Abstract
Methods for the global analysis of protein expression offer an approach to study the molecular basis of disease. Studies of protein expression in tissue, such as brain, are complicated by the need for efficient and unbiased digestion of proteins that permit identification of peptides by shotgun proteomic methods. In particular, identification and characterization of less abundant membrane proteins has been of great interest for studies of brain physiology, but often proteins of interest are of low abundance or exist in multiple isoforms. Parsing protein isoforms as a function of disease will be essential. In this study, we develop a digestion scheme using detergents compatible with mass spectrometry that improves membrane protein identification from brain tissue. We show the modified procedure yields close to 5,000 protein identifications from 1.8 mg of rat brain homogenate with an average of 25% protein sequence coverage. This procedure achieves a remarkable reduction in the amount of starting material required to observe a broad spectrum of membrane proteins. Among the proteins identified from a mammalian brain homogenate, 1897 (35%) proteins are annotated by GeneOntology as membrane proteins, and 1225 (22.6%) proteins are predicted to contain at least one transmembrane domain. Membrane proteins identified included neurotransmitter receptors and ion channels implicated in important physiological functions and disease.
Keywords: brain proteome, mass spectrometry, membrane proteins, proteolysis, shotgun proteomics
INTRODUCTION
The brain is a complex system of thousands of proteins that form a complicated molecular network. Large-scale proteomic analysis can help unravel the complexities of brain function as many of the activities of the brain involve intricate signaling networks and changes in post-translational modifications. Although, bottom up shotgun sequencing is useful for proteome analysis, the complexity of the mammalian brain presents technical and biological challenges. The brain consists of many distinct functional regions, each of which consists of different cell types. The structural morphology between these regions exacerbate the complexity of the biology. For example, the cell body and axon terminal of the same neuron are often located in two different functional regions. In addition, the brain has the highest lipid content of any other tissue (excluding adipose) making it particularly challenging for mass spectrometry-based proteomic analysis 1. Therefore, new approaches to prepare brain tissue are needed to improve the potential of mass spectrometry-based shotgun proteomic technology for the study of disease.
Gel-based proteomic methods, such as two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), have been used to characterize the brain proteome 2–4. Membrane proteins are typically underrepresented in 2D PAGE analyses due to poor solubility in the starting buffer 5. A proteomic analysis of whole mouse brain using 2D-PAGE required large amounts of starting material (1–15 g) and yielded less than 500 protein identifications 6. To increase coverage of the proteome and, in particular, the coverage of membrane proteins, shotgun strategies using multidimensional liquid chromatography separation coupled with tandem mass spectrometry (LC/LC-MS/MS) have been applied to the brain 7. By using high pH and proteinase K digestion of protein mixtures in combination with LC/LC-MS/MS, Wu et al. reported the identification of 1,610 proteins from 500 µg of rat brain homogenate 8. Nielsen et al. reported a novel method to enrich membrane proteins from 150 mg of mouse cortex for analysis by LC-MS/MS 9. 862 proteins were identified from sixteen cortex fractions. They also applied the same technique to analyze proteins extracted from 15 mg of mouse hippocampus and identified 1685 proteins. Recently, Wang et al. reported a method of analyzing both global and cysteinyl-enriched peptides from a whole brain sample 10. A total of 48,328 peptides were identified covering 7792 non-redundant proteins from 200–300 mg of brain homogenate. Among the 7792 proteins, 5636 proteins were identified at a ~1.5% false positive rate estimated by reverse database searching using a minimum of two or more peptides for identification, and 1332 proteins were annotated as membrane proteins. The identification of total and membrane proteins from brain tissue has increased dramatically using bottom up approaches, but a large amount of starting material is usually required for identification of hydrophobic membrane proteins. Although it might be feasible to obtain a large amount of starting material by pooling brain tissue from different animals, it poses a challenge to the number of biological replicates that can be performed for each study and studies of normal versus diseased tissues. A reduction of starting material without improving sample processing and/or analytical procedures usually results in a decrease of protein sequence coverage and thus confidence in identified proteins. Hence, an important issue to reducing the amount of starting material without decreasing protein identifications is to improve the processing of protein mixtures.
Previously, we demonstrated the utility of mass spectrometry (MS) compatible detergents for shotgun analysis of complex protein mixtures 11. We showed that a modified trypsin digestion protocol incorporating detergents compatible with mass spectrometry improves proteolytic processing of protein mixtures and results in a dramatic reduction in the amount of material required for proteome analysis. Three commercially available mass spectrometry compatible detergents were compared: RapiGest SF (RPG), Invitrosol (IVS), and PPS Silent Surfactant (PPS). RapiGest SF and PPS Silent Surfactant are acid-labile surfactants. Invitrosol (IVS) is a homogeneous surfactant that elutes in three peaks well separated from the elution times of most peptides. All three surfactants can act as mild denaturants to solubilize and unfold hydrophobic proteins at concentrations that do not inhibit common proteolytic enzymes such as trypsin. In addition to solubilizing and denaturing hydrophobic proteins in solution, we also observed a complimentarity to the proteins identified using the different conditions suggesting a strategy to increase coverage of a proteome. In this study, we use this approach to improve global profiling of brain tissue, and more importantly the detection of membrane proteins. As membrane proteins play an important role in brain function, the ability to observe this class of proteins is critical for the study of diseases in brain.
MATERIALS AND METHODS
Materials
Invitrosol™ was purchased from Invitrogen (Carlsbad, CA). RapiGest™ SF acid labile surfactant was purchased from Waters Corp. (Milford, MA). PPS™ Slient surfactant was provided by Dr. Norris from Protein Discovery Inc. (Knoxville, TN). Trypsin (modified, sequencing grade) was obtained from Roche. Other laboratory reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless noted otherwise.
Sample Preparation
A Sprague-Dawley rat at postnatal day 45 was sacrificed and the brain was quickly removed, and then frozen with liquid nitrogen. One entire rat brain was homogenized in a Teflon dounce grinder in 1–2 ml of 0.32M sucrose, 4mM HEPES, and protease inhibitors (Roche Applied Science). The protein concentration was determined using a BCA protein assay (Pierce).
Digestion protocols
300 µg of cell lysates were precipitated with methanol/chloroform and resuspended in different conditions for trypsin digestion. Each digestion condition (300µg) was run in triplicates (100µg).
Trypsin digestion with the MS detergents
40 µl of invitrosol (5X stock), 20 µl of RapiGest SF (1% stock), and 20 µl of PPS (1% stock) were used to resuspend the protein pellets. The concentration of each detergent used in this study was determined based on the maximum recommended concentration suggested by the manufactures. Then, proteins were incubated at 60°C for 5 minutes and the remaining solvent was added to the samples (200µl final volume). All samples were sonicated for 2 hours in a water bath and digested with trypsin for 16 hours at 37°C. For trypsin digestion in aqueous Tris buffer, samples were reduced in 5mM TCEP for 20 minutes at room temperature, and cysteines were alkylated in the dark in 25mM iodoacetamide (IAM) for 30 minutes at room temperature prior to the addition of trypsin. Trypsin was added to each sample at a ratio of 1:30 enzyme/protein along with 2 mM CaCl2.
Post-Digestion
Following digestion, all reactions were acidified with 90% formic acid (2% final) to stop proteolysis. Samples with RapiGest SF and PPS were acidified and incubated at 37°C for additional 4 hours to facilitate the hydrolysis of detergents. Then, samples were centrifuged for 30 minutes at 14,000 rpm to remove insoluble material. The soluble peptide mixtures were collected for LC-MS/MS analysis.
