs onto genomic sequence (14). Although this has often been attempted, a multiple alignment calculation should be redundant, since all the information needed to align the ESTs on the genome sequence is intrinsic to the BLAST output.
The specific requirements for searching EST databases impose the need to provide dedicated servers. The BLAST2-based EbEST server (http://rgd.mcw.edu/EBEST/) allows large queries, clusters the matching ESTs and provides graphical output summarising the matched exons (15). EbEST can integrate results from other gene prediction methods, providing a useful pointer to potential gene structure. However, the EbEST server does not provide the user with the raw findings that might be needed to judge the quality of the interpretation or reveal alternative processing. The GenSeqer dynamic programming software is available as a server for plant ESTs only (http://gremlin1.zool.iastate.edu/cgi-bin/gs.cgi) or may be downloaded for local use (16). Query size of the server is limited, probably reflecting the computational load imposed by the slow (but sensitive) dynamic programming algorithm, which incorporates a hidden Markov model for splice site detection: for databases of limited size, GeneSeqer is likely to outperform BLAST2 servers in the detection of divergent sequences as well as very short exons.
Although BLAST2 is probably not quite as sensitive as dynamic programming algorithms, it is much faster. For the rapid detection of exons in ESTs that are expressed from a given spliced gene, it would be difficult to improve substantially on the BLAST2 algorithm (17). It can detect multiple exons and tolerate sequencing error within an exon. The one obvious drawback is that it cannot detect very short matches and, therefore, the occasional very small exon (below ∼20 bases long) will be overlooked.
Based on feedback from experimentalists wishing to access ESTs, we felt that there was a need for an online resource for querying EST databases that could not only provide researchers with a graphical output summarising exon/intron structure, including any splice variants, but would also provide the raw BLAST results so that artefactual matches might be understood and eliminated. The main computational tools needed to build this server already exist; for example, RepeatMasker has become the standard tool for masking dispersed repeats in genomic sequence (18), while ‘gapped’ BLAST (17) is well suited to homology searching with ESTs. Therefore, to meet this need, we focussed on (i) the integration of these tools to give optimal search results, and (ii) parsing the BLAST output and designing appropriate presentation for the EST matches, which has been achieved by providing the user with new alignment and graphical display outputs, the latter using the Artemis sequence display program (19). This manuscript outlines the development and application of the Gene2EST server which, it is hoped, will prove to be of some use to the research community.
MATERIALS AND METHODS
Computational details
The Gene2EST server is freely accessible at http://woody.embl-heidelberg.de/gene2est/ with links from the EMBL home page http://www.embl-heidelberg.de/. The server and output parser scripts are written in the PYTHON programming language (20). The Web interface uses the cgimodel framework (21). Script modules will be made available through the BIOPYTHON project (http://www.biopython.org/).
RepeatMasker and Repbase are used to exclude dispersed repeats found in genomic sequence queries (11,18). RepeatMasker can be obtained from http://ftp.genome.washington.edu/RM/RepeatMasker.html and Repbase from http://www.girinst.org/Repbase_Update-Information.html. BLAST2.011 (17), used for the sequence similarity search, allows short gaps in HSPs (BLAST high scoring pieces), which is ideal for the high frequency of errors in ESTs. BLAST2 is available from the NCBI at http://www.ncbi.nlm.nih.gov/BLAST/index.html. For Gene2EST graphical display, Artemis must be installed on the user’s local computer. It can be downloaded from the Sanger Centre (http://www.sanger.ac.uk/). Version 4 of Artemis is required (as version 3 omitted the lines connecting joined feature elements).
Gene2EST currently runs on a LINUX server with dual 800 MHz INTEL Pentium III processors. RepeatMasker/BLAST2 jobs are queued to a single processor, leaving the second processor available for efficient output parsing and other interactive usage.
