Abstract
Low density lipoprotein is a heterogeneous group of lipoproteins that differs in lipid and protein composition. One copy of apolipoprotein (apo)B accounts for over 95% of the LDL protein, but the presence of minor proteins could disturb its biological behavior. Our aim was to study the content of minor proteins in LDL subfractions separated by anion exchange chromatography. Electropositive LDL [LDL(+)] is the native form, whereas electronegative LDL [LDL(−)] is a minor atherogenic fraction present in blood. LC-ESI MS/MS analysis of both LDL fractions identified up to 28 different proteins. Of these, 13 proteins, including apoB, were detected in all the analyzed samples. LDL(−) showed a higher content of most minor proteins. Statistical analysis of proteomic data indicated that the content of apoE, apoA-I, apoC-III, apoA-II, apoD, apoF, and apoJ was higher in LDL(−) than in LDL(+). Immunoturbidimetry, ELISA, or Western blot analysis confirmed these differences. ApoJ and apoF presented the highest difference between LDL(+) and LDL(−) (>15-fold). In summary, the increased content of several apolipoproteins, and specifically of apoF and apoJ, could be related to the physicochemical characteristics of LDL(−), such as apoB misfolding, aggregation, and abnormal lipid composition.
Keywords: apolipoproteins, atherosclerosis, modified LDL
LDL, the main cholesterol carrier in plasma, is composed of approximately 75% lipid (mainly cholesterol) and 25% protein. The major protein in LDL is apolipoprotein B-100 (apoB-100), a protein of 550 kDa that accounts for more than 95% of the total protein mass in LDL (1). However, minor amounts of other apolipoproteins associated to LDL have been reported. Several of these proteins, such as apoE, apoC-III, and platelet-activating factor acetylhydrolase (PAF-AH), have important roles in LDL metabolism and modulate its atherogenicity despite their low concentration (2–4). Several studies have made a proteomic approach using 2D-electrophoresis or SELDI-based analysis to detect minor proteins in LDL (5–9), including apoE, apoC-III, apoC-II, apoA-I, apoA-IV, apoM, apoJ, serum amyloid A4 (SAA4), calgranulin A, lysozyme C, apoD, apoH, α1-antitrypsin, orosomucoid-1, paraoxonase-1, retinol binding protein, and prenylcysteine lyase-1.
LDL is not a homogeneous entity but a group of particles that differs in density, size, electric charge, and composition. It is well established that small, dense LDL particles are more atherogenic than large, buoyant particles (10), and both of these subfractions differ in their protein content (6). Another property of LDL that confers enhanced atherogenicity is increased electronegative charge. Electronegative LDL [LDL(−)] is a minor subfraction of plasma LDL that is pro-inflammatory and induces apoptosis in endothelial cells and leukocytes (11–13). Its relative proportion is increased in subjects with a high cardiovascular risk (14, 15). Interestingly, LDL(−) has an increased content of several minor proteins, such as PAF-AH, apoE, apoC-III, and apoA-I, that could be involved in its inflammatory properties (11, 12, 16–19). Beyond these proteins, others, as yet unknown, are probably present in very small amounts in LDL(−). Our aim was to study differences in the content of minor proteins between native LDL [LDL(+)] and modified LDL [LDL(−)], using a gel free-approach based on liquid chromatography coupled to electrospray ionization tandem mass spectrometry (LC-ESI MS/MS).
METHODS
LDL subfraction isolation
Plasma samples were obtained after a 10 h overnight fast from healthy, normolipemic (total cholesterol < 5.5 mmol/l, triglyceride < 1 mmol/l, HDL cholesterol > 1 mmol/l), nonsmoking subjects from the hospital staff. All samples were obtained in fasting state (for at least 10 h) and body mass index of all subjects was < 25.5 kg/cm2. The study was approved by the institution's ethics committee, and all volunteers gave their informed consent. Total LDL (1.019 < d < 1.050 g/ml) was isolated from pooled plasma (at least 10 individuals in each pool with an equal plasma volume from each) by flotation sequential ultracentrifugation (20) at 4°C and in presence of 1 mM EDTA. Since LDL(−) accounts for less than 5% of total LDL, the use of pooled plasma was the only strategy that allowed us to obtain a sufficient amount of LDL(−) to perform proteomic analysis. Because the use of pooled plasma can mask inter-individual variability, we repeated the analysis using six independent pools to account for individual variability when comparing LDL to LDL(−) protein contents. Approximately 100–150 ml of plasma was used for each separation, and 60–80 mg apoB of total LDL was obtained (recovery 60–70%). LDL was then subfractionated into LDL(+) and LDL(−) by anion-exchange chromatography as previously described (17). LDL(−) proportion accounted for 4.5 ± 0.5%, and the recovery was approximately 50–55%, yielding a final amount of 1.5–2 mg apoB of LDL(−). Composition of LDL subfractions was assessed as previously described (11, 17). Major lipids, including total cholesterol, triglycerides (TG; Roche Diagnostics, Switzerland), phospholipids (Wako Chemicals, Germany), NEFA (Wako), and apoB (Roche Diagnostics, Switzerland) were measured by commercial methods in a Hitachi 911 autoanalyzer. Sphingomyelinase-modified LDL (SMase-LDL) was obtained by incubation of LDL(+) with SMase from Bacillus Cereus at 37°C for 2 h, as previously described (21).
Protein delipidation and solubilization
The method of Karlsson et al. (5) was used with minor modifications. Briefly, 1 ml of LDL (1 g/l apoB) was delipidated by mixing with 14 ml of ice-cold tributyl phosphate:acetone:methanol (1:12:1), incubating for 2 h at 4°C, and centrifuging for 15 min at 3000 g. Protein pellets were washed sequentially with 1 ml of tributyl phosphate, acetone, and methanol. Nitrogen-dried pellets were stored at –80°C until analysis (less than 1 month). Protein pellets were resuspended by intense vortexing and repeated aspiration using 400 μl of cold-buffer Tris 10 mM containing 6 M guanidinium chloride. The content of protein in the soluble material was measured by the BCA method (Pierce, Rockford, IL).