Multidimensional chromatography and tandem mass spectrometry
Peptide mixtures were pressure-loaded onto a 250 µm inner diameter (i.d.) fused-silica capillary packed first with 2.5 cm of 5 µm strong cation exchange material (Partisphere SCX, Whatman), followed by 5 cm of 10 µm C12 reverse phase (RP) particles (Jupitor, Phenomenex or Polaris 2000, Metachem Technologies). Loaded and washed microcapillaries were connected via a 2 µm filtered union (UpChurch Scientific) to a 100 µm i.d. column, which had been pulled to a 5 µm i.d. tip using a P-2000 CO2 laser puller (Sutter Instruments), then packed with 13 cm of 4 µm C12 reverse phase (RP) particles (Jupitor, Phenomenex or Polaris 2000, Metachem Technologies) and equilibrated in 5% acetonitrile, 0.1 % formic acid (Buffer A). This split-column was then installed in-line with a Quaternary Agilent 1100 series HPLC pump. Overflow tubing was used to decrease the flow rate from 0.1 ml/min to about 200–300 nl/min. Fully automated 13-step chromatography runs were carried out. 110-minute gradient was used for each chromatographic step. Three different elution buffers were used: 5% acetonitrile, 0.1 % formic acid (Buffer A); 80% acetonitrile, 0.1% formic acid (Buffer B); and 0.5 M ammonium acetate, 5% acetonitrile, 0.1% formic acid (Buffer C). In such sequences of chromatographic events, peptides are sequentially eluted from the SCX resin to the RP resin by increasing salt steps (increase in Buffer C concentration), followed by organic gradients (increase in Buffer B concentration). The last chromatography step consists in a high salt wash with 100% Buffer C followed by acetonitrile gradient. The application of a 2.5 kV distal voltage electrosprayed the eluting peptides directly into an LTQ ion trap mass spectrometer equipped with a nano-LC electrospray ionization source (Thermo Finnigan). Full MS spectra were recorded on the peptides over a 400 to 2000 m/z range, followed by five tandem mass (MS/MS) events sequentially generated in a data-dependent manner on the first, second, third, and fourth most intense ions selected from the full MS spectrum (at 35% collision energy). Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (Thermo Finnigan, San Jose, CA).
Database search and interpretation of MS/MS datasets
Tandem mass spectra were extracted from raw files, and a binary classifier - previously trained on a manually validated data set - was used to remove the low quality MS/MS spectra. The remaining spectra were searched against a Rat protein database containing 38,841 protein sequences downloaded as FASTA-formatted sequences from EBI-IPI (database version 3.17, released on May, 18, 2006) 12, and 124 common contaminant proteins, for a total of 38,965 target database sequences. To calculate confidence levels and false positive rates, we used a decoy database containing the reverse sequences of 38,965 proteins appended to the target database 13, and the SEQUEST algorithm 14, 15 to find the best matching sequences from the combined database.
SEQUEST searches were done on an Intel Xeon 80-processor cluster running under the Linux operating system. The peptide mass search tolerance was set to 3 Da. No differential modifications were considered. For the aqueous digestion, the mass of the amino acid Cysteine was statically modified by +57 Da, due to carboxyamidomethylation of the sample. No enzymatic cleavage conditions were imposed on the database search, so the search space included all candidate peptides whose theoretical mass fell within the 3 Da mass tolerance window, despite their tryptic status.
The validity of peptide/spectrum matches was assessed in DTASelect2 16 using SEQUEST-defined parameters, the cross-correlation score (XCorr) and normalized difference in cross-correlation scores (DeltaCN). The search results were grouped by charge state (+1, +2, and +3) and tryptic status (fully tryptic, half-tryptic, and non-tryptic), resulting in 9 distinct sub-groups. In each one of the sub-groups, the distribution of XCorr and DeltaCN values for (a) direct and (b) decoy database hits was obtained, and the two subsets were separated by quadratic discriminant analysis. Outlier points in the two distributions (for example, matches with very low Xcorr but very high DeltaCN) were discarded. Full separation of the direct and decoy subsets is not generally possible; therefore, the discriminant score was set such that a false positive rate of 1% was determined based on the number of accepted decoy database peptides. This procedure was independently performed on each data subset, resulting in a false positive rate independent of tryptic status or charge state.
In addition, a minimum sequence length of 7 amino acid residues was required, and each protein on the list was supported by at least two peptide identifications. These additional requirements – especially the latter - resulted in the elimination of most decoy database and false positive hits, as these tended to be overwhelmingly present as proteins identified by single peptide matches. After this last filtering step, the false identification rate was reduced to below 1%. Finally, the Kyte-Doolittle hydropathy score (KD) of all peptides passed the filters were also calculated and reported in the DTASelect2 output.
The identified proteins were categorized based on their cellular localizations and molecular functions accordingly to Gene Ontology (GO) information obtained from the European Bioinformatics Institute at http://www.ebi.ac.uk/pub/databases/GO/goa/rat. The TMHMM (www.cbs.dtu.dk/services/TMHMM/) algorithm was used to predict transmembrane domains (TMDs) from the identified proteins.
Gene Ontology Data Analysis
We assigned cellular components and molecular functions based on the Gene Ontology (GO) consortium using GoMiner. For each GO category, Fisher’s exact test was used to calculate the p-value that reflects the significance over-representation of proteins identified in one GO category compared to the proteins identified from the entire protein database in the same GO category. The categories with p-values less than 0.05 were considered significantly over representated or under representated.
RESULTS
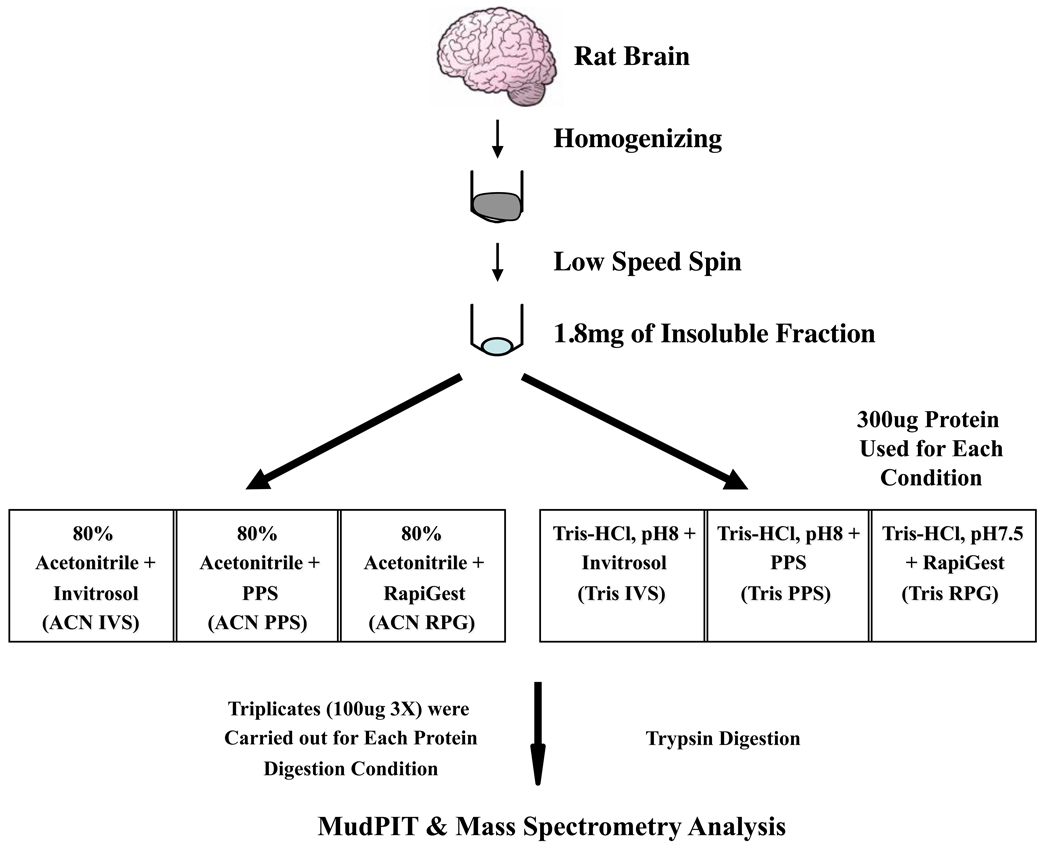
The initial step in shotgun proteomic experiments is proteolytic digestion of a complex protein mixture. The challenge is to disrupt existing protein-protein interactions, and denature and solubilize the proteins. Samples of tissue represent a bigger challenge because in addition to a diverse mixture of cell types there are extracellular components present. We showed previously that RPG, IVS, and PPS when used with organic or aqueous solvents can greatly increase protein coverage in complex cellular protein mixtures 11. In this study, we show this procedure can increase the identification of membrane proteins from brain tissue. Rat brain tissue was homogenized and the insoluble material was extracted. The material was divided into 6 aliquots of 300 µg each and dissolved in RPG (0.1%), IVS (1X) and PPS (0.2%) mixed with either mixed organic-aqueous (80% acetonitrile) or aqueous buffers (Tris-HCl, pH 8.0) for digestion. A total of 6 different digestion conditions were used for comparison, and each digestion condition was analyzed in triplicate (100 µg per replicate) with an identical experimental setup. A schematic illustration of the sample preparation is shown in Figure 1. The concentration of MS compatible detergents used in this study differs from our previous report 11. To increase the solubility of hydrophobic and membrane proteins in the rat brain homogenate in solution, we used the maximum concentration of each detergent recommended by the manufacture to solublize the protein pellet after the methanol/chloroform precipitation: 5X invitrosol, 1% RapiGest, and 1% PPS. This is a crucial step to ensure the inclusion of membrane proteins known to be important for the normal brain function. After the initial solubilization step, protein samples were diluted further with trypsin digestion buffer to achieve the optimal concentration of the MS compatible detergents for trypsin digestion. Detail protocols for sample preparation are described in the method section.