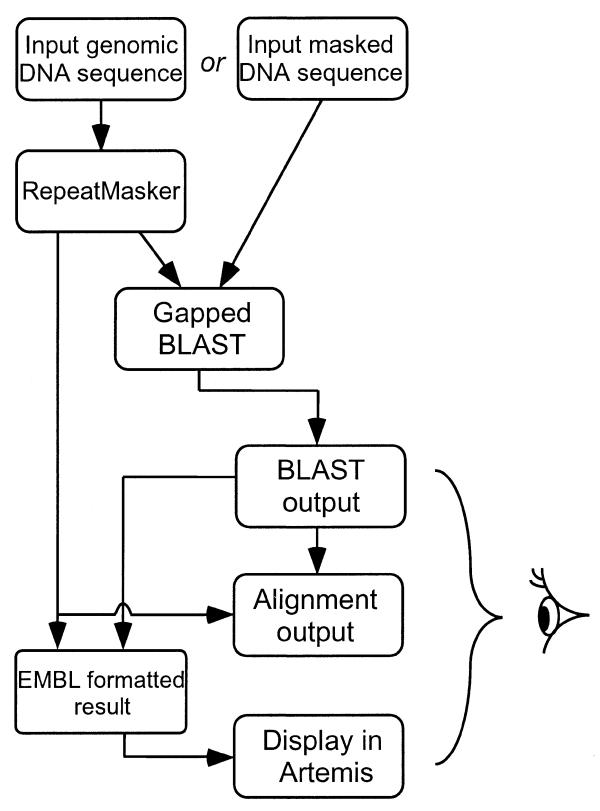
Figure 1 shows the steps used by Gene2EST, beginning with receiving the query input through to output generation. After first masking repeats in the query and then running BLAST, the user is sent an HTML-formatted BLAST output page. The user can then ask for the more derived outputs using buttons and parameter settings in the page header. These invoke the parser to process the BLAST output, providing alignment or EMBL format outputs. Online help is available for the Gene2EST server and users are recommended to consult this.
Figure 1.
Flowthrough of Gene2EST server. The user inputs a genomic sequence, which is screened for repeats before being submitted to a BLAST search. To save time, a previously masked sequence can be submitted, and RepeatMasker switched off. From the returned BLAST output the user can collect the parsed alignment and EMBL format output, the latter for display in Artemis. All three outputs should be examined by the user to properly establish the revealed expression products for the genomic sequence.
The Gene2EST BLAST parser is slightly adapted from the publicly available BLAST parser provided by Jeff Chang in the framework of the BIOPYTHON project (www.biopython.org). The parser is used to scan for HSPs with numbering, the name of the EST, the percent identity and the E-value. This information, together with RepeatMasker output, is processed by newly written PYTHON modules to provide either sequence alignment output for direct examination or EMBL formatted feature output suitable for graphical overview.
RESULTS AND DISCUSSION
Interpreting the results of EST database searches with eukaryotic genes is not always straightforward. Robust fully automated interpretation would be difficult to achieve given the arbitrary end points of clones, occasional very short exons and a high frequency of misleading matches such as intronic sequence primed from encoded poly(A) runs (22,23) and expressed dispersed repeats. Furthermore, although the expressed strand is usually thought to be known for an EST, there is no reliable way of determining whether this is true and most genes have ESTs matching in both strand orientations (22). Indeed, it is important to note that the 5′ and 3′ reads of an EST clone are conventionally stored in opposite orientations. Therefore, with Gene2EST, we have aimed to provide users of EST databases with output formats that will help in examining the matching ESTs. Gene2EST does not produce gene predictions, it provides EST matches mapped to gene sequences. Users should not assume that all EST matches found by Gene2EST belong to fully processed mRNAs or other mature and functional expression products. Instead, they should work through the three Gene2EST outputs (discussed below) and interpret which are the ‘true’ hits and which are not useful.
Artefacts in EST databases
The high throughput approach to generating ESTs inevitably limits the quality control that can be applied. ESTs are made from RNA purified over oligo(T) columns to select poly(A)-tailed mRNA but, as nuclear RNA cannot usually be eliminated, poly(A) stretches in unprocessed nuclear transcripts will also co-purify. Thus, EST clones generated from expressed pseudogenes (24) and intronic poly(A) runs may complicate interpretation (22,23). Other reported problems include contamination by vector and rRNA sequences (25,26). Results of EST searches may be checked against the following list of problems and artefacts in the EST databases:
• Highly biased gene representation.
Many gene products underrepresented or missing, especially if low expression.
• High sequence error including nucleotide deletions and insertions.
Problems for protein sequence queries due to translation frameshifts.
• Dispersed repeat sequences (Alus, etc.) abound in the EST databases.
