Abstract
Liver X receptor (LXR), a sterol-activated nuclear hormone receptor, has been implicated in cholesterol and fatty acid homeostasis via regulation of reverse cholesterol transport and de novo fatty acid synthesis. LXR is also involved in immune responses, including anti-inflammatory action and T cell proliferation. In this study, we demonstrated that activated LXR suppresses cell cycle progression and proliferation in certain cell types. Stimulation of LXR with synthetic ligand T0901317 or GW3965 inhibited cell growth rate and arrested the cell cycle at the G1/S boundary in several cells, such as RWPE1, THP1, SNU16, LNCaP, and HepG2. However, LXR ligands did not exhibit antiproliferative activity in PC3, HEK293, or HeLa cells. Interestingly, activated LXR-mediated cell cycle arrest is closely correlated with the lipogenic gene expression and triacylglyceride accumulation. In accordance with these findings, suppression of FAS via small-interference RNA (siRNA) partially alleviated the antiproliferative effect of LXR activation in RWPE1 cells. Together, these data suggest that LXR activation with its ligands inhibits cell proliferation and induces G1/S arrest through elevated lipogenic activity, thus proposing a novel effect of activated LXR on cell cycle regulation.
Keywords: liver X receptor, ligand, fatty acid synthesis
Liver X receptor (LXR)α and LXRβ, also known as NR1H3 and NR1H2, respectively, are members of a nuclear hormone receptor superfamily, which are implicated in metabolic homeostasis and inflammation (1). LXRα is highly expressed in several tissues, such as liver, adipose, and steroidogenic tissues, whereas LXRβ is expressed ubiquitously (2). LXR can be activated by certain oxygenated cholesterol derivatives, including 20(S)-hydroxycholesterol [20(S)-HC], 22(R)-HC, and 24HC, naturally occurring oxysterols that stimulate the expression of LXR target genes (3). For example, ATP-binding cassette transporter (ABC)A1, ABCG1, ABCG5, apolipoprotein (apo)E, cytochrome P-450 7A1 (CYP7A1), sterol response element binding protein 1c (SREBP1c), and fatty acid synthase (FAS) are directly upregulated by activated LXR, consistent with key roles in the regulation of cholesterol and lipid metabolism (1).
In the liver and intestine, LXR activation has been reported to regulate cholesterol homeostasis through the expression of certain target genes, such as CYP7A1 and ABCG5/8, which are responsible for cholesterol conversion into bile acid and excretion (4–7). Furthermore, activated LXR promotes the expression of several genes involved in cholesterol efflux, such as ABCA1, ABCG1, and apoE, to stimulate a reverse cholesterol transport from macrophage to liver (5). Consistent with these findings, LXR activation shows an anti-atherogenic effect in Ldlr and apoE knockout mice (8). Deletion of LXRα results in impaired cholesterol and bile acid metabolism in the liver, which increases peripheral cholesterol accumulation and leads to atherosclerosis (4, 9). Therefore, one of the key functions of LXR has been implicated in atherosclerosis and its related metabolic complications.
LXR activation governs not only cholesterol homeostasis but also fatty acid metabolism. For example, administration of T0901317, a synthetic LXR ligand, leads to hepatic steatosis and hypertriglyceridemia through the enhancement of de novo fatty acid synthesis, which is accomplished by the induction of key lipogenic genes, such as SREBP1c and FAS (10–12). Furthermore, it has been reported that chronic activation of LXR contributes to lipotoxicity and apoptosis in pancreatic β-cells through hyperactivation of lipogenesis (13). Due to unwanted potent lipogenic effect of T0901317, GW3965, another LXR ligand, has been developed (14). GW3965 exhibits a much milder effect on lipogenic activity of LXR, even though GW3965 selectively activates LXR to maintain cholesterol efflux.
Recently, other roles of LXR have been reported. Activation of LXR suppresses innate immunity by inhibiting the expression of inflammatory genes, such as inducible nitric oxide synthase (iNOS), cyclooxygenase 2 (COX2), and interleukin-6 (IL6), in response to bacterial infection or lipopolysaccharide (LPS) stimulation (15, 16). Moreover, LXRα/β-null macrophages reveal enhanced apoptosis after microbial infection due to defects of LXR-dependent target gene expression, implying that LXR would be important for macrophage survival and innate immune response (16).
Several reports suggest that LXR is involved in proliferation of several cell types, such as smooth muscle cell, insulin-secreting MIN6 cell, and prostate-originated cancer cell lines (17–21). Although it has been reported that LXR activation is associated with regulation of p27 and Smad3, the underlying molecular mechanism is largely unknown for cell cycle regulation. In the current study, we have extensively examined the effect of activated LXR on cell proliferation. Activation of LXR by its ligands induced G1/S arrest and attenuated cell proliferation in certain cells, such as RWPE1, THP1, SNU15, LNCaP, and HepG2 cells, but not in PC3, HeLa, and HEK293 cells. We further investigated the molecular mechanisms that are associated with LXR-induced cell cycle arrest. Collectively, our data suggest that lipogenic activity of activated LXR is important for the inhibition of cell proliferation and cell cycle progression.
MATERIAL AND METHODS
Reagents
T0901317 and GW3965 were purchased from Calbiochem and Sigma, respectively. Propodium iodide, Oil Red O, and mevalonic acid lactone was provided by Sigma. Mevalonic acid lactone was dissolved in 0.1 mol/l sodium hydroxide to convert lactone into sodium mevalonate. Then, the product was heated at 50°C for 2 h and adjusted to pH 7.4, as described previously (22, 23).
Cell culture
RWPE1, LNCaP, and PC3 cells were a kind gift from Dr. Sung Hee Baek (Seoul National University, Seoul, Republic of Korea). Each cell line was maintained in a medium as follows: RWPE1 in keratinocyte serum-free medium (SFM) supplemented with 5 ng/ml human recombinant EGF and 0.05 mg/ml bovine pituitary extract (Invitrogen); THP1 and SNU16 in RPMI-1640 supplemented with 10% FBS; HepG2 in Eagle's minimum essential medium alpha (MEMα) supplemented with 10% FBS; LNCaP in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% FBS; PC3 in Kaighn's F12K medium supplemented with 10% FBS; and HeLa and HEK293 in DMEM supplemented with 10% FBS. All cell lines were incubated in a humidified atmosphere of 5% CO2 at 37°C.
Cell proliferation assay with CCK-8 reagent
Cell proliferation rate was determined with Cell Counting Kit-8 (CCK-8) according to the manufacturer's protocol (Dojindo Laboratories), which is a sensitive colorimetric assay for viable cells.