Tryptic protein digestion
The delipidated protein sample (50 μg) in 20 μl of 6 M guanidinium chloride was adjusted to pH 8.0 by adding NaOH mixed with 20 μl of a 150 mM solution of DTT in 50 μM ammonium bicarbonate and then incubated for 30 min at room temperature. An amount of 20 μl of a 500 mM solution of iodoacetamide in 50 μM ammonium bicarbonate was added, and the mixture was further incubated for 1 h at room temperature. The cysteine-modified protein sample was then subjected to TCA/acetone precipitation (CleanUp Kit, GE Healthcare, Fairfield, CT). The protein pellet was dissolved in 30 μl of 1 M urea in 50 mM ammonium bicarbonate solution, and 1 μg of modified porcine trypsin (Trypsin-Gold, Promega, Madison, WI) was added. Digestion proceeded overnight at 37°C, and then the mixture was acidified by addition of 0.1% formic acid and frozen at −20°C until analyzed.
Proteomic analysis
Proteomic analysis was performed by LC-ESI MS/MS using 5 μg of protein. Six independent experiments using six different pools were performed. LC-ESI MS/MS analysis was performed on an Esquire HCT ion trap mass spectrometer (Bruker, Bremen, Germany) coupled to a nanoHPLC system (Ultimate, LcPackings, Netherlands). Samples were first concentrated on a 300 µm i.d. 1 mm PepMap nanotrapping column and then loaded onto a 75 µm i.d. 15 cm PepMap nanoseparation column (LC Packings, Netherlands). Peptides were eluted by an acetonitrile gradient (gradient: 0–60% B in 105 min, B = 80% acetonitrile, 0.1% formic acid in water; flow rate ∼300 nl/min) through a PicoTip emitter nano-spray needle (New Objective, Woburn, MA) onto the nanospray ionization source of the ion-trap mass spectrometer. MS/MS fragmentation (1.9 s, m/z 100–2,800) was performed on two of the most intense ions, as determined from a 1.2 s MS survey scan (m/z 310–1,500) and using a dynamic exclusion time of 1.2 min for precursor selection. An automated optimization of MS/MS fragmentation amplitude, starting from 0.60 V, was used. Proteins were identified using Mascot (Matrix Science, London, UK) to search the UniProt-SwissProt 57.0 human database. MS/MS spectra were searched with a precursor mass tolerance of 1.5 Da, fragment tolerance of 0.5 Da, trypsin specificity with a maximum of two missed cleavages, carbamidomethylation set as fixed modification, and methionine oxidation as variable modification. The positive identification criterion was set as an individual Mascot score for each peptide MS/MS spectrum higher than the corresponding homology threshold score.
Quantification of proteins
The content of non-apoB proteins in each LDL subfraction was calculated from the number of MS/MS spectra matching peptides corresponding to each protein detected (spectral count), divided by each protein molecular weight (number of peptides/Mw) to give a percentage molar content relative to the total non-apoB proteins. These values were averaged for all the samples analyzed of each of the two LDL fractions. To compare the relative levels of the non-apoB proteins between the two fractions, the values were normalized according to their total apoB content (there is one molecule of apoB per LDL particle) and divided by the protein molecular weight (number of peptides/Mw) to give a relative molar content. Results were expressed as arbitrary units of each protein after correction for apoB content.
Quantification of apolipoproteins by immunoturbidimetry
ApoA-I (Roche Diagnostics, Basel, Switzerland), apoA-II, apoE, apoC-III, and apoC-II (Kamiya, Seattle, WA) were quantified manually in LDL subfractions using commercial reagents for Hitachi, as follows: 100 μl of reagent 1 (Tris buffer) was mixed with 100 μl of LDL at 0.4 g/l and incubated at 37°C for 30 min. Afterwards, 50 μl of reagent 2 (antiserum) was added and incubated at 37°C for 30 min. Absorbance was measured in a microtiter plate reader at the wavelength recommended by the manufacturer. A standard curve was performed with an apolipoprotein multi-calibrator (Wako Chemicals, Neuss, Germany) diluted from 1/5 to 1/100. ApoJ was quantified by commercial ELISA (ALPCO Immunoassays, Salem, NH) using 1/20 dilution of LDL at 0.5 g/l apoB.
Western blot analysis
The content of apoJ and apoF in LDL subfractions was determined by Western blot. LDL subfractions (25 μg apoB/well) were submitted to SDS-PAGE electrophoresis in 12% commercial gels (BioRad, Hercules, CA). Electrophoresis was performed at 100 V for 2 h at 4°C. Proteins were transferred to a nitrocellulose membrane (1 h at 30 V, in Tris-glycine buffer containing 20% methanol and 0.1% SDS) and blocked overnight at 4°C with TTBS (Tris 20 mM, NaCl 150 mM, 0.1% Tween 20, pH 7.5) containing 2.5% nonfat milk. Western blot was performed using goat polyclonal antibody IgG anti-apoJ (PA1-26903, Affinity BioReagents, Golden, CO) or anti-apoF (C-13, Santa Cruz Biotechnology, Santa Cruz, CA) as a primary antibody (dilution 1/200 or 1/500 in TTBS containing 5% nonfat milk, 90 min). An HRP-conjugated anti-goat IgG (Jackson ImmunoResearch, Baltimore, PA) was used as a secondary antibody (dilution 1/1000 for apoJ or 1/10000 for apoF in TTBS containing 5% nonfat milk, 60 min). Nitrocellulose was revealed with Immun-Star HRP substrate kit (BioRad).
The association of apoA-I to LDL subfractions was studied by native gradient gel acrylamide electrophoresis (GGE) followed by Western blot. GGE was performed in 2–16% gradient gels, as described (19). In GGE, lipoproteins were run through the gel as complete particles. Lipoproteins were transferred to nitrocellulose, and Western blot was performed using a goat polyclonal antibody IgG anti-apoA-I (Acris, Hiddenhausen, Germany) as a primary antibody (dilution 1/1,000, 90 min) and HRP-conjugated anti-goat IgG as a secondary antibody (dilution 1/2,000, 60 min). Nitrocellulose was revealed with Immun-Star developing kit (BioRad).