• EST cDNA clones are mostly oligo(T) primed from the 3′ poly(A) tail.
5′ and internal exons are underrepresented.
For mRNAs with long 3′ non-coding regions, many ESTs may lack protein-coding sequence. [Note: some recent EST projects are choosing to internally prime EST clones, which will achieve better coverage of coding exons (27).]
• Expressed strand orientation is not reliably known.
• ESTs are not very useful for a given species unless the EST collection is large.
Only 80 000 Drosophila ESTs are of limited value (as of January 2001).
• ‘False’ ESTs may result from:
bacterial and yeast contaminants;
internal oligo(T) priming on intronic A-rich sequences in nuclear hnRNA (or possibly genomic DNA) is common;
‘mosaic ESTs’ arising from multiple ligations during cDNA library construction.
The Gene2EST BLAST output and the output parser buttons
When the BLAST2 run completes, the user is presented with a typical BLAST Web output with HTML links for retrieving the original EST database entries with SRS (28,29), e.g. important for pairing up 5′ and 3′ reads of the same clone and checking the tissue of origin. This output provides the user with the usual BLAST findings: E-value significance of the match, number and length of HSPs per EST, etc. For spliced mRNAs, HSPs should coincide with exons, though these may be incomplete at the ends of the ESTs if the ends do not correspond to the 5′ or 3′ ends of the mRNA.
In addition, the output header has several control fields to allow the user to retrieve the parsed Gene2EST alignment and EMBL outputs. Two cut-off values are used to control which ESTs and HSPs are collected by the parser. By default, only results above an E-value of 1.0e–4 and 93% identity are collected. The default settings provide protection against collecting sequences that do not come from the gene itself, including random matches, unmasked repeats, or paralogous gene sequences, while allowing for some sequencing error (or single nucleotide polymorphisms). For a given gene, these values may need to be modified, e.g. to collect very short exons.
The Gene2EST alignment output
The parser collects all instances of EST matches that exceed the specified cut-offs and aligns them to the gene sequence. The alignment output is used for examining the findings in detail at any given point in the gene. It can be saved as a text file that will be very large for large queries matching many ESTs. Not all HSPs that define exons stop exactly at the splice boundaries, since GT or AG sequences may be found in the adjacent exons. Where possible, internal exons are trimmed by the parser according to the GT/AG motifs: this cannot be done for the outside HSP match of an EST (or for single HSP matches).
The given sequence of a matching EST entry is aligned to the gene, allowing the user to examine sequence differences such as errors, including insertions/deletions, and perhaps also polymorphisms. Dispersed repeats recorded by RepeatMasker are shown above the genomic sequence. Figure 2 shows the detail from a typical Gene2EST alignment output spanning two exons and an intron which contains an Alu insertion.
Figure 2.
Alignment output from Gene2EST with features subsequently colour-coded for clarity. The query is a fragment of the human aldose reductase gene (EMBL accession no. AF032455) spanning exons 4 and 5. To save space, only a few of the HSPs from matching ESTs are shown. The intron contains a class Y Alu insertion (blue). Splice donor (red) and acceptor (green) signals abut the exon borders. Where possible, the parser has trimmed HSP overlaps to the GT/AG splice boundaries: pink bases show HSP ends that could not be trimmed by the parser, as there is no adjacent exon within the given EST.
Note that the BLAST2 algorithm will overlook any very short exons, although this is apparent to the parser from a numbering inconsistency. Missing exons are indicated in the alignment output by appending the requisite number of Xs to the preceding exon.
The EMBL format output for graphical display
The parser collects all instances of EST matches that exceed the specified cut-offs and assigns them as Feature (FT) records in an EMBL format sequence file (30). Multiple HSPs in one EST are linked by join statements. The file may be saved and read by any program that accepts EMBL format, but is particularly aimed at loading into the Artemis program (19) in order to get a graphical overview of the Gene2EST findings. Artemis is a JAVA-coded portable graphical display and annotation tool for genomic sequence. In favourable cases, loading Gene2EST output into Artemis can immediately reveal alternatively spliced gene products. A useful property of the Artemis display is that, regardless of the number of ESTs matching a given exon, they are effectively superposed as a single graphical object. Figure 3 shows an example of a Gene2EST output for an alternatively processed gene displayed in Artemis. Examination of the display is sufficient to reveal that the human fibrinogen γ-chain gene uses at least two transcription initiation sites, alternatively spliced internal exons, and two different 3′ terminal exons and poly(A) sites. ESTs may reveal whether the alternatives are used at the same frequency. For example, 12 ESTs were aligned at the minor poly(A) site at position 9831 while 119 ESTs aligned to the major poly(A) site at position 10 272, indicating an ∼1:10 difference in usage.
