Abstract
Generating reliable expression profiles from minute cell quantities is critical for scientific discovery and potential clinical applications. Here we present low-quantity digital gene expression (LQ-DGE), an amplification-free approach involving capture of poly(A)+ RNAs from cellular lysates onto poly(dT)-coated sequencing surfaces, followed by on-surface reverse transcription and sequencing. We applied LQ-DGE to profile malignant and nonmalignant mouse and human cells, demonstrating its quantitative power and potential applicability to archival specimens.
Recent advances in both cancer and stem cell biology have highlighted the biological importance of extraordinarily rare cells that may have fundamentally different gene expression patterns than surrounding cell types. Although next-generation sequencing technologies bring unprecedented power to gene expression studies, the inability to apply reliable molecular profiling analyses to such minimal numbers of cells has proven to be a major limitation of current sequencing-based strategies. Methods developed to date rely on multiple sample manipulation and amplification steps, which introduce errors and skew the original representation of the nucleic acid population1–4, rendering these methods unsuitable for applications requiring high fidelity.
To profile minute RNA quantities for gene expression patterns, we extended the single-molecule sequencing (SMS) technology5,6 to establish a low-quantity digital gene expression (LQ-DGE) application involving direct flow-cell capture of RNA. To optimize and test LQ-DGE, we selected two related yet distinct cell types: SM25 and 490. SM25 has been derived from a pancreatic intraepithelial neoplasia lesion in a genetically engineered mouse with pancreas-specific expression of the KrasG12D mutant7. Cell line 490 has been established from a malignant pancreatic ductal adenocarcinoma (PDAC) lesion, combining the KrasG12D mutation with conditional loss of the Trp53 gene8. Comparison of these two cell lines made it possible to model biologically important genetic differences among related cell types.
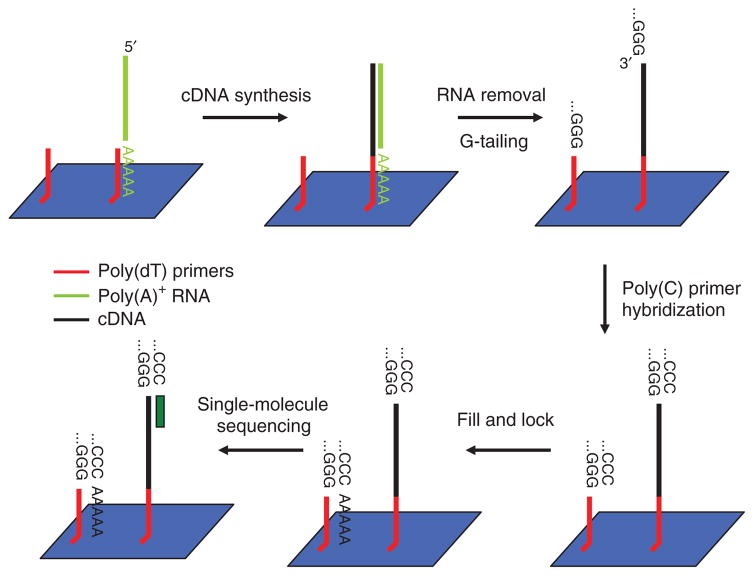
We performed LQ-DGE using 250 to 16,000 cells. Briefly, we captured poly(A)+ mRNA from cell lysates on the poly(dT)-coated sequencing flow cells. We initiated on-surface cDNA synthesis using SuperScript III reverse transcriptase, followed by terminal transferase–mediated on-surface guanine (G)-tailing of cDNAs covalently attached to surfaces to generate priming sites allowing sequencing from the ‘top’ (cDNA 3′ ends) (Fig. 1). We hybridized a poly(C) primer to the G-tailed templates, followed by a ‘fill-and-lock’ step6. Then we initiated SMS without additional modifications.
Figure 1.
LQ-DGE template capture, on-surface cDNA synthesis and sequencing workflow. After capturing the poly(A)+ RNA on surfaces coated with covalently attached poly(dT) primers, natural dNTPs and reverse transcriptases are used to synthesize cDNA. Indicated steps are followed to obtain cDNA sequence by synthesis (green rectangle).