Figure 1.
A schematic illustration of the experimental setup and conditions. We extracted proteins from mouse brain tissues using a combination of mixed organic and aqueous solvents with three MS-compatible detergents prior to the trypsin digestion. Proteins from each digestion condition were then analyzed by the MudPIT-based LC-MS/MS method.
A total of 18 MudPIT analyses were performed. Tandem mass spectra collected in each experiment were searched using SEQUEST against the rat IPI protein database plus a decoy database (see Methods section). A 1% peptide false discovery rate and a minimum of two-peptide matches to a protein were required for protein identification. Following database search and data filtering, a total of 60,712 peptides that corresponded to 4,797 proteins were identified from 1.8 mg of rat brain homogenate using significantly less starting material compared to similar studies reported previously (Supplementary table 1). By using our method, the number of proteins identified was much improved by combining the different specificities of the detergents used to solubilize the protein mixtures than the technical replicates with the same detergent. The optimized digestion protocol employing different types of detergents led to increased or equivalent protein identifications from less material than reported in the literature.
Increases in Overall Sequence Coverage and Representation of Proteins in the Brain Proteome
The complementary nature of this sample processing strategy increases the protein sequence coverage and thus the confidence of protein identification. In complex protein mixtures such as tissue homogenates, sequence coverage levels tend to be less overall (e.g. <10%) and even more so for less abundant proteins often having only one or two peptide matches 8, 10. In our study, a majority (71%) of proteins identified have more than 10% sequence coverage (Figure 2). We found it’s an effective strategy to increase protein sequence coverage by combining trypsin digestion with a combination of solvents and detergents. Over 57% of the proteins show at least a 10% increase in sequence coverage by using the three combinations of mixed solvents and detergents compared with triplicates of the same trypsin digestion conditions. Hence, we demonstrate that the modified digestion protocol can be used to improve protein identification in a proteome as complex as the brain compared to methods without detergent (three times less protein identifications, data not shown) and other shotgun-based brain proteome studies 8–10.
Figure 2.
Distribution of total sequence coverage in identified proteins. Figure 2 shows that over 60% of proteins identified have more than 10% protein sequence coverage.
Improved Identification of Brain Membrane Proteins
Many transmembrane and membrane-associated proteins in the brain form neurotransmitter receptors, ion channels, and G-proteins-couple receptors that are of interest because of their roles in the normal physiology of the brain and the neurological diseases 17. The hydrophobic nature of membrane proteins makes them difficult to study because of difficulty solubilizing and digesting the proteins. Gel-based proteomic studies using enriched brain membrane fractions identified mostly cytosolic proteins in the brain 2, 18. In other studies using a bottom-up approach, a range from 454 to 1447 (18.6%) transmembrane proteins have been identified from brain tissues 8–10.
In our previous study, the modified digestion protocol with MS compatible detergents yielded an increase in total number of proteins identified by improving the efficiency of proteolysis. In this study, we modified the digestion protocol to increase the solubility of membrane proteins. First, our results show good coverage of regions rich in transmembrane proteins. We observe roughly a 2-fold increase in proteins identified with molecular weight greater than 100kDa in our dataset (22%) as compared to the predicted rat proteome (12%). Next, we classified proteins identified from each modified trypsin digestion condition using the TMHMM algorithm 19 and GeneOntology (GO) 20 to determine the representation of membrane proteins in our data set (Table 1). After merging the data from all six proteolytic conditions, 4,797 proteins were identified from 1.8mg of rat brain homogenate, 1897 (40%) proteins are annotated as membrane proteins, and 1225 (26%) proteins predicted to contain at least one transmembrane domain. The improved digestion strategy increases the identification of transmembrane proteins with 10–100 fold less starting material compared to other shotgun brain proteome studies.
Table 1.
Categorization of proteins identified in 6 different trypsin digestion conditions with MS compatible detergents based on the cellular localization of Gene Ontology (GO).
| ACN IVS | ACN PPS | ACN RPG | Tris IVS | Tris PPS | Tris RPG | ||
|---|---|---|---|---|---|---|---|
| Predicted TM (%) | 423 (16.0) | 466 (16.9) | 450 (16.3) | 476 (17.8) | 431 (15.0) | 556 (16.9) | |
| No Predicted TM | |||||||
| Integral to membrane | 6 | 4 | 4 | 7 | 8 | 10 | |
| Plasma membrane | 30 | 27 | 26 | 29 | 27 | 34 | |
| Mitochondrial membrane | 27 | 30 | 33 | 33 | 32 | 37 | |
| Nuclear membrane | 18 | 14 | 17 | 15 | 6 | 8 | |
| Golgi membrane | 7 | 15 | 9 | 7 | 9 | 9 | |
| ER membrane | 6 | 10 | 5 | 6 | 5 | 9 | |
| Lysome membrane | 1 | 1 | 1 | 1 | 1 | 1 | |
| Membrane | 128 | 140 | 142 | 139 | 151 | 162 | |
| Cytoskeleton | 120 | 120 | 114 | 119 | 116 | 128 | |
| Cytoplasmic | 344 | 319 | 335 | 318 | 362 | 381 | |
| Nucleus | 162 | 168 | 173 | 157 | 286 | 204 | |
| Mitochondrion | 84 | 95 | 85 | 82 | 84 | 96 | |
| ER | 21 | 23 | 23 | 21 | 25 | 26 | |
| Endosome | 4 | 5 | 5 | 5 | 3 | 4 | |
| Lysosome | 4 | 6 | 5 | 4 | 5 | 6 | |
| Microsome | 2 | 2 | 2 | 2 | 2 | 3 | |
| Peroxisome | 2 | 4 | 5 | 4 | 4 | 6 | |
| Extracellular | 29 | 30 | 33 | 20 | 25 | 25 | |
| No assignment | 1227 | 1278 | 1302 | 1229 | 1295 | 1586 | |
| Total Protein (Spectra) Identified | 2645 (23779) | 2757 (22902) | 2769 (21873) | 2674 (22237) | 2877 (19642) | 3291 (25338) |
TM = transmembrane
We also observe that the use of different detergents yields complementary information from membrane proteins leading to higher overall sequence coverage. First, the highest number of brain transmembrane proteins was identified in an aqueous protein digestion with RapiGest (Tris RPG, 556) followed by an aqueous protein digestion with Invitrosol (Tris IVS, 423) (Table 1). Interestingly, the different solvent compositions used with RapiGest generated both the highest number of identified transmembrane proteins (Tris RPG, 556 proteins) and lowest (ACN RPG, 450 proteins). This result is consistent with our previous observation that there is a solvent preference for the detergents 11. Trypsin digestion in Tris buffer with the acid labile detergents RPG and PPS resulted in more identified transmembrane proteins than digestion in buffer with 80% acetonitrile. In this study, aqueous Tris RPG digestion increases transmembrane protein identifications by 24% over RPG digestion in 80% acetonitrile. Furthermore, we found interesting differences in peptide hydrophobicity. Aqueous digestions in combination with the MS-compatible detergents yielded more hydrophobic peptides (lower negative KD value) than organic solvent digestions (Figure 3). In particular, Tris PPS digestion yields peptides with the highest hydrophobicity (−2.86) followed by Tris RPG (−3.76) and Tri INV (−3.78). Therefore, the impact of buffer composition in combination with the detergents provides valuable information to identify the optimal digestion condition of brain membrane proteins.