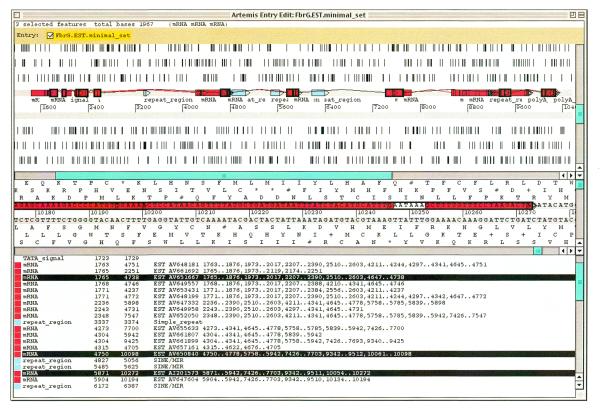
Figure 3.
Artemis display of Gene2EST output for the human fibrinogen γ-chain gene (EMBL accession no. M10014, containing 10 564 bases). ESTs span the complete gene, revealing alternative promoter, splice site and polyadenylation usage. The upper Artemis window has a graphical display of gene structure: expressed RNA exons are coloured red and connected by join lines, repeats are blue. Alternative processing is a likely explanation wherever two or more join lines overlap. By default, ESTs are all specified on the top DNA strand, regardless of actual orientation. The six translation frames show only stop codons (vertical lines) by default. The middle window displays sequence at the major polyadenylation site and the lower window lists the annotated features. The three selected EST features collectively span one of the mRNA variants, from the main promoter (TATA at base 1723) to the main poly(A) site (signal at base 10 247), and are edged in black in the upper window. For the figure, a minimal set of ESTs that cover the revealed set of alternative transcripts were selected from >500 matching human ESTs in EMBL release 64. TATA and poly(A) signals were added by hand.
The feature annotation tools in Artemis can be used to add value to the basic results. For example, nucleic acid signals such as TATAAAA and CCAAT boxes or AATAAA poly(A) signals can be labelled. Full-length, spliced mRNAs can be assembled from EST fragments and used to derive CDS (coding sequence) translations. Data from other sources such as experimental results can be used to provide additional annotation. Thus, as a project develops, Artemis can be used to keep track of new findings, continuing to provide graphical overview of features encoded in a DNA sequence.
Strategy for using Gene2EST
Gene2EST may be useful in a number of ways. Traditionally, a new gene product has first been reported from a single cDNA sequence, which may give a misleading picture of the spectrum of expression products. When the gene sequence corresponding to a given cDNA becomes available, Gene2EST can be used to rapidly screen for alternatively spliced products, as in Figure 3.
It is anticipated that Gene2EST will also be useful in the investigation of crudely characterised genes from genomic sequencing projects. For example, the ENSEMBL server may reveal a new human gene by homology to a previously known paralogous gene, although the new gene prediction is likely to be incomplete. By submitting the gene sequence to Gene2EST, it should be possible to establish a more complete description of the gene, provided that sufficient ESTs exist in the databases. In this way Gene2EST should be useful for experimentalists wishing to follow up newly revealed genes in the human and mouse genome projects.
Acknowledgments
ACKNOWLEDGEMENTS
We thank our beta testers, especially Rein Aasland and Bertran Séraphin for useful input. Gene2EST could not have been provided without the prior development and availability of the component software tools. This work was supported in part by HFSP grant RG-231/98.