Cholesteryl ester transfer protein activity
Cholesteryl ester transfer protein (CETP) activity was measured as the transfer of labeled cholesteryl oleate from HDL to LDL(+) or LDL(−) using lipoproteins isolated from pools of human normolipemic individuals and cynomolgus monkey CETP-Tg mice (line UCTP-20) as source of CETP previously described (22). Rates of labeled cholesteryl oleate transfer from HDL to LDL were linear up to 1,500 μM/h.
Statistical analysis
Results are expressed as mean ± SD. SPSS 11.5.2 statistical package was used. Differences between LDL subfractions were tested with Wilcoxon's t-test for paired data. A P value < 0.05 was considered significant.
RESULTS
Lipid and protein composition of LDL fractions was similar to that previously described (10, 16). LDL(−) had higher triglyceride and NEFA content and lesser apoB content than LDL(+) (Table 1). The total protein content recovered after delipidation was very low, 5.3 ± 2.6% in LDL(+) and 8.8 ± 4.6% in LDL(−). This was due to the high hydrophobicity of apoB, which is poorly solubilized.
TABLE 1.
Lipid and apoB composition of LDL subfractions
| LDL(+) | LDL(−) | |
|---|---|---|
| Cholesterol (%) | 41.4 ± 2.8 | 41.7 ± 3.3 |
| Triglyceride (%) | 6.9 ± 1.7 | 9.5 ± 2.3a |
| Phospholipid (%) | 26.8 ± 3.1 | 25.7 ± 2.7 |
| NEFA (mol/mol apoB) | 15.6 ± 3.4 | 27.8 ± 6.7a |
| ApoB (%) | 24.9 ± 2.0 | 23.1 ± 2.6a |
Apo, apolipoprotein.
P < 0.05 versus LDL(+) using the Wilcoxon t-test.
Six independent LC ESI-MS/MS experiments were performed using LDL subfractions isolated from six independent plasma pools from different subjects. A total of 28 proteins, including apoB, were detected in at least one of the experiments (Table 2). The details on protein identification, including the different peptide sequences identified for each protein, are given in supplementary Tables I and II [proteins in LDL(−) and proteins in LDL(+)]. Fourteen proteins were observed in three or fewer experiments in very low concentrations (1–3 peptides detected; supplementary Tables I and II). Most of these proteins are involved in coagulation, inflammation, or innate immunity. Thirteen proteins other than apoB were detected in five or six experiments, in one or both samples. These proteins were quantified relative to apoB, and statistical analysis indicated that the content of apoE, apoA-I, apoC-III, apoA-II, apoD, apoF, and apoJ was higher in LDL(−) than in LDL(+) (Table 3). Apo(a) was detected only in LDL(−), whereas platelet basic protein content was found only in LDL(+). No significant differences were observed in apoC-I, apoC-II, albumin, and SAA4, although a trend to an increased content of all these proteins, with the exception of apoC-I, was observed in LDL(−)compared with LDL(+).
TABLE 2.
Summary of proteins detected in LDL subfractions
| LDL(+) | LDL(−) | ||
|---|---|---|---|
| Protein | Number of experiments in which a protein was detected | Function | |
| Apolipoprotein B | 6 | 6 | Lipid transport, receptor binding |
| Apolipoprotein E | 6 | 6 | Receptor binding |
| Apolipoprotein A-I | 6 | 6 | Lipid transport, receptor binding |
| Apolipoprotein C-III | 6 | 6 | Lipid metabolism |
| Apolipoprotein D | 5 | 6 | Lipid metabolism |
| Apolipoprotein A-II | 5 | 6 | Lipid metabolism |
| Apolipoprotein F (Lipid transfer inhibitor protein) | 2 | 6 | Lipid metabolism |
| Apolipoprotein J (Clusterin) | 1 | 6 | Lipid metabolism, inflammation, apoptosis |
| Apolipoprotein C-I | 6 | 5 | Lipid metabolism |
| Albumin | 5 | 5 | Blood homeostasis |
| Serum amyloid A-4 protein | 5 | 5 | Innate immunity |
| Apolipoprotein C-II | 5 | 5 | Lipid metabolism |
| Apolipoprotein(a) | 0 | 5 | Lipid transport |
| Platelet basic protein (NAP2, CXCL7) | 5 | 0 | Inflammation |
| Apolipoprotein A-IV | 3 | 3 | Lipid metabolism |
| Apolipoprotein C-IV precursor | 2 | 3 | Lipid metabolism |
| Prothrombin precursor (Fragment) | 3 | 2 | Coagulation |
| Platelet factor 4 variant precursor (CXCL4) | 1 | 2 | Inflammation |
| Dermcidin precursor | 3 | 1 | Innate immunity |
| SH3 and cysteine-rich domain-containing protein | 1 | 1 | Cell senescence |
| Protein S100-A9 (Calgranulin B) | 1 | 1 | Inflammation |
| Platelet-activating factor acetylhydrolase precursor | 0 | 1 | Inflammation |
| Coagulation factor X precursor | 0 | 1 | Coagulation |
| Chromosome 1 open reading frame 56 | 0 | 1 | Unknown |
| IG Lambda chain | 0 | 1 | Immunity |
| Apolipoprotein-L1 | 1 | 0 | Innate immunity |
| Serum amyloid A2 | 1 | 0 | Innate immunity |
| Fibrinogen α chain | 1 | 0 | Coagulation, acute phase response |
| Antibacterial protein FALL-39 | 1 | 0 | Innate immunity |
TABLE 3.