PI staining and FACS analysis
Trypsinized cells were washed with 1× PBS and kept in 70% ethanol for one day for fixation. Fixed cells were washed with 1× PBS twice and incubated with propodium iodide (PI) solution (0.1% Nonidet P-40, 100 μg/ml RNase, and 200 μg/l PI) for 10 min. Stained cells were analyzed by FACSCaliburTM (BD Biosciences), and the number of the cells in each stage was calculated with the ModFit LTTM cell cycle analysis program (Verity Software House) according to the manufacturer's instructions.
Oil Red O staining
Oil Red O stock solution was prepared in propylene glycol (0.7 g/100ml) and heated to 100°C for 10 min. DMSO or T0901317-treated cells were fixed with 10% formaldehyde-containing PBS for 15 min and washed with PBS twice. Then cells were soaked in 100% propylene glycol and stained with Oil Red O stock solution for 30 min. Stained cells were washed with 85% propylene glycol and distilled water until the background became clear. Images were captured with EVOS® ORIGINAL microscope (Advanced Microscopy Group) and NIKON TMS inverted microscope (NIKON).
RNA isolation and real-time quantitative PCR
As described earlier (24), total RNA was isolated with TRIzol reagent (Invitrogen). Then equal amounts of RNA were subjected to cDNA synthesis by using RevertAid M-MuLV reverse transcriptase (Fermentas). The relative amount of mRNA was evaluated by MyIQ real-time quantitative PCR detection system (Bio-Rad Laboratories) and calculated following normalization to the GAPDH mRNA. The primer sequences that were used for the real-time quantitative PCR analyses are described in supplementary Table I.
Small-interference RNA transfection
Small-interference RNA (siRNA) duplexes for SREBP1 and FAS were designed by Dharmacon and produced from GenePharma. RWPE1 cells were transfected with Lipofectamine2000TM reagent (Invitrogen) according to the manufacturer's protocol. The sequence information for siRNA is described in supplementary Table II.
Protein isolation and Western blotting
The cells were lysed with NETN buffer [100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl (pH 8.0), 0.5% Nonidet P-40 and CompleteTM protease inhibitor cocktail (Roche)] and subjected to Western blotting as described previously (13). Briefly, equal amounts of the proteins were separated by SDS-PAGE and transferred to PVDF (polyvinylidene difluoride) membranes (Millipore). After transfer, the membranes were blocked with 4% nonfat milk and probed with primary antibodies. The primary antibodies used in Western blotting were β-tubulin, cyclin A, cyclin B1, cyclin D1, cyclin E, cyclin H, and Rb 1, which were provided by Sigma. The SREBP1 antibody was produced by LabFrontier using 1-403 construct of SREBP1c as an immunogen. The results were visualized with horseradish peroxidase-conjugated secondary antibodies (Sigma) and enhanced chemiluminescence (Pierce) by using LAS-3000 imaging system (Fujifilm).
RESULTS
LXR activation suppressed cell proliferation
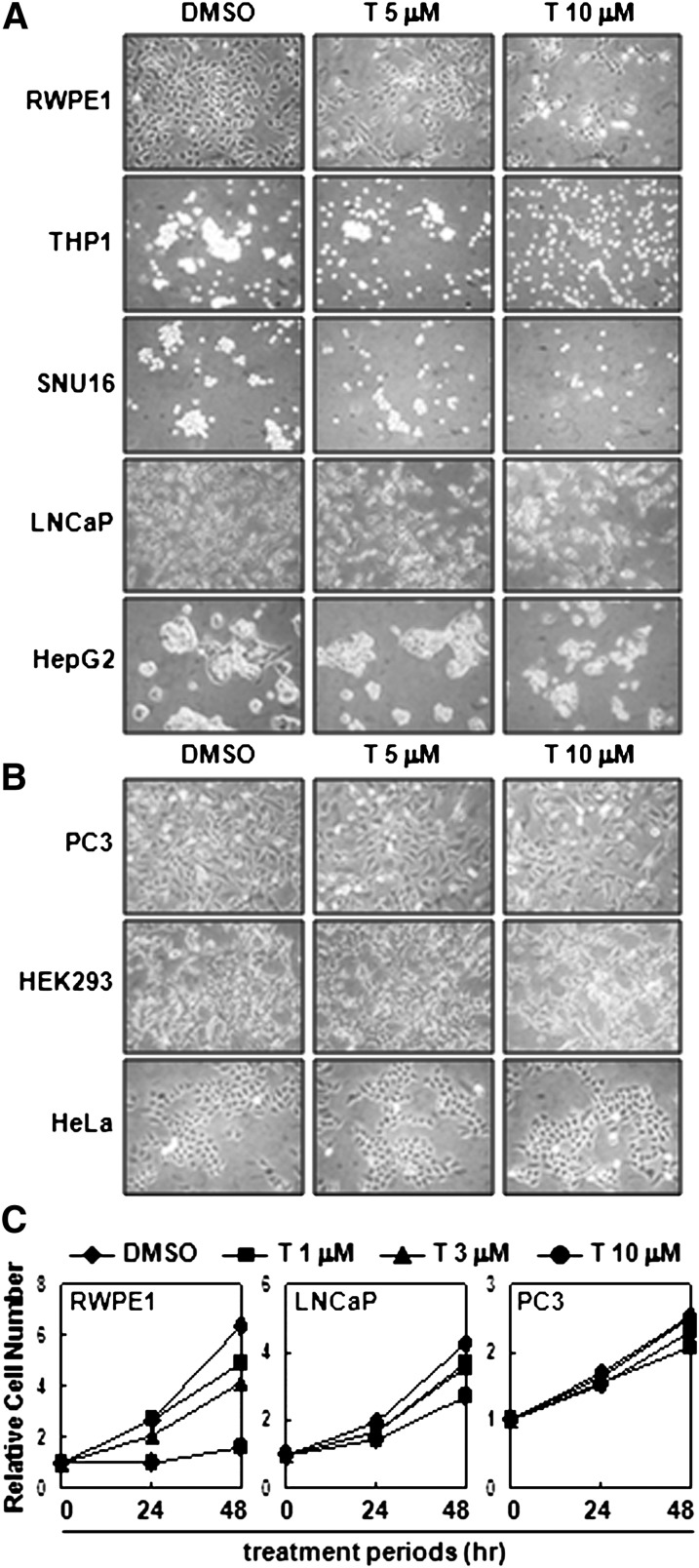
To test whether activated LXR is involved in cell proliferation, T0901317, a synthetic LXR ligand, was treated into various cell lines, and we monitored their proliferation rate. As shown in Fig. 1A, the proliferation of certain cells, such as RWPE1, THP1, SNU16, LNCaP, and HepG2 was dose-dependently inhibited by activated LXR with T0901317. On the contrary, proliferation of PC3, HEK293, and HeLa was not affected by T0901317 (Fig. 1B). Consistently, results of CCK-8 assay, which is a colorimetric method to measure the number of viable cells, also revealed that cell proliferation of RWPE1 and LNCaP, but not PC3, was diminished by LXR ligand (Fig. 1C), implying that inhibitory effect of LXR activation on cell proliferation appears to be limited in certain cell types.