Figure 3.
A summary of averaged hydrophobicity (KD) from peptides identified in six proteolytic conditions. The Kyte-Doolittle hydropathy score (KD) of peptides identified in each digestion condition was calculated and plotted in histograms. The average KD of peptides from each digestion condition is summarized in a table. The result shows that aqueous digestions in combination with the MS-compatible detergents yield more hydrophobic peptides (lower negative KD value) than organic solvent digestions.
Improving protein sequence coverage is particularly important for identification of brain transmembrane proteins because they are usually less abundant, hard to digest, and have a variety of isoforms. In table 2, we list twelve identified GABA-A receptors/transporters and sixteen glutamate receptors. These proteins are important for normal brain functions and are multi-span transmembrane proteins. The percentage sequence coverage of each protein identified using each set of digestion conditions (triplicate runs) is listed and compared. We notice that the amino acid sequence coverage of these membrane proteins varies in different digestion conditions after adding together peptides from triplicate runs and may reflect protein solubility in combinations of solvents and MS compatible detergents (Table 2). For example, Gamma-aminobutyric-acid receptor gamma-1 subunit and Gamma-aminobutyric-acid type B receptor subunit 1 (spliced isoform 1E) were only identified in the PRG and PPS digestions in the aqueous solution. On the other hand, Gamma-aminobutyric-acid receptor alpha-5 subunit was identified exclusively in the INV and RPG digestions in the 80% acetonitrile (Table 2).
Table 2.
List of identified GABA-A receptors/transporters and glutamate receptors. Percentage sequence coverage (SeqCov) of each protein is listed in each of digestion condition. Total sequence coverage is calculated based on the collective peptides identified in all six conditions for each protein. x indicates no peptide identification for the listed protein.
| Locus | Description | L | MW | pI | ACN INV SeqCov |
ACN PPS SeqCov |
ACN RPG SeqCov |
Tris INV SeqCov |
Tris PPS SeqCov |
Tris RPG SeqCov |
|---|---|---|---|---|---|---|---|---|---|---|
| GABA-A receptors and transporters | ||||||||||
| IPI00192642.1 | Gamma-aminobutyric-acid receptor alpha-1 subunit precursor | 455 | 51754 | 9.3 | 10.3 | 13 | 11.2 | x | 6.6 | 11.2 |
| IPI00197343.1 | Gamma-aminobutyric-acid receptor alpha-3 subunit precursor | 493 | 55433 | 8.8 | 4.9 | x | 11 | 6.3 | x | 8.5 |
| IPI00325359.3 | Gamma-aminobutyric-acid receptor alpha-5 subunit precursor | 464 | 52337 | 9.1 | 6.7 | x | 4.1 | x | x | x |
| IPI00209268.1 | Gamma-aminobutyric-acid receptor beta-1 subunit precursor | 474 | 54072 | 8.8 | 11.2 | 8.6 | 6.1 | 11.2 | 10.1 | x |
| IPI00209269.1 | Gamma-aminobutyric-acid receptor beta-2 subunit precursor | 474 | 54634 | 9.3 | 11.2 | x | x | 13.7 | 10.8 | 16.7 |
| IPI00327083.3 | Gamma-aminobutyric-acid receptor beta-3 subunit precursor | 473 | 54166 | 9.1 | 18.8 | 14.2 | x | x | x | 19.7 |
| IPI00211960.1 | Gamma-aminobutyric-acid receptor gamma-1 subunit precursor | 465 | 53551 | 8 | x | x | x | x | 6.5 | 8.4 |
| IPI00192646.1 | Gamma-aminobutyric-acid receptor gamma-2 subunit precursor | 466 | 54077 | 8.5 | 9.9 | 15.9 | 9 | 6.4 | x | 13.1 |
| IPI00331966.6 | Gamma-aminobutyric acid type B receptor, subunit 2 precursor | 940 | 1E+05 | 8.7 | 3 | 2.9 | 2.7 | 3.8 | x | 3 |
| IPI00208182.1 | Gamma-aminobutyric acid type B receptor, subunit 1 precursor, Splice Isoform 1E | 991 | 1E+05 | 8 | x | x | x | 11.9 | 8.2 | 11 |
| IPI00187596.1 | Sodium- and chloride-dependent GABA transporter 1 | 599 | 67002 | 8 | 13.2 | 13.2 | 12.4 | 13.4 | 3.5 | 8.5 |
| IPI00214462.1 | Sodium- and chloride-dependent GABA transporter 3 | 627 | 69947 | 7 | 12.4 | 7.5 | 11.8 | 18 | 6.1 | 7.5 |
| Glutamate Receptors | ||||||||||
| IPI00326054.1 | Glutamate [NMDA] receptor subunit epsilon 1 precursor | 1464 | 2E+05 | 7 | x | x | 2.3 | 2.7 | 4.9 | 4.6 |
| IPI00326061.1 | Glutamate [NMDA] receptor subunit epsilon 2 precursor | 1482 | 2E+05 | 6.9 | 7.3 | 5.7 | 4.5 | 6.6 | 3.9 | 7.6 |
| IPI00198625.5 | Glutamate [NMDA] receptor subunit zeta 1 precursor, Splice Isoform A | 938 | 1E+05 | 8.8 | 3.7 | 5.5 | 7.7 | 3.4 | 5.4 | 7.1 |
| IPI00207091.1 | Glutamate receptor delta-1 subunit precursor | 1009 | 1E+05 | 6.7 | x | x | x | x | 5.7 | 2.7 |
| IPI00206854.2 | Glutamate receptor delta-2 subunit precursor | 1007 | 1E+05 | 6.1 | x | 3.2 | x | 7.1 | x | 10 |
| IPI00231012.2 | Glutamate receptor 1 precursor, Splice Isoform Flip | 907 | 1E+05 | 7.7 | 8.6 | x | x | 10.9 | 6 | 8.6 |
| IPI00231061.5 | Glutamate receptor 2 precursor, Splice Isoform Flip | 883 | 98745 | 7.6 | 18.7 | 20.5 | x | 32.7 | 23.8 | 26 |
| IPI00231095.1 | Glutamate receptor 3 precursor, Splice Isoform Flip | 888 | 1E+05 | 8.3 | x | 5.6 | 3.8 | 19.3 | x | x |
| IPI00195445.1 | Glutamate receptor 4 precursor, Splice Isoform 1 | 902 | 1E+05 | 7.9 | x | x | x | 9 | x | 4.9 |
| IPI00210260.1 | Metabotropic glutamate receptor 1 precursor, Splice Isoform 1A | 1199 | 1E+05 | 6.9 | 2.7 | 5.6 | x | 3.7 | 2 | 2.7 |
| IPI00212618.1 | Metabotropic glutamate receptor 2 precursor | 872 | 95774 | 7.8 | 2.3 | 5 | 2.2 | 7.3 | 10.7 | 4.8 |
| IPI00212619.2 | Glutamate receptor, metabotropic 3 | 936 | 1E+05 | 8.2 | 5.8 | 6.9 | x | 14 | 15 | 11.6 |
| IPI00212621.1 | Metabotropic glutamate receptor 5 precursor, Splice Isoform 2 | 1203 | 1E+05 | 7.8 | 3.8 | 3.3 | 5.6 | 7.6 | 4.1 | 3.6 |
| IPI00198587.1 | Metabotropic glutamate receptor 7 precursor | 915 | 1E+05 | 7.9 | x | 3 | 3.9 | 3.1 | 5.6 | 5.7 |
| IPI00393591.2 | Metabotropic glutamate receptor 8 precursor | 910 | 1E+05 | 8.2 | x | x | x | x | x | 5.3 |
| IPI00231277.4 | Splice Isoform GluR7A of Glutamate receptor, ionotropic kainate 3 precursor | 910 | 1E+05 | 7.4 | X | X | X | X | X | 4.4 |
| Neurotransmitter Receptors | ||||||||||
| IPI00187680.3 | 5-hydroxytryptamine 2C receptor | 460 | 51917 | 9.3 | x | x | x | 5.2 | x | x |
| IPI00421955.1 | Dopamine-and cAMP-regulated phosphoprotein DARPP-32 | 205 | 22913 | 4.6 | 46.3 | 37.1 | 59 | 44.4 | 38 | 61 |
| Ion Channels | ||||||||||