Relative content of proteins detected by LC-ESI MS/MS
| Protein | LDL(+) | LDL(−) | Ratio (−)/(+) |
|---|---|---|---|
| Apolipoprotein E | 42 ± 16 | 217 ± 51a | 5.2 |
| Apolipoprotein A-I | 39 ± 27 | 146 ± 32a | 3.8 |
| Apolipoprotein C-III | 90 ± 55 | 374 ± 278a | 4.2 |
| Apolipoprotein C-I | 76 ± 50 | 74 ± 54 | 1.0 |
| Apolipoprotein C-II | 28 ± 34 | 49 ± 49 | 1.8 |
| Apolipoprotein A-II | 29 ± 9 | 141 ± 56a | 4.8 |
| Apolipoprotein D | 26 ± 15 | 61 ± 24a | 2.4 |
| Apolipoprotein F (inhibitor CETP) | 2 ± 3 | 63 ± 26a | 31.5 |
| Apolipoprotein J (Clusterin) | 1 ± 1 | 11 ± 5a | 11.0 |
| Apolipoprotein(a) | — | 6 ± 2 | — |
| Albumin | 5 ± 4 | 11 ± 10 | 2.2 |
| Serum amyloid A-4 protein | 15 ± 13 | 32 ± 24 | 2.1 |
| Platelet basic protein | 1 ± 1 | — | — |
Five or six experiments were conducted. Values are mean ± SD expressed in arbitrary molar units. CETP, cholesteryl ester transfer protein.
P < 0.05 versus LDL(+) using the Wilcoxon t-test.
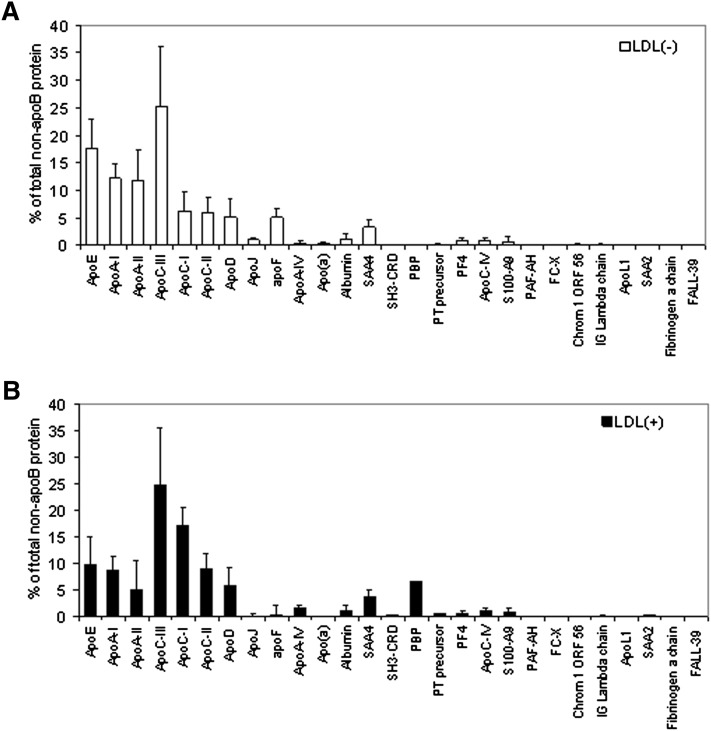
The most abundant proteins (expressed in a molar basis) in both LDL fractions (higher than 8% of minor proteins) were apoC-III > apoE > apoA-I > apoA-II in LDL(−) (Fig. 1A) and apoC-III > apoC-I > apoE > apoC-II > apoA-I in LDL(+) (Fig. 1B). They accounted for more than 85% of minor proteins. Although the error of the quantification method used is relatively high (see error bars in Fig. 1 and SD values in Table 3), the analysis allowed the identification of some statistically significant major differences between the two LDL types.
Fig. 1.
Relative content of minor proteins in LDL(−) (upper panel) and LDL(+) (lower panel). Data are expressed as mean ± SD of the percentage of total non-apoB protein on a molar basis (n = 6). apo, apolipoprotein; LDL(+), electropositive low-density lipoprotein; LDL(−), electronegative low-density lipoprotein.
To confirm the increased content in LDL(−) of some apolipoproteins detected by proteomic analysis, apoA-I, apoA-II, apoE, apoC-III, and apoC-II were quantified in LDL subfractions by immunoturbidimetry, adapting commercially available methods to detect very low concentrations. Table 4 shows the content of these apolipoproteins in LDL(−). The ratio between LDL(+) and LDL(−) was similar to that observed by proteomic analysis (Table 3), and the absolute amount of apoC-III and apoE in both LDL subfractions concurred with previously reported data (11, 17).
TABLE 4.
Content of apolipoproteins quantified by immunoturbidimetry or ELISA in LDL subfractions
| Apolipoprotein | LDL(+) | LDL(−) | Ratio (−)/(+) | Ratio from LC-ESI MS/MS |
|---|---|---|---|---|
| ApoA-I (mmol/mol apoB) | 68 ± 15 | 233 ± 17a | 3.4 | 3.8 |
| ApoA-II (mmol/mol apoB) | 10 ± 8 | 56 ± 12a | 5.4 | 4.8 |
| ApoE (mmol/mol apoB) | 23 ± 21 | 93 ± 30a | 4.0 | 5.2 |
| ApoC-III (mmol/mol apoB) | 49 ± 27 | 210 ± 145a | 4.3 | 4.2 |
| ApoC-II (mmol/mol apoB) | 33 ± 5 | 43 ± 5 | 1.3 | 1.8 |
| ApoJ (mmol/mol apoB) | 1.6 ± 1.5 | 10.2 ± 8.4 | 6.4 | 11.0 |
All apolipoproteins were quantified by commercial immunoturbidimetric methods as described in “Methods,” except apoJ, which was measured by ELISA. Values are mean ± SD expressed in mmol/mol apoB. Apo, apolipoprotein.
P < 0.05 versus LDL(+) using the Wilcoxon t-test.