Fig. 1.
LXR activation suppresses the proliferation of certain cell lines. A: RWPE1, THP1, SNU16, LNCaP, and HepG2 cells were incubated with LXR ligand T0901317 (T) for two days, and microscopic images were taken. B: Similar experiments were performed with PC3, HEK293, and HeLa cells. C: RWPE1, LNCaP, and PC3 cells were treated with T0901317, and the proliferation rates were monitored by CCK-8 assay. CCK-8, Cell Counting Kit-8; LXR, liver X receptor.
Recent studies have reported that a high dose of T0901317 is able to unexpectedly stimulate other nuclear hormone receptors, such as FXR and PXR, although T0901317 is more potent in activating LXR (25, 26). To ascertain whether the inhibitory effect of T0901317 on cell proliferation is mediated by activated LXR, we used another LXR agonist, GW3965. GW3965 has been developed as a more specific ligand for LXR, and it hardly influences the transcriptional activities of other nuclear hormone receptors, including FXR and PXR (14). When GW3965 was treated to RWPE1 cells, cell proliferation was significantly suppressed in a dose-dependent manner (supplementary Fig. I). Similarly, we observed antiproliferative activity of GW3965 in LNCaP cells (supplementary Fig. II), suggesting that activation of LXR would repress cell growth and proliferation in certain cells.
LXR activation induces G1/S arrest
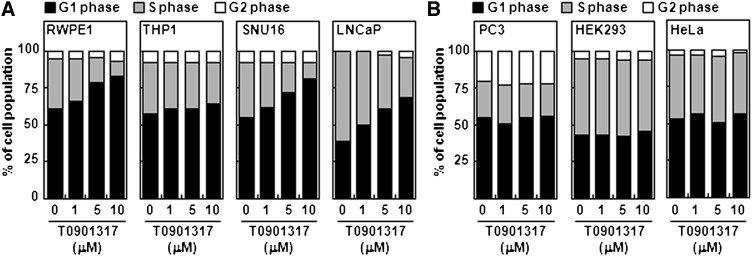
Because LXR activation mitigates the proliferation rate in certain cells, we further investigated whether LXR activation would modulate cell cycle progression. When we analyzed the cell cycle distribution via flow cytometry analysis, we discovered that LXR ligand T0901317 increased the population in the G1 phase and decreased the population in the S phase in RWPE1, THP1, SNU16, and LNCaP cells (Fig. 2A). Both T0901317 and GW3965 arrested LNCaP cells at G1/S boundary in a dose-dependent manner (supplementary Fig. II-C). However, LXR activation did not change the cell cycle status in PC3, HEK293, or HeLa cells (Fig. 2B). These results suggest that activation of LXR in certain cell types would prevent cell cycle progression into the S phase by inducing G1/S arrest, eventually leading to downregulation of cell proliferation in certain cell types.
Fig. 2.
LXR activation induces G1 arrest in certain cell lines. A: RWPE1, THP1, SNU16, and LNCaP cells were treated with T0901317 at the indicated concentrations for one day. After harvesting, cells were trypsinized and subjected to FACS analysis as described in Materials and Methods. The percentage of cells in each phase was calculated with the ModFit LTTM analysis program. B: Similar to (A), PC3, HEK293, and HeLa cells were treated with T0901317, and FACS analyses were carried out. Similar experiments were repeated three times, and representative results are presented. LXR, liver X receptor.
Differential regulation of target gene expression by LXR
Because activation of LXR exhibited antiproliferative effects in certain cells, but not in all cell types, we hypothesized that activated LXR might differentially regulate target gene expression depending on cell type. This idea was explored by examining the patterns of target gene expression on LXR ligands in various cells.
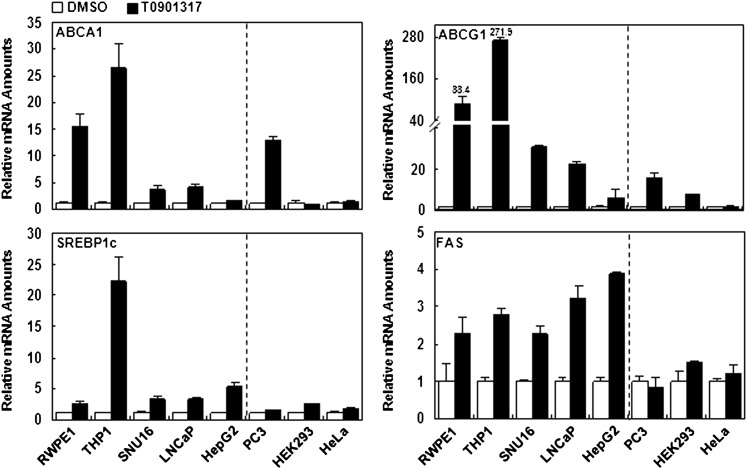
In RWPE1, THP1, SNU16, LNCaP, and HepG2 cells whose proliferations were sensitively suppressed by activated LXR, most LXR target genes, including ABCA1, ABCG1, SREBP1c, and FAS, were significantly upregulated by the LXR ligand. However, in HEK293 and HeLa cells, expression of LXR target genes was barely changed by LXR activation (Fig. 3). Interestingly, although proliferation of PC3 cells was not significantly altered by the LXR ligand, expression of ABCA1 and ABCG1 mRNAs was greatly stimulated by the LXR ligand in PC3 cells. However, the levels of SREBP1c and FAS mRNA were not significantly influenced by the LXR ligand in PC3 cells. These results imply that expression pattern of fatty acid metabolism-related genes, such as SREBP1c and FAS, appears to be more tightly associated with the suppression of cell proliferation by LXR activation rather than cholesterol metabolism-related genes.
Fig. 3.