| IPI00365813.2 | Voltage-dependent anion channel 1 | 234 | 25729 | 8.8 | 27.8 | 22.6 | 22.6 | 22.6 | 22.6 | 22.6 |
| IPI00421874.3 | Voltage-dependent anion-selective channel protein 1 | 282 | 30624 | 8.5 | 75.9 | 76.2 | 70.2 | 82.3 | 79.1 | 77.7 |
| IPI00198327.2 | Voltage-dependent anion-selective channel protein 2 | 295 | 31746 | 7.5 | 44.7 | 40.3 | 36.6 | 68.5 | 68.1 | 69.8 |
| IPI00207891.2 | Voltage-dependent anion-selective channel protein 3 (VDAC-3) (rVDAC3) | 284 | 30912 | 8.7 | 29.2 | 36.6 | 38.7 | 50.4 | 43.7 | 50 |
| IPI00190644.1 | Potassium voltage-gated channel subfamily A member 1 | 495 | 56379 | 5.1 | x | 11.3 | x | 8.3 | x | 6.1 |
| IPI00208365.1 | Potassium voltage-gated channel subfamily A member 2 | 499 | 56701 | 4.9 | x | 11.4 | x | 16.2 | 10 | 11.6 |
| IPI00208362.1 | Potassium voltage-gated channel subfamily A member 4 | 655 | 73390 | 5.1 | x | x | x | x | 4.9 | 5.3 |
| IPI00190053.1 | Potassium voltage-gated channel subfamily A member 6 | 530 | 58884 | 5.1 | x | x | x | 7.7 | x | x |
| IPI00394218.1 | Potassium voltage-gated channel subfamily D member 2 | 630 | 70549 | 8.1 | 10.6 | 9.5 | 12.9 | 6.7 | x | x |
| IPI00389372.1 | Potassium voltage-gated channel subfamily D member 3, Splice Isoform 1 | 655 | 73513 | 8.2 | x | 4.1 | x | x | x | x |
| IPI00207012.1 | Voltage-gated potassium channel beta-1 subunit | 401 | 44710 | 9.3 | x | x | x | 9 | x | 13.2 |
| IPI00211012.1 | Voltage-gated potassium channel beta-2 subunit | 367 | 41021 | 9 | 16.9 | 20.2 | 19.1 | 29.4 | 21.3 | 22.6 |
| IPI00197991.5 | Kv3.3c voltage gated potassium channel subunit splice variant C | 770 | 82068 | 6.9 | x | x | x | 3.6 | x | 5.2 |
| IPI00391769.1 | Voltage-gated calcium channel alpha2/delta-1 subunit | 1091 | 1E+05 | 5.3 | 20.7 | 25.4 | 22.3 | x | x | x |
| IPI00365653.2 | Putative voltage-gated calcium channel alpha(2)delta-4 subunit | 1463 | 2E+05 | 6 | x | x | x | x | 3.6 | x |
| IPI00211876.1 | Voltage-dependent L-type calcium channel beta-3 subunit | 484 | 54564 | 6.3 | x | x | x | 6.4 | x | x |
| IPI00211870.1 | Voltage-dependent P/Q-type calcium channel alpha-1A subunit, Splice Isoform 1 | 2212 | 3E+05 | 8.6 | 4 | 3.5 | x | x | x | 2.8 |
| IPI00200639.2 | Voltage-dependent N-type calcium channel alpha-1B subunit | 2356 | 3E+05 | 8.6 | 2.2 | 1.1 | 2.4 | x | x | x |
| IPI00199192.3 | Voltage-dependent T-type calcium channel alpha-1G subunit | 2254 | 3E+05 | 7.3 | 1.4 | x | x | x | x | x |
| IPI00207426.1 | Voltage-dependent calcium channel gamma-8 subunit | 421 | 43269 | 9.2 | 23 | 14 | 23 | 12.1 | 12.1 | 7.8 |
| IPI00400699.3 | RIIA Voltage-gated sodium channel | 1956 | 2E+05 | 5.6 | 6.7 | 5.1 | x | 7.4 | 6 | 6.5 |
| IPI00213193.3 | Voltage-gated sodium channel variant rPN4a | 1988 | 2E+05 | 6.3 | x | 1.7 | x | x | x | 2.3 |
| IPI00210089.1 | sodium channel, voltage-gated, type 10, alpha polypeptide | 1957 | 2E+05 | 6.2 | x | x | x | x | 1.5 | x |
| Transporters | ||||||||||
| IPI00421430.1 | Zinc transporter ZnT-3 | 388 | 41898 | 7.3 | 14.4 | 8.5 | 8.5 | 9.5 | 9.8 | 17.5 |
| IPI00214787.1 | Monocarboxylate transporter 1 | 494 | 53238 | 8.3 | 4.9 | x | 5.3 | 7.9 | x | 7.1 |
| IPI00191391.1 | Solute carrier family 2, facilitated glucose transporter member 1 | 492 | 53963 | 8.7 | 10.8 | 8.5 | 6.9 | x | 6.9 | 6.9 |
| IPI00198723.1 | Solute carrier family 2, facilitated glucose transporter member 3 | 493 | 53581 | 5 | 11.8 | 8.3 | 6.5 | 10.5 | x | 2.4 |
| IPI00395291.1 | Neutral amino acid transporter ASCT1 | 529 | 55900 | 6.3 | 19.1 | 18.5 | 9.8 | 15.9 | 4.5 | 13.4 |
| IPI00204778.1 | Large neutral amino acids transporter small subunit 1 | 512 | 55903 | 7.9 | 2.9 | x | 8.2 | 8.2 | x | 10 |
| IPI00324377.1 | Splice Isoform GLAST-1 of Excitatory amino acid transporter 1 | 543 | 59698 | 8.4 | 25.6 | 15.3 | 15.3 | 19.9 | x | x |
| IPI00214057.5 | Excitatory amino acid transporter 4 | 561 | 60715 | 9.1 | 4.6 | 8.4 | 4.6 | 7.5 | x | x |
In addition to providing more confident identification, high levels of sequence coverage are necessary to differentiate isoforms of proteins. For example, an analysis of peptides used to identify three transmembrane proteins, Sodium- and choloride-dependent GABA transporter 1, GP145-TrkB receptor, and excitatory amino acid transporter 1, shows extensive sequence coverage of the extracellular ligand binding domains and the intracellular signaling domains as a result of the unique and complementary mixture of peptides generated by digestion in combinations of detergents and solvents. Additionally, peptides generated by digesting the excitatory amino acid transporter 1 with Invitrosol in 80% acetonitrile extend into the predicted transmembrane domains of the protein (Supplementary figure 1). In fact, we found 2.8% of total peptides identified contain amino acids extending into the transmembrane region. We find enrichment of peptides containing the transmembrane domains with a significant increase in the number of peptides containing transmembrane regions compared to Wu et al using Proteinase K digestion 8. Most of these peptides are found in the aqueous digestion with RapiGest (Tris RPG, 88 peptides), followed by the organic digestion with Invitrosol (ACN IVS, 86 peptides), and the aqueous protein digestion with Invitrosol (Tris IVS, 73 peptides) (Table 3). We observe that use of organic solvents with Invitrosol yields the highest percentage of peptides containing transmembrane sequences over the total number of transmembrane proteins identified (20.3%) (Table 3). In other words, trypsin digestion of transmembrane proteins with invitrosol in 80% acentonitrile shows an increasing trend of generating peptides containing transmembrane regions comparing to other digestion conditions. This trend has been previously observed 21. It suggests that perturbing the lipid bilayer near the transmembrane region in the presence of high percentage organic solvent facilitates the proteolysis of the helical regions of transmembrane proteins. In summary, our results demonstrate that MS compatible detergents provide a valuable tool to increase the identification and sequence coverage of brain membrane proteins, including proteins localized to the synapse (discussed below).