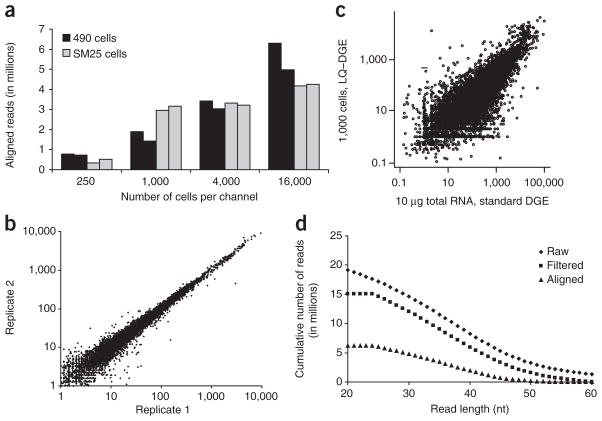
To determine the effect of cell quantity on the number of usable reads obtained per channel, we performed a titration experiment (Fig. 2a). As few as 250 cells generated sufficient usable reads for digital gene expression (DGE) profiling (Supplementary Fig. 1). The measurements were highly reproducible, as demonstrated by profiling 1,000 cells, using lysates prepared at separate times in two independent runs (Fig. 2b). Our comparison of four different commercial cell lysis conditions showed a high correlation between transcript counts obtained (r = 0.937–0.946; Supplementary Fig. 2). However, these correlations were lower than the correlation obtained from profiles generated with the same lysis condition, suggesting that the LQ-DGE profiles may be slightly dependent on cell-lysis methods.
Figure 2.
Throughput, reproducibility and counting power of LQ-DGE. (a) Usable read yields obtained per channel in two sequencing runs for each of 490 and SM25 cells. (b) Reproducibility of the approach across independent runs on one thousand 490 cells (r = 0.991). (c) Expression profiles obtained with LQ-DGE analysis of a thousand 490 cells were compared to those obtained with the standard DGE approach5 performed with 10 μg of 490 RNA isolated from four million cells. The two datasets had high agreement (r = 0.901). In b and c, x and y axes indicate log10 counts obtained per gene, and log10-transformed transcript counts were used for Pearson correlation calculations. Ribosomal and mitochondrial elements were not included in the graphs and correlation coefficient calculations. (d) Cumulative read length distribution of raw, quality filtered and aligned reads in a single channel. The mean aligned read length was 36 nt.
To determine the correlation of LQ-DGE with standard DGE methods5, we compared DGE profiling of four million 490 cells (10 μg of RNA) versus 1,000 cells profiled by LQ-DGE. Using LQ-DGE, we identified 8,620 transcripts exhibiting counts ≥10 transcripts per million reads (t.p.m.) in 490 cells, whereas the standard DGE identified 8,801 transcripts with ≥10 t.p.m. The two datasets had a positive correlation (Fig. 2c). The rRNA sequence amount using DGE profiling was 2–3%, compared to 0.01–0.02% with LQ-DGE, whereas the fraction of mitochondrial sequences was comparable for both methods (2–3%). Some rRNA species are in fact polyadenylated9,10, and the ~100-fold lower rRNA amounts obtained with the LQ-DGE approach suggest that their abundance in standard oligo(dT)-primed DGE assays may be due to mispriming events.
We examined the overall sequencing parameters to determine whether the modified sample-preparation and sequencing strategy altered the SMS performance. Over 97% of the reads were 24–60 nucleotides (nt) in length, with a median length of 36 nt (Fig. 2d). This length is 3 nt greater than the reported SMS performance involving sequencing of poly(A)-tailed DNA and cDNA prepared without on-surface reverse transcription of templates5,6. Although we cannot readily determine the underlying mechanism, it is conceivable that the slightly longer read lengths might have resulted from the positioning of sequencing-by-synthesis steps further away from the flow-cell surface, thus enhancing accessibility to enzymes and other components of the SMS chemistry. The average error rate was 4.7–5.1% per base, comparable for the two techniques.