LXR activation differentially affects the expression of LXR target genes. Each cell line was incubated with T0901317 (5 μM) for 18 h, and total RNA was isolated. After synthesizing cDNA from total RNA, the relative expression level of LXR target genes, such as ABCA1, ABCG1, SREBP1c, and FAS, was analyzed by real-time quantitative PCR. Each value was normalized with GAPDH. Fold induction of ABCG1 expression by T0901317 is noted. ABC, ATP-binding cassette; LXR, liver X receptor; SREBP1c, sterol regulatory element-binding protein 1c.
Mevalonate treatment does not affect ligand-induced cell cycle inhibition
LXR plays a key role in both cholesterol and fatty acid metabolism. Recently, it has been shown that LXR is able to repress T cell proliferation through regulation of LXR-dependent sterol metabolism (27). In T cells, induction of ABCG1 is essential for inhibitory effects of LXR, and treatment with mevalonate, a precursor for cholesterol, nullifies antiproliferative property of LXR activation.
Because proliferation of RWPE1 cells were sensitively repressed by LXR ligands, we treated RWPE1 cells with mevalonate in the absence or presence of LXR ligands, such as T0901317 and GW3965, to test whether control of cholesterol metabolism might be involved in the inhibitory effect of LXR activation on cell proliferation. Unlike T cells, mevalonate did not overcome the inhibitory effect of the LXR ligands on cell proliferation (Fig. 4A, C). Further, CCK-8 assays indicated that treatment of mevalonate did not reverse the cell cycle arrest by LXR ligands, even in a high concentration (Fig. 4B, D). These data suggest that regulation of cholesterol metabolism by LXR activation might be independent of cell proliferation, except for T cells.
Fig. 4.
Mevalonate does not relieve the LXR-induced suppression of cell proliferation. RWPE1 cells were treated with mevalonate in the absence or presence of 3 μM T0901317 (A and B) and 2 μM GW3965 (C and D). After two days of incubation, microscopic pictures were taken, and CCK-8 assays were performed. CCK-8, Cell Counting Kit-8; GW, GW3965; LXR, liver X receptor; MVA, mevalonate; T, T0901317.
Control of lipogenesis is involved in antiproliferative effect of activated LXR
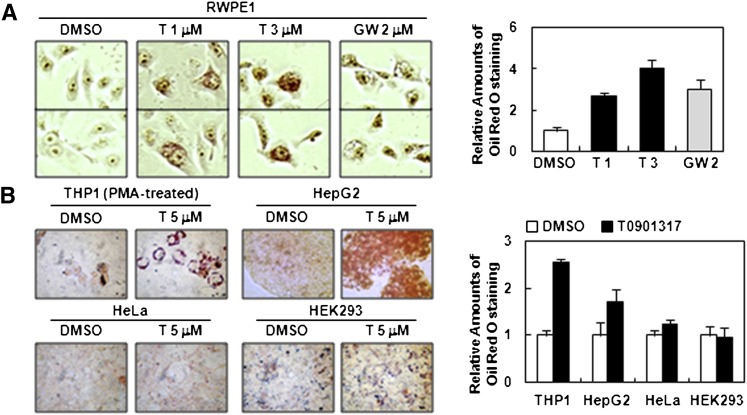
Next, we examined whether lipogenic activity of activated LXR is linked with an antiproliferative property. In RWPE1 cells, both T0901317 and GW3965 treatment greatly promoted positive cells stained with Oil Red O (Fig. 5A), reflecting the induction of de novo fatty acid synthesis by elevating the levels of accumulated neutral lipids, such as triacylglycerides and cholesterol esters. Similarly, other cell lines, including THP1 and HepG2, remarkably increased triacylglyceride accumulation upon LXR activation (Fig. 5B). On the contrary, HeLa and HEK293 cells, which are insensitive cell types for the antiproliferative action of stimulated LXR, did not elevate lipid accumulation (Fig. 5B), suggesting that lipogenic activity induced by LXR activation appears to play a key role in the antiproliferative effect.
Fig. 5.
Activation of LXR leads to lipid accumulation in RWPE1, THP1, and HepG2 cells. A: RWPE1 cells were treated with T0901317 (1 and 3 μM) and GW3965 (2 μM). After two days of incubation, cells were fixed and intracellular triacylglycerides were visualized with Oil Red O staining (see Materials and Methods). B: THP1, HepG2, HeLa, and HEK293 cells were treated with T0901317 and subjected to Oil Red O staining. Attachment of THP1 cells to the culture dish was induced by the treatment of phorbol 12-myristate 13-acetate (PMA, 0.1 μM) for 12 h. Accumulated lipid amount was quantified by ImageJ software. GW, GW3965; LXR, liver X receptor; T, T0901317.