Table 3.
A summary of identified proteins and peptides containing transmembrane (TM) regions in six digestion conditions. Proteins containing at least one transmembrane domain (TMD) are predicted by the TMHMM algorithm (see method). Peptides containing the sequences of the transmembrane region are identified and summarized in the second row. The last row of data represents the percentage of TMD peptides identified from the identified TMD proteins. Higher ratio of TMD peptide vs. TMD protein indicates higher representation of the transmembrane region in the particular digestion condition.
| Tris RPG | Tris IVS | Tris PPS | ACN IVS | ACN PPS | ACN RPG | |
|---|---|---|---|---|---|---|
| TMD Protein |
556 | 476 | 430 | 422 | 324 | 213 |
| TMD Peptide |
88 | 73 | 23 | 86 | 23 | 39 |
| TMD peptide/ TMD Protein |
15.8% | 15.3% | 5.3% | 20% | 4.9% | 8.7% |
Physiological Relevance of the Identified Brain Proteome
Annotation of the data using GeneOntologies revealed that around 35% (1897 proteins) of the identified proteins are classified as membrane proteins. To explore the physiological relevance of the identified proteins, we clustered proteins identified in our data based on GeneOntology (GO) and identify several GO categories with a significant enrichment (p<0.05) compared to the entire rat proteome. For example, we observed a three to four fold enrichment of proteins localized to the presynaptic membrane, synaptic vesicle membrane, synapse, and synaptosome (Supplementary figure 2A). This brain-specific compartment receives signals, in the form of neurotransmitters, from the presynaptic terminal of an adjacent neuron. The postsynaptic and presynaptic termini are the signaling center between two neurons, and consist of cell-surface receptors and a large variety of signaling pathways 22, 23. In addition to proteins with synaptic functions, we also observed a six and five-fold increase in proteins involved in glutamate transmission and transport activities, which is consistent with the fact that the major excitatory neurotransmitter in the brain is glutamate (Supplementary figure 2B). Glutamate receptors can be broadly divided into ligand-gated ion channels and G-protein coupled receptors. The three major subtypes of ionotropic receptors are AMPA (alpha-amino-3-hydroxy-5-methyl-ioxyzole-4-propionic acid), kainate, and NMDA (N-methyl-D-aspartate). In addition, there are two orphan receptors. In total, this family is represented by seventeen genes, and in our analysis, we identified ten of these ionotropic glutamate receptors (Table 2). In addition, we identified six of the eight G-protein coupled glutamate receptors (Table 2). Besides the glutamate receptors, we also identified other classes of membrane proteins essential for maintaining brain functions such as neurotransmitter receptors, ion channels, and transporters (Supplementary table 2). Together, our results demonstrate the utility of our method in identifying membrane proteins essential for the function of the brain.
DISCUSSION
Using total brain homogenate, we identified 5,422 proteins, including 1225 (22.6%) proteins that contained at least one transmembrane domain and 1897 (35%) proteins annotated as membrane proteins. Among these proteins, we identified many proteins currently under intense investigation in neuroscience. For example, we identified numerous proteins involved in synaptic function. The most notable proteins identified in this category are the glutamate and GABA receptors. Direct modulators of GABA receptors include benzodiazepines, barbiturates, neurosteroids and anesthetics, and altered GABA receptor expression has been strongly implicated in insomnia, anxiety, premenstrual syndrome, alcoholism, schizophrenia, depression and epilepsy 51–54. The glutamate receptor family plays important roles brain development and in some forms of synaptic plasticity that may underlie higher order processes, such as learning and memory. In terms of neurological disorders, glutamate is crucial to the process of neurodegeneration, and potential therapeutic effects of glutamate neuromodulators have been reported for Alzheimer’s disease, Parkinson’s disease, ischemic stroke, and neuropathic pain 55–61. In addition, we identified numerous membrane proteins, such as ion channels and amino acid transporters, known to play important roles in brain physiology.
In conclusion, we demonstrate that modification of trypsin digestion protocols with MS compatible detergents improves global and membrane protein identification in the mammalian brain. Furthermore, by coupling the trypsin digestion protocol with MS compatible detergents and high sensitivity LC-MS/MS analysis, it is possible to profile and quantify proteins from specific regions of the brain without pooling animal tissues, which often masks the natural variability between individual animals. Lastly, it is also possible to further increase the protein coverage by incorporating other protein digestion strategies such as proteinase K digestion. The combination of simple sample preparation, optimized trypsin digestion technique, and highly sensitive multidimensional LC-MS/MS analysis opens up new possibilities to proteomic-driven biological discoveries in normal brain physiology and neurological abnormality.
Supplementary Material
Supplementary Figure 1. Peptide mapping of three brain transmembrane proteins: sodium- and choloride-dependent GABA transporter 1, splice isoform GP145-TrkB of BDNF/NT-3 growth factors receptor, splice isoform GLAST-1 of excitatory amino acid transporter 1.
Supplementary Figure 2. Comparison of GO categories between the identified MS dataset and the entire rat proteome. (A) Significantly enriched (> 4 fold) GO categories from the MS dataset based on the cellular localization. (B) Significantly enriched (> 3 fold) GO categories from the MS dataset based on the molecular function.
ACKNOWLEDGMENT
This research was supported by an NIAID sub-contract grant UCSD/MCB0237059 and partially supported by NIH P41RR011823 and NIH 5R01 MH067880.