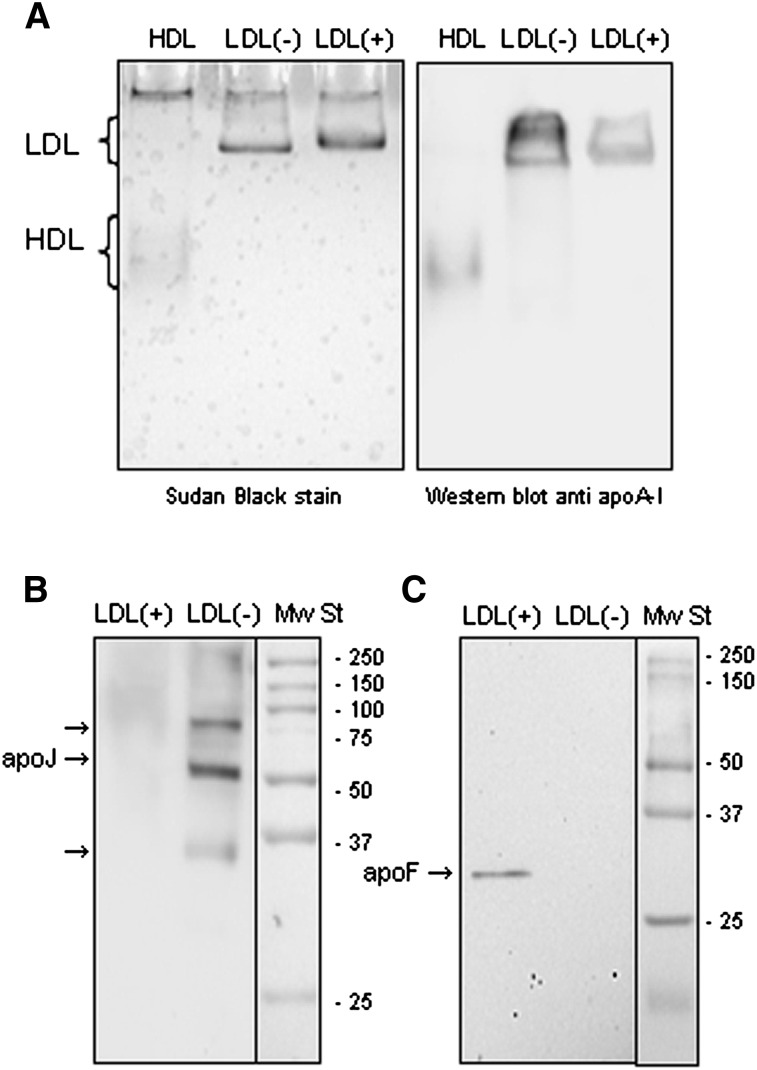
Several of the apolipoproteins detected in LDL subfractions are typical of high-density lipoproteins (HDL). This raises the question of whether LDL subfractions could be contaminated with HDL. To rule out this possibility, a Western blot was performed to detect apoA-I, the main protein constituent of HDL, in LDL electrophoresed under native conditions. Fig. 2A shows a representative GGE in which apoA-I was detected in the band corresponding to LDL size (24–28 nm). This band was much more intense in LDL(−) than in LDL(+). No apoA-I was observed in the region where HDL should be (size 8–15 nm).
Fig. 2.
Western blots of apoA-I (A), apoJ (B) and apoF (C). A: LDL(+) (20 μg apoB/lane), LDL(−) (20 μg apoB/lane) and HDL (1 μg apoA-I/lane) were submitted to GGE electrophoresis, and Western blot was performed as described in “Methods.” Right: Sudan Black staining (lipid staining). Left: anti apoA-I Western blot. B, C: LDL(+) (20 μg apoB/lane) and LDL(−) (20 μg apoB/lane) were submitted to SDS-PAGE in 12% acrylamide gels, and Western blots were performed as described in “Methods.” apo, apolipoprotein; GGE, non-denaturing gradient gel electrophoresis; LDL(+), electropositive low-density lipoprotein; LDL(−), electronegative low-density lipoprotein; Mw St, molecular weight standard.
ApoJ and apoF proteins presented the highest difference between LDL(+) and LDL(−). To confirm this different content, Western blots to detect apoF and apoJ were performed in both LDL subfractions. Fig. 2B and 2C show representative Western blots of LDL(+) and LDL(−) and confirm a much higher content of apoF and apoJ in LDL(−) than in LDL(+). With regard to apoF, an intense band of 33 kDa corresponding to apoF was detected in LDL(−), whereas no signal was observed in LDL(+) (Fig. 2C). With regard to apoJ, a major band of 75–80 kDa, corresponding to the highly glycosylated form of apoJ, and further minor bands of lower molecular weight, corresponding to deglycosylated and reduced forms (60 and 35 kDa), were detected (Fig. 2B). Indeed, apoJ content quantified by ELISA gave results comparable to those from the proteomic analysis (Tables 3 and 4) and confirmed the high content in LDL(−).
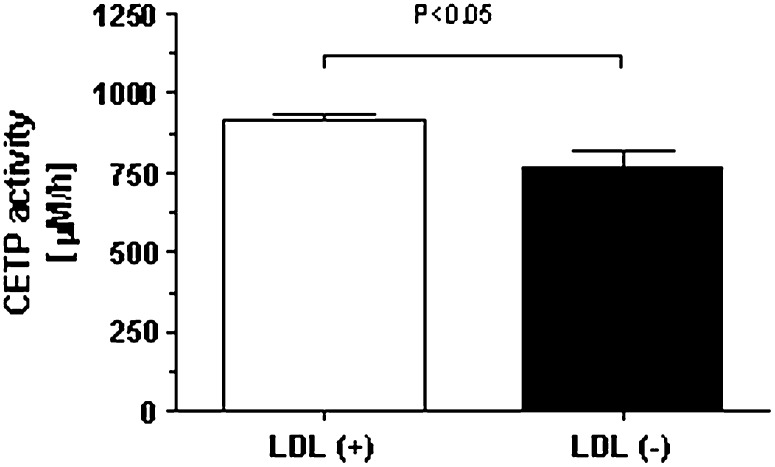
The role of apoF in LDL(−) is unclear, but it could be involved the abnormal lipid composition of this subfraction. The function of apoF was evaluated by measuring the activity of CETP, as apoF is a specific inhibitor of such lipid transfer activity. Fig. 3 shows that, compared with LDL(+), LDL(−) presents a lower ability to accept labeled cholesteryl esters (CE) from HDL.
Fig. 3.
In vitro effect of LDL(+) and LDL(−) on CETP activity. Results are expressed as mean ± SD of six independent experiments using different pools of LDL isolated from normolipemic subjects. Significant difference between LDL subfractions is shown. apo, apolipoprotein; CETP, cholesteryl ester transfer protein; LDL(+), electropositive low-density lipoprotein; LDL(−), electronegative low-density lipoprotein.