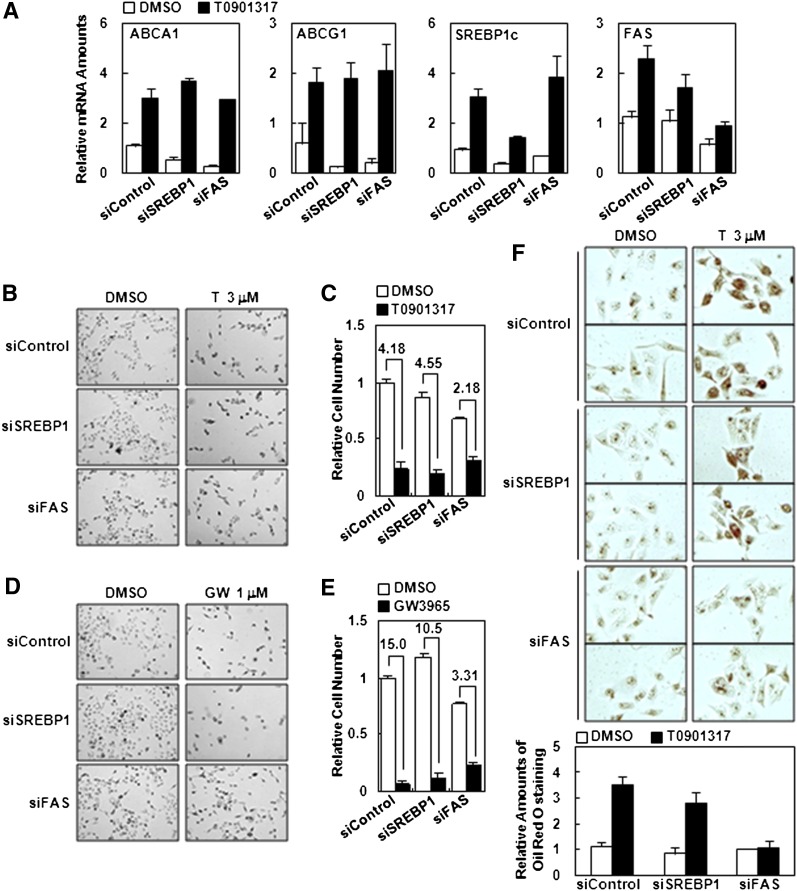
To mediate lipogenic stimulation upon LXR activation, it is well known that there are two crucial LXR target genes. One is SREBP1c which is a key transcription factor for several downstream lipogenic genes, such as FAS, ACC, and SCD1, as well as insulin signaling (28–31). The other is FAS, which directly facilitates the synthesis of long chain fatty acids, and known to be modulated in both LXR-dependent and LXR-independent manners (11). To test whether SREBP1c and/or FAS might be a key mediator for cell cycle arrest by LXR activation, we suppressed these genes via siRNA. Each RNAi construct exhibited ∼40–50% decrease of SREBP1 and FAS mRNA levels (Fig. 6A and supplementary Fig. III). Although suppression of SREBP1 via siRNA (hSREBP1_2394) did not significantly affect the antiproliferative property of activated LXR, suppression of FAS via siRNA (hFAS_6089) partially, but substantially, recovered the cell proliferation activity in the presence of LXR ligands ( ). Consistent with these results, the population of positive cells stained with Oil Red O was decreased in FAS-knockdown cells, but not significantly in SREBP1c-suppressed cells (Fig. 6F). Further, when other siRNA against SREBP1 (hSREBP1_3278 and hSREBP1_2493) and FAS (hFAS_6124 and hFAS_6428) were tested on target gene expression, cell proliferation, and lipid accumulation, we observed similar results (supplementary Fig. III). These data imply that FAS might be more crucial to mediate the effect of activated LXR-dependent cell cycle arrest in certain cells. Despite these data, we could not completely exclude the possibility that SREBP1c is not involved in activated LXR-dependent antiproliferation due to the technical limitation of achieving sufficient suppression of SREBP1 via siRNA.
Fig. 6.
Effect of SREBP1 and FAS knockdown on activated LXR-induced cell cycle arrest. A: RWPE1 cells were transfected with siRNA (control), SREBP1 (hSREBP1_2394), and FAS (hFAS_6089), and then treated with T0901317 (1 μM) for 18 h. Total RNA was isolated, and target gene expression was analyzed by real-time quantitative PCR. Transfected RWPE1 cells were incubated with 3 μM of T0901317 (B and C) and 1 μM of GW3965 (D and E). After two days of incubation, microscopic images were taken, and relative cell numbers were determined by CCK-8 assay. C and E: Fold of inhibition. F: Transfected RWPE1 cells with each siRNA were incubated with 3 μM T0901317 for two days, and Oil Red O staining was carried out. Accumulated lipid amount was quantified by ImageJ software. GW, GW3965; LXR, liver X receptor; siRNA, small-interference RNA; SREBP, sterol regulatory element-binding protein; T, T0901317.
Expression level of cyclin A was decreased by LXR ligands
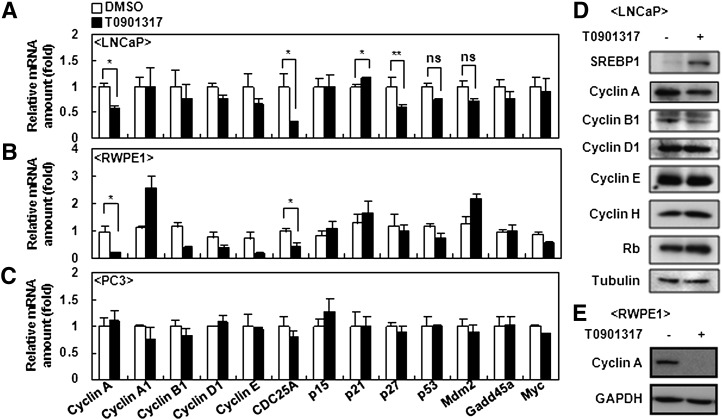
Cell cycle regulation is affected by key cell cycle-related genes, such as cyclins and cdk inhibitors. To investigate the molecular mechanism of LXR-dependent cell cycle arrest in selective cells, we intensively examined the levels of several cyclins, cdc25a phosphatase, cdk inhibitors, cell cycle-related transcription factors, and their target genes by analyses with cDNA microarrays and qPCR. For assessing gene expression profiles, we primarily compared LNCaP with PC3 cells, because these cells represent activated LXR-sensitive and LXR-insensitive antiproliferation, respectively. The mRNA level of cyclin A and cdc25a were selectively decreased by activated LXR in LNCaP cells without significant changes in mRNA levels of other cyclins, such as A1, B1, D1, and E, whereas none of them were altered in PC3 cells (Fig. 7A, C). In accordance with these findings, the protein level of cyclin A was decreased by the LXR ligand in LNCaP cells, but the protein level was not decreased in other cyclins, such as B1, D1, E, and H (Fig. 7D). Simultaneously, when we examined the expression level of cdk inhibitor genes, such as p15, p21, and p27, there were minor changes in mRNA level of LNCaP cells. To ascertain these data, we also examined gene expression profiles in RWPE1 cells and obtained results similar to LNCaP cells (Fig. 7B, E). These findings indicate that cyclin A and cdc25a might be potential candidates to mediate activated LXR-induced antiproliferation in certain cell types.
Fig. 7.
Expression profiles of cell cycle-regulating genes in LNCaP and PC3 cells. LNCaP(A), RWPE1(B), and PC3(C) cells were incubated with 5 μM T0901317 for 18 h, and total RNA were isolated. After synthesizing cDNA from total RNA, mRNA expression levels of cyclin A, cyclin A1, cyclin B1, cyclin D1, cyclin E, cdc25a, p15, p21, p27, p27, p53, mdm2, gadd45a, and myc were analyzed by real-time quantitative PCR and normalized with GAPDH control. LNCaP(D) and RWPE1(E) cells were treated with 5 μM T0901317 for one day, and total protein was subjected to Western blotting analysis. *P < 0.05; **P = 0.146; ns, not significant.