REFERENCE
- 1.Yehuda S, Rabinovitz S, Mostofsky DI. Essential fatty acids are mediators of brain biochemistry and cognitive functions. J Neurosci Res. 1999;56(6):565–570. doi: 10.1002/(SICI)1097-4547(19990615)56:6<565::AID-JNR2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 2.Krapfenbauer K, Fountoulakis M, Lubec G. A rat brain protein expression map including cytosolic and enriched mitochondrial and microsomal fractions. Electrophoresis. 2003;24(11):1847–1870. doi: 10.1002/elps.200305401. [DOI] [PubMed] [Google Scholar]
- 3.Beranova-Giorgianni S, Pabst MJ, Russell TM, Giorgianni F, Goldowitz D, Desiderio DM. Preliminary analysis of the mouse cerebellum proteome. Brain Res Mol Brain Res. 2002;98(1–2):135–140. doi: 10.1016/s0169-328x(01)00333-3. [DOI] [PubMed] [Google Scholar]
- 4.Stevens SM, Jr, Zharikova AD, Prokai L. Proteomic analysis of the synaptic plasma membrane fraction isolated from rat forebrain. Brain Res Mol Brain Res. 2003;117(2):116–128. doi: 10.1016/s0169-328x(03)00282-1. [DOI] [PubMed] [Google Scholar]
- 5.Rabilloud T. Use of thiourea to increase the solubility of membrane proteins in two-dimensional electrophoresis. Electrophoresis. 1998;19(5):758–760. doi: 10.1002/elps.1150190526. [DOI] [PubMed] [Google Scholar]
- 6.Klose J, Nock C, Herrmann M, Stuhler K, Marcus K, Bluggel M, Krause E, Schalkwyk LC, Rastan S, Brown SD, Bussow K, Himmelbauer H, Lehrach H. Genetic analysis of the mouse brain proteome. Nat Genet. 2002;30(4):385–393. doi: 10.1038/ng861. [DOI] [PubMed] [Google Scholar]
- 7.Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J Neurochem. 2004;88(3):759–768. doi: 10.1046/j.1471-4159.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- 8.Wu CC, MacCoss MJ, Howell KE, Yates JR., 3rd A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21(5):532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen PA, Olsen JV, Podtelejnikov AV, Andersen JR, Mann M, Wisniewski JR. Proteomic mapping of brain plasma membrane proteins. Mol Cell Proteomics. 2005;4(4):402–408. doi: 10.1074/mcp.T500002-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Qian WJ, Chin MH, Petyuk VA, Barry RC, Liu T, Gritsenko MA, Mottaz HM, Moore RJ, Camp Ii DG, Khan AH, Smith DJ, Smith RD. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J Proteome Res. 2006;5(2):361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen EI, Cociorva D, Norris JL, Yates JR., 3rd Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J Proteome Res. 2007;6(7):2529–2538. doi: 10.1021/pr060682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Besson MT, Soustelle L, Birman S. Selective high-affinity transport of aspartate by a Drosophila homologue of the excitatory amino-acid transporters. Curr Biol. 2000;10(4):207–210. doi: 10.1016/s0960-9822(00)00339-0. [DOI] [PubMed] [Google Scholar]
- 13.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 14.Eng JK, McCormack AL, Yates, r. JR. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. r. [DOI] [PubMed] [Google Scholar]
- 15.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67(8):1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 16.Tabb DL, McDonald WH, Yates, r. JR. DTASelect and Contrast: Tools for Assembling and Comparing Protein Identification from Shotgun Proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant KJ, Wu CC. Advances in neuromembrane proteomics: efforts towards a comprehensive analysis of membrane proteins in the brain. Brief Funct Genomic Proteomic. 2007;6(1):59–69. doi: 10.1093/bfgp/elm001. [DOI] [PubMed] [Google Scholar]
- 18.Yang JW, Czech T, Lubec G. Proteomic profiling of human hippocampus. Electrophoresis. 2004;25(7–8):1169–1174. doi: 10.1002/elps.200305809. [DOI] [PubMed] [Google Scholar]
- 19.Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17(7):646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer F, Wolters D, Rogner M, Poetsch A. Toward the complete membrane proteome: high coverage of integral membrane proteins through transmembrane peptide detection. Mol Cell Proteomics. 2006;5(3):444–453. doi: 10.1074/mcp.M500234-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Pozdniakov OM, Babakova LL. The plasticity of the neuromuscular synapse in pathology. Zh Nevrol Psikhiatr Im S S Korsakova. 1998;98(3):50–53. [PubMed] [Google Scholar]
- 23.Pocklington AJ, Armstrong JD, Grant SG. Organization of brain complexity--synapse proteome form and function. Brief Funct Genomic Proteomic. 2006;5(1):66–73. doi: 10.1093/bfgp/ell013. [DOI] [PubMed] [Google Scholar]
- 24.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51(1):7–61. [PubMed] [Google Scholar]
- 25.Michels G, Moss SJ. GABAA receptors: properties and trafficking. Crit Rev Biochem Mol Biol. 2007;42(1):3–14. doi: 10.1080/10409230601146219. [DOI] [PubMed] [Google Scholar]
- 26.Bezanilla F. Voltage-gated ion channels. IEEE Trans Nanobioscience. 2005;4(1):34–48. doi: 10.1109/tnb.2004.842463. [DOI] [PubMed] [Google Scholar]
- 27.Sands Z, Grottesi A, Sansom MS. Voltage-gated ion channels. Curr Biol. 2005;15(2):R44–R47. doi: 10.1016/j.cub.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 28.Jurkat-Rott K, Lehmann-Horn F. Human muscle voltage-gated ion channels and hereditary disease. Curr Opin Pharmacol. 2001;1(3):280–287. doi: 10.1016/s1471-4892(01)00050-9. [DOI] [PubMed] [Google Scholar]
- 29.Lehmann-Horn F, Jurkat-Rott K. Voltage-gated ion channels and hereditary disease. Physiol Rev. 1999;79(4):1317–1372. doi: 10.1152/physrev.1999.79.4.1317. [DOI] [PubMed] [Google Scholar]
- 30.Forte M, Guy HR, Mannella CA. Molecular genetics of the VDAC ion channel: structural model and sequence analysis. J Bioenerg Biomembr. 1987;19(4):341–350. doi: 10.1007/BF00768537. [DOI] [PubMed] [Google Scholar]
- 31.Clapham DE, Runnels LW, Strubing C. The TRP ion channel family. Nat Rev Neurosci. 2001;2(6):387–396. doi: 10.1038/35077544. [DOI] [PubMed] [Google Scholar]
- 32.Yiangou Y, Facer P, Chessell IP, Bountra C, Chan C, Fertleman C, Smith V, Anand P. Voltage-gated ion channel Na(v)1.7 innervation in patients with idiopathic rectal hypersensitivity and paroxysmal extreme pain disorder (familial rectal pain) Neurosci Lett. 2007;427(2):77–82. doi: 10.1016/j.neulet.2007.09.027. [DOI] [PubMed] [Google Scholar]
- 33.Gordon D, Merrick D, Auld V, Dunn R, Goldin AL, Davidson N, Catterall WA. Tissue-specific expression of the RI and RII sodium channel subtypes. Proc Natl Acad Sci U S A. 1987;84(23):8682–8686. doi: 10.1073/pnas.84.23.8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hebert TE, Monette R, Stone JC, Drapeau P, Dunn RJ. Insertion mutations of the RIIA Na+ channel reveal novel features of voltage gating and protein kinase A modulation. Pflugers Arch. 1994;427(5–6):500–509. doi: 10.1007/BF00374267. [DOI] [PubMed] [Google Scholar]