To determine whether LQ-DGE can be used to profile minute quantities of degraded RNA species, we used RNAs from matched fresh and formalin-fixed, paraffin-embedded (FFPE) mouse PDAC tissue samples8. Analyses of fresh tissue and of FFPE tissue revealed 9,617 and 9,849 transcripts exhibiting counts ≥10 t.p.m., respectively. LQ-DGE profiles revealed strong correlation (r = 0.86; Supplementary Fig. 3) between the fresh and FFPE RNA samples, with 84% of genes exhibiting ≤ twofold variance between the two specimens. The fraction of reads from rRNAs was only 0.03% in the FFPE RNA sample, suggesting that LQ-DGE avoids the highly abundant rRNA species. Thus, LQ-DGE profiles were largely preserved whether the cell or tissue samples were frozen or FFPE. Additional studies are needed to determine the effectiveness of using LQ-DGE to identify differential expression in FFPE samples.
Although LQ-DGE offers an alternative RNA quantitation approach that is advantageous for analysis of minute cell quantities, this method may still suffer from common cDNA synthesis artifacts, such as spurious second-strand formation and reverse transcriptase–related biases owing to RNA structure. For instance, 1.5–1.9% of reads mapping to known genes were aligned opposite to their known transcription direction (Supplementary Table 1). This percentage was lower than the 4.6% obtained with the standard DGE approach, and some of these may in fact represent bona fide antisense transcription events11. However, given that a reverse transcriptase is used for on-surface reverse transcription, LQ-DGE may not be strand-specific because a portion of reads may contain both first- and second-strand cDNA sequences. The reported 1.5–1.9% strandedness of LQ-DGE may be an underestimate. We observed no substantial biases originating from transcript length and sequencing priming site with LQ-DGE (Supplementary Discussion and Supplementary Figs. 4–10). We initially selected the SuperScript III reverse transcriptase because it is reported to give satisfactory performance for bead-based on-surface applications4. To test for potential reverse transcriptase–specific effects, we compared the profiles obtained with SuperScript III reverse transcriptase to those obtained using HIV reverse transcriptase (Supplementary Fig. 11) and observed high concordance between the transcript counts obtained using either of these enzymes (r = 0.993).
To determine whether LQ-DGE could be used to identify differentially expressed genes, we compared profiles of 490 and SM25 cells (8,620 and 8,771 transcripts with counts ≥10 t.p.m., respectively). We identified 2,088 genes exhibiting both a ≥ twofold difference in expression between the two cell types and a minimal count of 10 t.p.m in at least one of the cell lines. From these, we selected several genes exhibiting differential expression for validation using quantitative real-time PCR (qRT-PCR) and observed high agreement between LQ-DGE and qRT-PCR measurements (Supplementary Figs. 12–13). Scanning all differentially expressed genes identified by LQ-DGE using gene ontology analysis demonstrated significant differences among genes implicated in cell-cycle regulation (P = 2.4 × 10−12), cell differentiation (P = 2.2 × 10−11) and cellular response to stress (P = 8.00 × 10−10), pathways that are consistent with the difference in tumorigenic properties between these cell types. Thus, LQ-DGE can be used to distinguish expression profiles between closely related cells, a property that is critical to its application to rare cells, such as circulating tumor cells and stem cells.
From a technical standpoint, the ability to minimize input cell quantities using LQ-DGE stems from the integration of multiple sample-preparation steps. Unlike our other approach for low-quantity RNA profiling12, LQ-DGE does not require RNA isolation or mRNA selection and takes advantage of the higher-affinity RNA-DNA hybridization kinetics, relative to DNA-DNA, allowing poly(A)+ templates to be captured on flow-cell surfaces while avoiding RNA species such as rRNA that are undesired in DGE-type applications. It also does not involve fragmentation or size-selection steps, thus minimizing bias, particularly against short transcripts. Because each transcript captured on the surface can give rise to only one read, LQ-DGE expression data do not require normalization for transcript length or other factors13. The absence of manipulation steps known to cause representational bias, such as ligation, restriction digestion or amplification, also minimizes potential artifacts. For instance, duplicate reads, a common artifact resulting from amplification, have been reported to be 39.7–94.1% of reads obtained with the Illumina standard RNA-seq strategy and 6.1–7.2% with an improved RNA-seq strategy requiring 250 ng of poly(A)+ RNA14. This percentage is only 0.01–0.03% with LQ-DGE profiles generated from 1,000 cells. Finally, as with other sequencing-based approaches, LQ-DGE can also be used to identify new transcriptional units (Supplementary Fig. 14).