DISCUSSION
In this study, we investigated the inhibitory effect of LXR activation on cell proliferation and cell cycle progression. We revealed that liganded LXR attenuated proliferation of certain cells through G1/S arrest, whereas the proliferation of other cell lines, such as PC3, HEK293, and HeLa, was not affected by LXR stimulation (Figs. 1 and 2). Gene expression pattern analyses in particular indicated that expression of lipogenic genes was closely associated with the antiproliferative effect of activated LXR (Fig. 3). Furthermore, knockdown of FAS partly relieved activated LXR-induced cell growth arrest (Fig. 6). However, the exact mechanism(s) involved in LXR-dependent cell cycle attenuation remains to be further investigated.
Interestingly, some of our data suggest that the regulation of fatty acid metabolism by LXR activation contributes to cell cycle inhibition. First, PC3, HEK293, and HeLa cells, which are insensitive to activated LXR-dependent antiproliferative effect, failed to induce expression of lipogenic genes such as SREBP1c and FAS, even though they were able to promote expression of cholesterol efflux genes (Fig. 3). Second, the number of Oil Red O-stained positive cells in the presence of LXR ligands was only elevated in activated LXR-dependent cell cycle-arrested cells (Fig. 5). Third, treatment of mevalonate, which is a precursor of cholesterol, did not rescue the RWPE1 cells from activated LXR-induced cell cycle arrest (Fig. 4). Lastly, suppression of FAS via siRNA partially diminished the inhibitory effect of activated LXR on cell proliferation (Fig. 6), implying that upregulation of FAS might be important for activated LXR-induced cell cycle arrest, at least in part. To ascertain whether inhibition of FAS would relieve LXR-induced cell cycle arrest, we treated the cells with cerulenin, a well-known FAS inhibitor. However, we failed to optimize experimental conditions because cerulenin exhibited potent cytotoxicity, which is well-known in many cells (32–35). Consistently, previous reports have suggested that cerulenin would be a lead compound for an anticancer drug due to its potent antiproliferative property (32–35).
It appears that LXR-dependent antiproliferation is regulated by several mechanisms based on cell type and context. Consistent with our data, recent studies have reported that LXR is involved in antiproliferation in several cells (17–21). In T cells, LXR activation inhibits cell proliferation via cholesterol metabolism (27). In addition, LXR affects cell cycle arrest by increasing p27 in smooth muscle cells, pancreatic β-cells, and prostate cancer cells (17, 21, 36). In this study, we did not observe upregulation of p27 mRNA or protein in the presence of LXR ligand (Fig. 7; data not shown). Rather, we noticed that increase of de novo fatty acid synthesis appeared to be crucial for cell cycle arrest, supported by the results of differential regulation of LXR target genes and Oil Red O staining as well as FAS knockdown experiments. It is well-established that elevated fatty acid level confers lipotoxicity and even cell death, concomitant with G1/S arrest in certain cells (37–39). For instance, RWPE1, macrophage, and pancreatic β-cells are very active for lipid uptake and synthesis (37, 40, 41), and uncontrolled lipogenic activity leads to antiproliferation and apoptosis in these cells (data not shown). Therefore, it is feasible that dysregulated lipid accumulation accompanied by increased lipogenic activity would be one of key mechanisms to mediate the antiproliferative effect of LXR activation.
The antiproliferative property of activated LXR is also linked to decreased levels of cyclin A and cdc25a. Cyclin A is essential for controlling DNA replication during the S phase and timing mitosis entry during the G2 phase (42–44). Accordingly, the level of cyclin A is elevated in a variety of tumors, such as breast cancer, leukemia, and prostate cancer (42). It remains to be investigated whether there is any correlation between lipotoxicity and cyclin A regulation upon LXR activation in certain cells. Cdc25a is another cell cycle-regulating gene encoding a tyrosine phosphatase, which is involved in the regulation of G1/S transition by activating cyclin A/Cdk2 and cyclin E/Cdk2 complexes through removal of inhibitory phosphorylations (45, 46). Thus, it is possible that reduction of cdc25a also contributes to the inhibition of cell cycle progression at the G1/S checkpoint upon LXR activation. However, the exact mechanism for cell cycle inhibition by LXR activation remains to be elucidated.
To explore the inhibition of cell proliferation by LXRα and LXRβ, we examined the expression levels of LXRα and LXRβ in various cell lines (supplementary Fig. IV). Because most cells expressed substantial amounts of both LXRα and LXRβ mRNA, we did not find any correlation between LXRα or LXRβ expression and cell cycle inhibition. Additionally, we investigated the effect of LXRα and LXRβ suppression via siRNA in RWPE1 cells. Neither LXRα nor LXRβ knockdown in RWPE1 cells failed to relieve antiproliferation in the presence of LXR ligands. Although we tested several LXRα and LXRβ siRNA constructs (hLXRα_934, hLXRα_542, hLXRβ_508, and hLXRβ_1176), the knockdown efficiency of LXRα and LXRβ siRNA was only ∼40–50%, respectively (supplementary Fig. V). Moreover, combined treatment of LXRα and LXRβ siRNA did not improve the technical limitations of individual LXR knockdown (data not shown). When we examined the antiproliferative effect of LXR activation in mouse embryonic fibroblasts (MEF) from LXRα-knockout (αKO), LXRβ-knockout (βKO), and LXRα/β-double knockout (DKO) mice, LXR activation with its ligands did not suppress proliferation rates in those MEF (supplementary Fig. VI-A). Under the same conditions, we did not detect any dramatic change of lipid contents on LXR ligands in MEF (data not shown). However, we noted that expression of several LXR target genes was upregulated by LXR ligand in wild-type and LXRα KO MEF (supplementary Fig. VI-B). Thus, even though we propose that LXR activation with elevated lipogenic activity appears to play a role in mediating antiproliferation in certain cell types, future studies are essential to clarify whether LXRα and/or LXRβ is crucial for LXR-dependent antiproliferation.
Taken together, we propose that LXR may play a role in the regulation of cell proliferation as well as energy homeostasis. LXR activation actively attenuates cell cycle progression in certain cell types, such as RWPE1, THP1, SNU16, LNCaP, and HepG2, but not in others, such as PC3, HEK293, and HeLa cells. Further, we found that the inhibition of cell proliferation is associated with the lipogenic activity of LXR. These results provide new insight for the role of LXR on cell cycle control with lipogenic activity.