DISCUSSION
Current results show that a number of plasma proteins can bind to LDL in circulation. Five of these proteins (apolipoproteins AI, A-II, C-II, C-III, and E), whose amounts were relatively high were also quantified by immunoturbidimetry in both LDL subfractions. Albeit in a lower concentration, other proteins that were always detected in one or both LDL fractions were apoC-I, apoD, apoJ, apoF, SAA4, albumin, apo(a), and platelet basic protein. With the exception of the platelet basic protein, the presence of all these proteins has been previously described in LDL (4–9).
Several other proteins were detected in three or fewer experiments in the current study. Some of them, such as apoC-IV, apoA-IV, PAF-AH, calgranulin, and SAA2, have been previously reported to be associated with lipoproteins (3–9, 23). Others are known to be involved in innate immunity, inflammation, and coagulation. It has been reported that acute phase proteins and coagulation factors bind to lipoproteins (24). It is difficult to appraise the relevance of these proteins in LDL function, but it is probably low. The exception is PAF-AH, as LDL is its main transporter in plasma (4) and this protein plays a major role in the pro/anti-inflammatory balance induced by LDL (16). Interestingly, none of the previous proteomic studies detected PAF-AH in LDL, even though 70% of plasma PAF-AH is attached to LDL and, preferentially, to LDL(−) (4). The high affinity binding between PAF-AH and apoB (25) could explain why PAF-AH was detected only in one sample in the present study and never in previous studies using a 2-DE proteomic approach. It is possible that most PAF-AH was lost during sample treatment due to its strong attachment to the highly insoluble apoB.
The variable presence of some proteins is due to limitations in the proteomic analysis. Differences in the protein content observed in the current and previous studies should be attributed to differences in the isolation and delipidation methods. Ultracentrifugation could result in the loss of some proteins from LDL due to high shear forces and ionic strength. The interaction of apolipoproteins and enzymes with lipoproteins varies throughout their lifetime, and most of these proteins bind only temporarily to LDL. Thus, if a protein is detected in all analyzed samples, it can be assumed that it—at least for a short period of time—forms part of some LDL particles and could have a role in their metabolic processing in blood.
Current data confirm that LDL(−) has a higher content of minor proteins than LDL(+). The content of most of the analyzed proteins was higher in LDL(−), although statistical differences were achieved only in seven proteins. In addition to the previously described higher content of apoE, apoC-III, and apoA-I in LDL(−) (11, 12, 17, 26), our results indicate that apo(a), apoD, apoJ, apoF, and apoA-II were also significantly increased in LDL(−). Some of these proteins (apoA-I, apoA-II, apoD, and apoJ) are commonly bound to HDL particles. The presence in LDL of proteins commonly bound to HDL has been reported (3, 23), and it could be argued that the presence of these proteins in LDL is due to the presence of contaminating HDL. However, the likelihood of contamination is very low because the density range chosen in the present study to isolate LDL (1.019–1.050 g/ml) also contributed to minimizing the possibility of contamination. To rule out this possibility, Western blot analysis to detect apoA-I after native GGE demonstrated that apoA-I was specifically bound to LDL.
The increased content of apo(a) is due to contamination with Lp(a) during LDL isolation. Because the density ranges of LDL (1.019–1.063 g/ml) and Lp(a) (1.050–1.100 g/ml) partially overlap, LDL was isolated in the density range 1.019–1.050 g/ml in the present study. Despite this, minor contamination was always detected in total LDL. Lp(a) content, measured by commercial immunoturbidimetry method (Roche) in all LDL preparations, was always lower than 2%, which is in good agreement with results of proteomic analysis. Apo(a) has a very high content of sialic acid, which confers enhanced electronegativity to the Lp(a) particle. Consequently, contamination of Lp(a) in total LDL always leads to its coelution with LDL(−). However, the content of apo(a) in LDL(−) was lower than 2% (Fig. 1).
In our opinion, apoJ and apoF could play a key role in the function of LDL(−), as both proteins were detected in much higher concentrations in LDL(−) than in LDL(+). The biological characteristics of these proteins could explain some properties of LDL(−). ApoJ, also known as clusterin, has been involved in ageing, cancer, diabetes, kidney and neuron degeneration, and vascular damage. However, mechanisms involved in these pathologic processes are not well understood, as apoJ shows multiple functions, including apoptosis inhibition (27) and regulation of the complement complex (28). Besides, it has been reported that apoJ inhibits the oxidative modification of LDL (29). The association of apoJ with LDL(−), but not LDL(+), could be explained by its recently described chaperone function. ApoJ is the first identified extracellular chaperone, and it binds to a wide variety of partially unfolded proteins that present hydrophobic regions (30), including amyloid-like structures (31). The binding of apoJ to unfolded proteins promotes its recognition by LDL-receptor-related protein 2 (LRP2) and favors its clearance and degradation (30). Interestingly, LDL(−) presents some structural abnormalities, including particle aggregation (32) and apoB misfolding (33, 34), that could be related to the presence of apoJ in this subfraction.
In contrast with apoJ, apoF (also called lipid transfer inhibitor protein) has a well-defined function as a physiological regulator of CETP (35). ApoF is mainly associated with LDL, and its inhibitory action is due to the suppression of the interaction between CETP and LDL (36). CETP mediates the flux of CE and TG between lipoproteins. LDL gives CE to and accepts TG from VLDL. In turn, LDL accepts CE from and gives TG to HDL. The fact that most apoF is bound to LDL(−) suggests that lipid exchange between this LDL subfraction and other lipoproteins could be impaired. Our data show that lipid exchange between HDL and LDL(−) is abnormally low due to the high content of apoF. As the preferred substrate of CETP in plasma is HDL (36), this impairment could be involved with the increased relative content of triglyceride in LDL(−). Indeed, the increased content of apoC-III, the physiologic inhibitor of lipoprotein lipase (LpL), in combination with similar apoC-II, the physiologic activator of LpL, could also be related with increased triglyceride content of LDL(−).