References
- 1.Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1656. [DOI] [PubMed] [Google Scholar]
- 2.Gibson T.J. and Spring,J. (1998) Genetic redundancy in vertebrates: polyploidy and persistence of genes encoding multidomain proteins. Trends Genet., 14, 46–49. [DOI] [PubMed] [Google Scholar]
- 3.Shastry B.S. (1995) Genetic knockouts in mice: an update. Experientia, 51, 1028–1039. [DOI] [PubMed] [Google Scholar]
- 4.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 5.Smith C.W. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 6.Claverie J.M. (1997) Computational methods for the identification of genes in vertebrate genomic sequences. Hum. Mol. Genet., 6, 1735–1744. [DOI] [PubMed] [Google Scholar]
- 7.Claverie J.M. (1999) Computational methods for the identification of differential and coordinated gene expression. Hum. Mol. Genet., 8, 1821–1832. [DOI] [PubMed] [Google Scholar]
- 8.C.elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 9.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 10.Reese M.G., Hartzell,G., Harris,N.L., Ohler,U., Abril,J.F. and Lewis,S.E. (2000) Genome annotation assessment in Drosophila melanogaster. Genome Res., 10, 483–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurka J. (1998) Repeats in genomic DNA: mining and meaning. Curr. Opin. Struct. Biol., 8, 333–337. [DOI] [PubMed] [Google Scholar]
- 12.Schmid C.W. (1998) Does SINE evolution preclude Alu function? Nucleic Acids Res., 26, 4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smit A.F. (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev., 9, 657–663. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J. and Jacob,H.J. (1998) EbEST: an automated tool using expressed sequence tags to delineate gene structure. Genome Res., 8, 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Usuka J., Zhu,W. and Brendel,V. (2000) Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics, 16, 203–211. [DOI] [PubMed] [Google Scholar]
- 17.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurka J. (2000) Repbase update: a database and an electronic journal of repetitive elements. Trends Genet., 16, 418–420. [DOI] [PubMed] [Google Scholar]
- 19.Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.-A. and Barrell,B. (2000) Artemis: sequence visualisation and annotation. Bioinformatics, 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 20.Watters A., van Rossum,G. and Ahlstrom,J. (1996) Internet Programming with Python. MIS Press/Henry Holt publishers, New York.
- 21.Chenna R. and Gemünd,C. (2000) cgimodel: CGI Programming Made Easy with Python. Linux J., 75, 142–149. [Google Scholar]
- 22.Aaronson J.S., Eckman,B., Blevins,R.A., Borkowski,J.A., Myerson,J., Imran,S. and Elliston,K.O. (1996) Toward the development of a gene index to the human genome: an assessment of the nature of high-throughput EST sequence data. Genome Res., 6, 829–845. [DOI] [PubMed] [Google Scholar]
- 23.Liang F., Holt,I., Pertea,G., Karamycheva,S., Salzberg,S.L. and Quackenbush,J. (2000) Gene index analysis of the human genome estimates approximately 120,000 genes. Nature Genet., 25, 239–240. [DOI] [PubMed] [Google Scholar]
- 24.Mighell A.J., Smith,N.R., Robinson,P.A. and Markham,A.F. (2000) Vertebrate pseudogenes. FEBS Lett., 468, 109–114. [DOI] [PubMed] [Google Scholar]
- 25.Miller C., Gurd,J. and Brass,A. (1999) A RAPID algorithm for sequence database comparisons: application to the identification of vector contamination in the EMBL databases. Bioinformatics, 15, 111–121. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez I.L. and Sylvester,J.E. (1997) Incognito rRNA and rDNA in databases and libraries. Genome Res., 7, 65–70. [DOI] [PubMed] [Google Scholar]
- 27.Dias Neto E., Garcia Correa,R., Verjovski-Almeida,S., Briones,M.R., Nagai,M.A., da Silva,W.,Jr, Zago,M.A., Bordin,S., Costa,F.F., Goldman,G.H. et al. (2000) Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proc. Natl Acad. Sci. USA, 97, 3491–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Etzold T., Ulyanov,A. and Argos,P. (1996) SRS: information retrieval system for molecular biology data banks. Methods Enzymol., 266, 114–128. [DOI] [PubMed] [Google Scholar]
- 29.Etzold T. and Argos,P. (1993) SRS—an indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci., 9, 49–57. [DOI] [PubMed] [Google Scholar]
- 30.Baker W., van den Broek,A., Camon,E., Hingamp,P., Sterk,P., Stoesser,G. and Tuli,M.A. (2000) The EMBL nucleotide sequence database. Nucleic Acids Res., 28, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]