Supplementary Material
Footnotes
Abbreviations:
- ABC
- ATP-binding cassette
- apo
- apolipoprotein
- CCK-8
- Cell Counting Kit-8
- COX2
- cyclooxygenase 2
- CYP7A1
- cholesterol 7 alpha-hydroxylase
- FXR
- farnesoid X receptor
- IL6
- interleukin-6
- LPS
- lipopolysaccharide
- LXR
- liver X receptor
- MEF
- mouse embryonic fibroblast
- iNOS
- inducible nitric oxide synthase
- PI
- propodium idodide
- PXR
- pregnane X receptor
- siRNA
- small-interference RNA
- SREBP
- sterol regulatory element-binding protein
This work was supported by Stem Cell Research Center Grant SC-3230 of the First Century Frontier Research Program; National Research Foundation Grant 2009-0091913 from the Ministry of Education, Science and Technology (MEST) of Korea; Research Center for Functional Cellulomics Grant 2010-0001492 of the Science Research Center Program; and World Class University Project Grant R31-2009-000-100320.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and six figures.
REFERENCES
- 1.Steffensen K. R., Gustafsson J. A. 2004. Putative metabolic effects of the liver X receptor (LXR). Diabetes. 53(Suppl. 1): S36–S42. [DOI] [PubMed] [Google Scholar]
- 2.Annicotte J. S., Schoonjans K., Auwerx J. 2004. Expression of the liver X receptor alpha and beta in embryonic and adult mice. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 277: 312–316. [DOI] [PubMed] [Google Scholar]
- 3.Janowski B. A., Willy P. J., Devi T. R., Falck J. R., Mangelsdorf D. J. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 383: 728–731. [DOI] [PubMed] [Google Scholar]
- 4.Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., Mangelsdorf D. J. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 93: 693–704. [DOI] [PubMed] [Google Scholar]
- 5.Repa J. J., Mangelsdorf D. J. 2002. The liver X receptor gene team: potential new players in atherosclerosis. Nat. Med. 8: 1243–1248. [DOI] [PubMed] [Google Scholar]
- 6.Naik S. U., Wang X., Da Silva J. S., Jaye M., Macphee C. H., Reilly M. P., Billheimer J. T., Rothblat G. H., Rader D. J. 2006. Pharmacological activation of liver X receptors promotes reverse cholesterol transport in vivo. Circulation. 113: 90–97. [DOI] [PubMed] [Google Scholar]
- 7.Plosch T., Kok T., Bloks V. W., Smit M. J., Havinga R., Chimini G., Groen A. K., Kuipers F. 2002. Increased hepatobiliary and fecal cholesterol excretion upon activation of the liver X receptor is independent of ABCA1. J. Biol. Chem. 277: 33870–33877. [DOI] [PubMed] [Google Scholar]
- 8.Ohashi R., Mu H., Yao Q., Chen C. 2004. Cellular and molecular mechanisms of atherosclerosis with mouse models. Trends Cardiovasc. Med. 14: 187–190. [DOI] [PubMed] [Google Scholar]
- 9.Tangirala R. K., Bischoff E. D., Joseph S. B., Wagner B. L., Walczak R., Laffitte B. A., Daige C. L., Thomas D., Heyman R. A., Mangelsdorf D. J., et al. 2002. Identification of macrophage liver X receptors as inhibitors of atherosclerosis. Proc. Natl. Acad. Sci. USA. 99: 11896–11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darimont C., Avanti O., Zbinden I., Leone-Vautravers P., Mansourian R., Giusti V., Mace K. 2006. Liver X receptor preferentially activates de novo lipogenesis in human preadipocytes. Biochimie. 88: 309–318. [DOI] [PubMed] [Google Scholar]
- 11.Joseph S. B., Laffitte B. A., Patel P. H., Watson M. A., Matsukuma K. E., Walczak R., Collins J. L., Osborne T. F., Tontonoz P. 2002. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 277: 11019–11025. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa T., Shimano H., Amemiya-Kudo M., Yahagi N., Hasty A. H., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., et al. 2001. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell. Biol. 21: 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe S. S., Choi A. H., Lee J. W., Kim K. H., Chung J. J., Park J., Lee K. M., Park K. G., Lee I. K., Kim J. B. 2007. Chronic activation of liver X receptor induces beta-cell apoptosis through hyperactivation of lipogenesis: liver X receptor-mediated lipotoxicity in pancreatic beta-cells. Diabetes. 56: 1534–1543. [DOI] [PubMed] [Google Scholar]
- 14.Collins J. L., Fivush A. M., Watson M. A., Galardi C. M., Lewis M. C., Moore L. B., Parks D. J., Wilson J. G., Tippin T. K., Binz J. G., et al. 2002. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J. Med. Chem. 45: 1963–1966. [DOI] [PubMed] [Google Scholar]
- 15.Joseph S. B., Castrillo A., Laffitte B. A., Mangelsdorf D. J., Tontonoz P. 2003. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat. Med. 9: 213–219. [DOI] [PubMed] [Google Scholar]
- 16.Joseph S. B., Bradley M. N., Castrillo A., Bruhn K. W., Mak P. A., Pei L., Hogenesch J., O'Connell R. M., Cheng G., Saez E., et al. 2004. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 119: 299–309. [DOI] [PubMed] [Google Scholar]
- 17.Blaschke F., Leppanen O., Takata Y., Caglayan E., Liu J., Fishbein M. C., Kappert K., Nakayama K. I., Collins A. R., Fleck E., et al. 2004. Liver X receptor agonists suppress vascular smooth muscle cell proliferation and inhibit neointima formation in balloon-injured rat carotid arteries. Circ. Res. 95: e110–e123. [DOI] [PubMed] [Google Scholar]
- 18.Wente W., Brenner M. B., Zitzer H., Gromada J., Efanov A. M. 2007. Activation of liver X receptors and retinoid X receptors induces growth arrest and apoptosis in insulin-secreting cells. Endocrinology. 148: 1843–1849. [DOI] [PubMed] [Google Scholar]
- 19.Chuu C. P., Hiipakka R. A., Kokontis J. M., Fukuchi J., Chen R. Y., Liao S. 2006. Inhibition of tumor growth and progression of LNCaP prostate cancer cells in athymic mice by androgen and liver X receptor agonist. Cancer Res. 66: 6482–6486. [DOI] [PubMed] [Google Scholar]
- 20.Fukuchi J., Kokontis J. M., Hiipakka R. A., Chuu C. P., Liao S. 2004. Antiproliferative effect of liver X receptor agonists on LNCaP human prostate cancer cells. Cancer Res. 64: 7686–7689. [DOI] [PubMed] [Google Scholar]