In summary, our results show a number of proteins that are associated with LDL subfractions, some of which have not been identified previously. LDL(−) presents a higher content of most of these proteins than does LDL(+). The increased content of some of these proteins, such as apoC-III, apoE, and apoA-I, has been described previously in LDL(−), but increases in apoA-II, apoD, apoJ, and apoF in LDL(−) are described here for the first time. ApoF and apoJ, whose content in LDL(−) is much higher than in LDL(+), are of particular interest as they could be involved in some of the atherogenic properties of LDL(−). However, further studies are needed to determine the exact roles that apoF and apoJ could play in the abnormal characteristics of LDL(−).
Supplementary Material
Acknowledgments
The authors thank Carolyn Newey for editorial assistance.
Footnotes
Abbreviations:
- apo
- apolipoprotein
- BHT
- butylated hydroxytoluene
- CE
- cholesteryl ester
- CETP
- cholesteryl ester transfer protein
- GGE
- non-denaturing gradient gel electrophoresis
- LDL(+)
- electropositive low-density lipoprotein
- LDL(−)
- electronegative low-density lipoprotein
- Lp(a)
- lipoprotein (a)
- LRP2
- low-density lipoprotein receptor-related protein 2
- PAF-AH
- platelet-activating factor-acetylhydrolase
- SAA4
- serum amyloid A4
- TG
- triglyceride
- TTBS
- Tween-containing tris buffer saline
This work was supported by Ministerio de Sanidad/Instituto de Salud Carlos III/FIS (ISCIII/FIS) Grants PI060500, CP040110 (S.B.), and CP060220 (JLS-Q.); and Ministerio de Educación y Ciencia Grant AP2004-1468 (C.B). The Proteomics Laboratory at Vall d'Hebron Research Institute is a member of the Instituto Nacional de Proteómica (ProteoRed) funded by Fundación Genoma España. CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM) is a project of the ISCIII/FIS.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables.
REFERENCES
- 1.Segrest J. P., Jones M. K., De Loof H., Dashti N. 2001. Structure of apolipoprotein B-100 in low density lipoproteins. J. Lipid Res. 42: 1346–1367. [PubMed] [Google Scholar]
- 2.Alaupovic P., Knight-Gibson C., Wang C. S., Downs D., Koren E., Brewer H. B., Jr., Gregg R. E. 1991. Isolation and characterization of an apoA-II-containing lipoprotein (LP-A-II:B complex) from plasma very low density lipoproteins of patients with Tangier disease and type V hyperlipoproteinemia. J. Lipid Res. 32: 9–19. [PubMed] [Google Scholar]
- 3.Fruchart J. C. 1990. Lipoprotein heterogeneity and its effect on apolipoprotein assays. Scand. J. Clin. Lab. Invest. Suppl. 198: 51–57. [PubMed] [Google Scholar]
- 4.Stafforini D. M., McIntyre T. M., Zimmerman G. A., Prescott S. M. 1997. Platelet-activating factor acetylhydrolases. J. Biol. Chem. 272: 17895–17898. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson H., Leanderson P., Tagesson C., Lindahl M. 2005. Lipoproteomics I: mapping of proteins in low-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 5: 551–565. [DOI] [PubMed] [Google Scholar]
- 6.Davidsson P., Hulthe J., Fagerberg B., Olsson B. M., Hallberg C., Dahllof B., Camejo G. 2005. A proteomic study of the apolipoproteins in LDL subclasses in patients with the metabolic syndrome and type 2 diabetes. J. Lipid Res. 46: 1999–2006. [DOI] [PubMed] [Google Scholar]
- 7.Stahlman M., Davidsson P., Kanmert I., Rosengren B., Boren J., Fagerberg B., Camejo G. 2008. Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/sucrose or in KBr. J. Lipid Res. 49: 481–490. [DOI] [PubMed] [Google Scholar]
- 8.Banfi C., Brioschi M., Barcella S., Wait R., Begum S., Galli S., Rizzi A., Tremoli E. 2009. Proteomic analysis of human low-density lipoprotein reveals the presence of prenylcysteine lyase, a hydrogen peroxide-generating enzyme. Proteomics. 9: 1344–1352. [DOI] [PubMed] [Google Scholar]
- 9.Sun H. Y., Chen S. F., Lai M. D., Chang T. T., Chen T. L., Li P. Y., Shieh D. B., Young K. C. 2010. Comparative proteomic profiling of plasma very-low-density and low-density lipoproteins. Clin. Chim. Acta. 411: 336–344. [DOI] [PubMed] [Google Scholar]
- 10.Krauss R. M. 1995. Dense low density lipoproteins and coronary artery disease. Am. J. Cardiol. 75: 53B–57B. [DOI] [PubMed] [Google Scholar]
- 11.De Castellarnau C., Sanchez-Quesada J. L., Benitez S., Rosa R., Caveda L., Vila L., Ordonez-Llanos J. 2000. Electronegative LDL from normolipemic subjects induces IL-8 and monocyte chemotactic protein secretion by human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 20: 2281–2287. [DOI] [PubMed] [Google Scholar]
- 12.Chen H. H., Hosken B. D., Huang M., Gaubatz J. W., Myers C. L., Macfarlane R. D., Pownall H. J., Yang C. Y. 2007. Electronegative LDLs from familial hypercholesterolemic patients are physicochemically heterogeneous but uniformly proapoptotic. J. Lipid Res. 48: 177–184. [DOI] [PubMed] [Google Scholar]
- 13.Benitez S., Bancells C., Ordonez-Llanos J., Sanchez-Quesada J. L. 2007. Pro-inflammatory action of LDL(−) on mononuclear cells is counteracted by increased IL10 production. Biochim. Biophys. Acta. 1771: 613–622. [DOI] [PubMed] [Google Scholar]