- 35.Dietrich PS, McGivern JG, Delgado SG, Koch BD, Eglen RM, Hunter JC, Sangameswaran L. Functional analysis of a voltage-gated sodium channel and its splice variant from rat dorsal root ganglia. J Neurochem. 1998;70(6):2262–2272. doi: 10.1046/j.1471-4159.1998.70062262.x. [DOI] [PubMed] [Google Scholar]
- 36.Musarella M, Alcaraz G, Caillol G, Boudier JL, Couraud F, Autillo-Touati A. Expression of Nav1.6 sodium channels by Schwann cells at neuromuscular junctions: role in the motor endplate disease phenotype. Glia. 2006;53(1):13–23. doi: 10.1002/glia.20252. [DOI] [PubMed] [Google Scholar]
- 37.Noda M, Ikeda T, Kayano T, Suzuki H, Takeshima H, Kurasaki M, Takahashi H, Numa S. Existence of distinct sodium channel messenger RNAs in rat brain. Nature. 1986;320(6058):188–192. doi: 10.1038/320188a0. [DOI] [PubMed] [Google Scholar]
- 38.Amaya F, Wang H, Costigan M, Allchorne AJ, Hatcher JP, Egerton J, Stean T, Morisset V, Grose D, Gunthorpe MJ, Chessell IP, Tate S, Green PJ, Woolf CJ. The voltage-gated sodium channel Na(v)1.9 is an effector of peripheral inflammatory pain hypersensitivity. J Neurosci. 2006;26(50):12852–12860. doi: 10.1523/JNEUROSCI.4015-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devor M. Sodium channels and mechanisms of neuropathic pain. J Pain. 2006;7(1 Suppl 1):S3–S12. doi: 10.1016/j.jpain.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Shen H, Wang F, Zhang Y, Xu J, Long J, Qin H, Liu F, Guo J. Zinc distribution and expression pattern of ZnT3 in mouse brain. Biol Trace Elem Res. 2007;119(2):166–174. doi: 10.1007/s12011-007-0056-2. [DOI] [PubMed] [Google Scholar]
- 41.Jo SM, Danscher G, Schroder HD, Suh SW. Depletion of vesicular zinc in dorsal horn of spinal cord causes increased neuropathic pain in mice. Biometals. 2007 doi: 10.1007/s10534-007-9103-x. [DOI] [PubMed] [Google Scholar]
- 42.Nakai M, Chen L, Nowak RA. Tissue distribution of basigin and monocarboxylate transporter 1 in the adult male mouse: a study using the wild-type and basigin gene knockout mice. Anat Rec A Discov Mol Cell Evol Biol. 2006;288(5):527–535. doi: 10.1002/ar.a.20320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rayner DV. Glucose transporters in the brain. Proc Nutr Soc. 1996;55(1B):209–219. doi: 10.1079/pns19960022. [DOI] [PubMed] [Google Scholar]
- 44.Simpson IA, Vannucci SJ, Maher F. Glucose transporters in mammalian brain. Biochem Soc Trans. 1994;22(3):671–675. doi: 10.1042/bst0220671. [DOI] [PubMed] [Google Scholar]
- 45.Kanai Y, Hediger MA. The glutamate and neutral amino acid transporter family: physiological and pharmacological implications. Eur J Pharmacol. 2003;479(1–3):237–247. doi: 10.1016/j.ejphar.2003.08.073. [DOI] [PubMed] [Google Scholar]
- 46.Kanai Y, Hediger MA. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch. 2004;447(5):469–479. doi: 10.1007/s00424-003-1146-4. [DOI] [PubMed] [Google Scholar]
- 47.Sakai K, Shimizu H, Koike T, Furuya S, Watanabe M. Neutral amino acid transporter ASCT1 is preferentially expressed in L-Ser-synthetic/storing glial cells in the mouse brain with transient expression in developing capillaries. J Neurosci. 2003;23(2):550–560. doi: 10.1523/JNEUROSCI.23-02-00550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boado RJ, Li JY, Nagaya M, Zhang C, Pardridge WM. Selective expression of the large neutral amino acid transporter at the blood-brain barrier. Proc Natl Acad Sci U S A. 1999;96(21):12079–12084. doi: 10.1073/pnas.96.21.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amara SG, Fontana AC. Excitatory amino acid transporters: keeping up with glutamate. Neurochem Int. 2002;41(5):313–318. doi: 10.1016/s0197-0186(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 50.Furuta A, Martin LJ, Lin CL, Dykes-Hoberg M, Rothstein JD. Cellular and synaptic localization of the neuronal glutamate transporters excitatory amino acid transporter 3 and 4. Neuroscience. 1997;81(4):1031–1042. doi: 10.1016/s0306-4522(97)00252-2. [DOI] [PubMed] [Google Scholar]
- 51.Walsh JK. Understanding GABA and its relation to insomnia and therapeutics. J Clin Sleep Med. 2006;2(2):S5–S6. [PubMed] [Google Scholar]
- 52.Hetmar O, Nielsen M. GABA receptor function and manic depressive disease. Nord Med. 1986;101(2):62–65. [PubMed] [Google Scholar]
- 53.Bruening S, Oh E, Hetzenauer A, Escobar-Alvarez S, Westphalen RI, Hemmings HC, Jr, Singewald N, Shippenberg T, Toth M. The anxiety-like phenotype of 5-HT receptor null mice is associated with genetic background-specific perturbations in the prefrontal cortex GABA-glutamate system. J Neurochem. 2006;99(3):892–899. doi: 10.1111/j.1471-4159.2006.04129.x. [DOI] [PubMed] [Google Scholar]
- 54.Peng Z, Huang CS, Stell BM, Mody I, Houser CR. Altered expression of the delta subunit of the GABAA receptor in a mouse model of temporal lobe epilepsy. J Neurosci. 2004;24(39):8629–8639. doi: 10.1523/JNEUROSCI.2877-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lujan-Miras R. Metabotropic glutamate receptors: new molecular targets in the treatment of neurological and psychiatric diseases. Rev Neurol. 2005;40(1):43–53. [PubMed] [Google Scholar]
- 56.Gubellini P, Pisani A, Centonze D, Bernardi G, Calabresi P. Metabotropic glutamate receptors and striatal synaptic plasticity: implications for neurological diseases. Prog Neurobiol. 2004;74(5):271–300. doi: 10.1016/j.pneurobio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 57.Sela BA. Neurological significance of glutamate receptors in the central nervous system. Harefuah. 1998;134(9):710–716. [PubMed] [Google Scholar]
- 58.Obrenovitch TP, Urenjak J, Zilkha E, Jay TM. Excitotoxicity in neurological disorders--the glutamate paradox. Int J Dev Neurosci. 2000;18(2–3):281–287. doi: 10.1016/s0736-5748(99)00096-9. [DOI] [PubMed] [Google Scholar]
- 59.Obrenovitch TP, Urenjak J. Altered glutamatergic transmission in neurological disorders: from high extracellular glutamate to excessive synaptic efficacy. Prog Neurobiol. 1997;51(1):39–87. doi: 10.1016/s0301-0082(96)00049-4. [DOI] [PubMed] [Google Scholar]
- 60.Rogers SW, Twyman RE, Gahring LC. The role of autoimmunity to glutamate receptors in neurological disease. Mol Med Today. 1996;2(2):76–81. doi: 10.1016/1357-4310(96)88742-9. [DOI] [PubMed] [Google Scholar]
- 61.Prusiner SB. Disorders of glutamate metabolism and neurological dysfunction. Annu Rev Med. 1981;32:521–542. doi: 10.1146/annurev.me.32.020181.002513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Peptide mapping of three brain transmembrane proteins: sodium- and choloride-dependent GABA transporter 1, splice isoform GP145-TrkB of BDNF/NT-3 growth factors receptor, splice isoform GLAST-1 of excitatory amino acid transporter 1.
Supplementary Figure 2. Comparison of GO categories between the identified MS dataset and the entire rat proteome. (A) Significantly enriched (> 4 fold) GO categories from the MS dataset based on the cellular localization. (B) Significantly enriched (> 3 fold) GO categories from the MS dataset based on the molecular function.