- 21.Meng Z. X., Nie J., Ling J. J., Sun J. X., Zhu Y. X., Gao L., Lv J. H., Zhu D. Y., Sun Y. J., Han X. 2009. Activation of liver X receptors inhibits pancreatic islet beta cell proliferation through cell cycle arrest. Diabetologia. 52: 125–135. [DOI] [PubMed] [Google Scholar]
- 22.Choi M., Rolle S., Rane M., Haller H., Luft F. C., Kettritz R. 2003. Extracellular signal-regulated kinase inhibition by statins inhibits neutrophil activation by ANCA. Kidney Int. 63: 96–106. [DOI] [PubMed] [Google Scholar]
- 23.Wagner A. H., Gebauer M., Guldenzoph B., Hecker M. 2002. 3-hydroxy-3-methylglutaryl coenzyme A reductase-independent inhibition of CD40 expression by atorvastatin in human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 22: 1784–1789. [DOI] [PubMed] [Google Scholar]
- 24.Kim K. H., Choi S. H., Lee T. S., Oh W. K., Kim D. S., Kim J. B. 2006. Selective LXRalpha inhibitory effects observed in plant extracts of MEH184 (Parthenocissua tricuspidata) and MEH185 (Euscaphis japonica). Biochem. Biophys. Res. Commun. 349: 513–518. [DOI] [PubMed] [Google Scholar]
- 25.Houck K. A., Borchert K. M., Hepler C. D., Thomas J. S., Bramlett K. S., Michael L. F., Burris T. P. 2004. T0901317 is a dual LXR/FXR agonist. Mol. Genet. Metab. 83: 184–187. [DOI] [PubMed] [Google Scholar]
- 26.Mitro N., Vargas L., Romeo R., Koder A., Saez E. 2007. T0901317 is a potent PXR ligand: implications for the biology ascribed to LXR. FEBS Lett. 581: 1721–1726. [DOI] [PubMed] [Google Scholar]
- 27.Bensinger S. J., Bradley M. N., Joseph S. B., Zelcer N., Janssen E. M., Hausner M. A., Shih R., Parks J. S., Edwards P. A., Jamieson B. D., et al. 2008. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 134: 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J. B., Sarraf P., Wright M., Yao K. M., Mueller E., Solanes G., Lowell B. B., Spiegelman B. M. 1998. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J. Clin. Invest. 101: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J. B., Spiegelman B. M. 1996. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 10: 1096–1107. [DOI] [PubMed] [Google Scholar]
- 30.Tontonoz P., Kim J. B., Graves R. A., Spiegelman B. M. 1993. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13: 4753–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim K. H., Song M. J., Yoo E. J., Choe S. S., Park S. D., Kim J. B. 2004. Regulatory role of glycogen synthase kinase 3 for transcriptional activity of ADD1/SREBP1c. J. Biol. Chem. 279: 51999–52006. [DOI] [PubMed] [Google Scholar]
- 32.Menendez J. A., Vellon L., Mehmi I., Oza B. P., Ropero S., Colomer R., Lupu R. 2004. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in cancer cells. Proc. Natl. Acad. Sci. USA. 101: 10715–10720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thupari J. N., Pinn M. L., Kuhajda F. P. 2001. Fatty acid synthase inhibition in human breast cancer cells leads to malonyl-CoA-induced inhibition of fatty acid oxidation and cytotoxicity. Biochem. Biophys. Res. Commun. 285: 217–223. [DOI] [PubMed] [Google Scholar]
- 34.Kuhajda F. P., Pizer E. S., Li J. N., Mani N. S., Frehywot G. L., Townsend C. A. 2000. Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proc. Natl. Acad. Sci. USA. 97: 3450–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okawa Y., Hideshima T., Ikeda H., Raje N., Vallet S., Kiziltepe T., Yasui H., Enatsu S., Pozzi S., Breitkreutz I., et al. 2008. Fatty acid synthase is a novel therapeutic target in multiple myeloma. Br. J. Haematol. 141: 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Chuu C. P., Kokontis J. M., Hiipakka R. A., Liao S. 2007. Modulation of liver X receptor signaling as novel therapy for prostate cancer. J. Biomed. Sci. 14: 543–553. [DOI] [PubMed] [Google Scholar]
- 37.Aronis A., Madar Z., Tirosh O. 2008. Lipotoxic effects of triacylglycerols in J774.2 macrophages. Nutrition. 24: 167–176. [DOI] [PubMed] [Google Scholar]
- 38.Malhi H., Bronk S. F., Werneburg N. W., Gores G. J. 2006. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J. Biol. Chem. 281: 12093–12101. [DOI] [PubMed] [Google Scholar]
- 39.Feldstein A. E., Werneburg N. W., Canbay A., Guicciardi M. E., Bronk S. F., Rydzewski R., Burgart L. J., Gores G. J. 2004. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha expression via a lysosomal pathway. Hepatology. 40: 185–194. [DOI] [PubMed] [Google Scholar]
- 40.Wrede C. E., Dickson L. M., Lingohr M. K., Briaud I., Rhodes C. J. 2002. Protein kinase B/Akt prevents fatty acid-induced apoptosis in pancreatic beta-cells (INS-1). J. Biol. Chem. 277: 49676–49684. [DOI] [PubMed] [Google Scholar]
- 41.Bollheimer L. C., Skelly R. H., Chester M. W., McGarry J. D., Rhodes C. J. 1998. Chronic exposure to free fatty acid reduces pancreatic beta cell insulin content by increasing basal insulin secretion that is not compensated for by a corresponding increase in proinsulin biosynthesis translation. J. Clin. Invest. 101: 1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yam C. H., Fung T. K., Poon R. Y. 2002. Cyclin A in cell cycle control and cancer. Cell. Mol. Life Sci. 59: 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Boer L., Oakes V., Beamish H., Giles N., Stevens F., Somodevilla-Torres M., Desouza C., Gabrielli B. 2008. Cyclin A/cdk2 coordinates centrosomal and nuclear mitotic events. Oncogene. 27: 4261–4268. [DOI] [PubMed] [Google Scholar]
- 44.Girard F., Strausfeld U., Fernandez A., Lamb N. J. 1991. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 67: 1169–1179. [DOI] [PubMed] [Google Scholar]
- 45.Katich S. C., Zerfass-Thome K., Hoffmann I. 2001. Regulation of the Cdc25A gene by the human papillomavirus Type 16 E7 oncogene. Oncogene. 20: 543–550. [DOI] [PubMed] [Google Scholar]
- 46.Sexl V., Diehl J. A., Sherr C. J., Ashmun R., Beach D., Roussel M. F. 1999. A rate limiting function of cdc25A for S phase entry inversely correlates with tyrosine dephosphorylation of Cdk2. Oncogene. 18: 573–582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.