Whereas the traditional SMS approach involves hybridization of poly(A)-tailed templates on the flow-cell surface, the LQ-DGE cDNAs are synthesized on the flow-cell surface and remain covalently attached. Each cDNA can thus be sequenced multiple times by melting away the strands synthesized during sequencing, and repeating the poly(C) primer hybridization and ‘fill-and-lock’ steps. This resequencing capability can be useful because, with the SMS chemistry, only 15–25% of the templates give rise to reads that can be aligned back to reference sequences. Such a multipass sequencing capability may allow sufficient read depth as LQ-DGE is adapted for lower-cell-quantity applications. Our proof-of-concept experiment revealed that second-pass sequencing resulted in threefold lower alignable read yields compared to the first-pass sequencing (Supplementary Discussion and Supplementary Table 2). The efficiency of the melt step was 82% (Supplementary Fig. 15), suggesting that the decrease in aligned reads in the second-pass sequencing can be partly explained by the inefficiencies of the melting process. Optimizations of this resequencing strategy may improve the efficiency of the LQ-DGE multipass sequencing.
Although this technology at its present state does not accomplish the ultimate goal of reliable single-cell analyses, it raises the possibility that improvements in the components of LQ-DGE, including flow-cell design, nucleic-acid capture and sequencing chemistry, may succeed at this goal, providing new avenues in understanding the heterogeneity and dynamics of complex tissues and cell populations.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemethods/.
Supplementary Material
Acknowledgments
We thank K. Kerouac for technical assistance. D.T.T. is supported by the Pancreatic Cancer Action Network–American Association for Cancer Research Research Fellowship, and D.A.H. is supported by the Howard Hughes Medical Institute. This work was supported by US National Human Genome Research Institute grant 1R44HG005279-01 to F.O. and P.M.M.
Footnotes
Accession codes. National Center for Biotechnology Information (NCBI) Sequence Read Archive: SRA010077.
Note: Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
F.O. conceived and designed the study; F.O., D.T.T., B.W.B. and S.P. performed the experiments; N.B. provided the cells; F.O., B.S.W. and S.R. performed the computational analyses; F.O., D.T.T., P.M.M. and D.A.H. wrote the paper, which all authors edited.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturemethods/.
Published online at http://www.nature.com/naturemethods/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Linsen SE, et al. Nat Methods. 2009;6:474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- 2.Pinard R, et al. BMC Genomics. 2006;7:216. doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subkhankulova T, Livesey FJ. Genome Biol. 2006;7:R18. doi: 10.1186/gb-2006-7-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniguchi K, Kajiyama T, Kambara H. Nat Methods. 2009;6:503–506. doi: 10.1038/nmeth.1338. [DOI] [PubMed] [Google Scholar]
- 5.Lipson D, et al. Nat Biotechnol. 2009;27:652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- 6.Pushkarev D, Neff NF, Quake SR. Nat Biotechnol. 2009;27:847–852. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguirre AJ, et al. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardeesy N, et al. Proc Natl Acad Sci USA. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozsolak F, et al. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 10.Slomovic S, Laufer D, Geiger D, Schuster G. Nucleic Acids Res. 2006;34:2966–2975. doi: 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perocchi F, Xu Z, Clauder-Munster S, Steinmetz LM. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozsolak F, et al. Genome Res. 2010;20:519–525. doi: 10.1101/gr.102129.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshlack A, Wakefield MJ. Biol Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mamanova L, et al. Nat Methods. 2010;7:130–132. doi: 10.1038/nmeth.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.