- 14.Benitez S., Ordonez-Llanos J., Franco M., Marin C., Paz E., Lopez-Miranda J., Otal C., Perez-Jimenez F., Sanchez-Quesada J. L. 2004. Effect of simvastatin in familial hypercholesterolemia on the affinity of electronegative low-density lipoprotein subfractions to the low-density lipoprotein receptor. Am. J. Cardiol. 93: 414–420. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Quesada J. L., Perez A., Caixas A., Rigla M., Payes A., Benitez S., Ordonez-Llanos J. 2001. Effect of glycemic optimization on electronegative low-density lipoprotein in diabetes: relation to nonenzymatic glycosylation and oxidative modification. J. Clin. Endocrinol. Metab. 86: 3243–3249. [DOI] [PubMed] [Google Scholar]
- 16.Benitez S., Sanchez-Quesada J. L., Ribas V., Jorba O., Blanco-Vaca F., Gonzalez-Sastre F., Ordonez-Llanos J. 2003. Platelet-activating factor acetylhydrolase is mainly associated with electronegative low-density lipoprotein subfraction. Circulation. 108: 92–96. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez-Quesada J. L., Camacho M., Anton R., Benitez S., Vila L., Ordonez-Llanos J. 2003. Electronegative LDL of FH subjects: chemical characterization and induction of chemokine release from human endothelial cells. Atherosclerosis. 166: 261–270. [DOI] [PubMed] [Google Scholar]
- 18.Yang C. Y., Chen H. H., Huang M. T., Raya J. L., Yang J. H., Chen C. H., Gaubatz J. W., Pownall H. J., Taylor A. A., Ballantyne C. M., et al. 2007. Pro-apoptotic low-density lipoprotein subfractions in type II diabetes. Atherosclerosis. 193: 283–291. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Quesada J. L., Benitez S., Otal C., Franco M., Blanco-Vaca F., Ordonez-Llanos J. 2002. Density distribution of electronegative LDL in normolipemic and hyperlipemic subjects. J. Lipid Res. 43: 699–705. [PubMed] [Google Scholar]
- 20.Havel R. J., Eder H. A., Bragdon J. H. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oorni K., Hakala J. K., Annila A., Ala-Korpela M., Kovanen P. T. 1998. Sphingomyelinase induces aggregation and fusion, but phospholipase A2 only aggregation, of low density lipoprotein (LDL) particles. Two distinct mechanisms leading to increased binding strength of LDL to human aortic proteoglycans. J. Biol. Chem. 273: 29127–29134. [DOI] [PubMed] [Google Scholar]
- 22.Escola-Gil J. C., Julve J., Marzal-Casacuberta A., Ordonez-Llanos J., Gonzalez-Sastre F., Blanco-Vaca F. 2001. ApoA-II expression in CETP transgenic mice increases VLDL production and impairs VLDL clearance. J. Lipid Res. 42: 241–248. [PubMed] [Google Scholar]
- 23.Alaupovic P. 1991. Apolipoprotein composition as the basis for classifying plasma lipoproteins. Characterization of ApoA- and ApoB-containing lipoprotein families. Prog. Lipid Res. 30: 105–138. [DOI] [PubMed] [Google Scholar]
- 24.Chait A., Han C. Y., Oram J. F., Heinecke J. W. 2005. Thematic review series: the immune system and atherogenesis. Lipoprotein-associated inflammatory proteins: markers or mediators of cardiovascular disease? J. Lipid Res. 46: 389–403. [DOI] [PubMed] [Google Scholar]
- 25.Tjoelker L. W., Eberhardt C., Wilder C., Dietsch G., Trong H. L., Cousens L. S., Zimmerman G. A., McIntyre T. M., Stafforini D. M., Prescott S. M., et al. 1996. Functional and structural features of plasma platelet-activating factor acetylhydrolase. Adv. Exp. Med. Biol. 416: 107–111. [DOI] [PubMed] [Google Scholar]
- 26.Demuth K., Myara I., Chappey B., Vedie B., Pech-Amsellem M. A., Haberland M. E., Moatti N. 1996. A cytotoxic electronegative LDL subfraction is present in human plasma. Arterioscler. Thromb. Vasc. Biol. 16: 773–783. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H., Kim J. K., Edwards C. A., Xu Z., Taichman R., Wang C. Y. 2005. Clusterin inhibits apoptosis by interacting with activated Bax. Nat. Cell Biol. 7: 909–915. [DOI] [PubMed] [Google Scholar]
- 28.Chauhan A. K., Moore T. L. 2006. Presence of plasma complement regulatory proteins clusterin (Apo J) and vitronectin (S40) on circulating immune complexes (CIC). Clin. Exp. Immunol. 145: 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navab M., Anantharamaiah G. M., Reddy S. T., Van Lenten B. J., Wagner A. C., Hama S., Hough G., Bachini E., Garber D. W., Mishra V. K., et al. 2005. An oral apoJ peptide renders HDL antiinflammatory in mice and monkeys and dramatically reduces atherosclerosis in apolipoprotein E-null mice. Arterioscler. Thromb. Vasc. Biol. 25: 1932–1937. [DOI] [PubMed] [Google Scholar]
- 30.Lakins J. N., Poon S., Easterbrook-Smith S. B., Carver J. A., Tenniswood M. P., Wilson M. R. 2002. Evidence that clusterin has discrete chaperone and ligand binding sites. Biochemistry. 41: 282–291. [DOI] [PubMed] [Google Scholar]
- 31.Calero M., Rostagno A., Frangione B., Ghiso J. 2005. Clusterin and Alzheimer's disease. Subcell. Biochem. 38: 273–298. [PubMed] [Google Scholar]
- 32.Bancells C., Benitez S., Villegas S., Jorba O., Ordonez-Llanos J., Sanchez-Quesada J. L. 2008. Novel phospholipolytic activities associated with electronegative low-density lipoprotein are involved in increased self-aggregation. Biochemistry. 47: 8186–8194. [DOI] [PubMed] [Google Scholar]
- 33.Parasassi T., Bittolo-Bon G., Brunelli R., Cazzolato G., Krasnowska E. K., Mei G., Sevanian A., Ursini F. 2001. Loss of apoB-100 secondary structure and conformation in hydroperoxide rich, electronegative LDL(−). Free Radic. Biol. Med. 31: 82–89. [DOI] [PubMed] [Google Scholar]
- 34.Parasassi T., De Spirito M., Mei G., Brunelli R., Greco G., Lenzi L., Maulucci G., Nicolai E., Papi M., Arcovito G., et al. 2008. Low density lipoprotein misfolding and amyloidogenesis. FASEB J. 22: 2350–2356. [DOI] [PubMed] [Google Scholar]
- 35.Wang X., Driscoll D. M., Morton R. E. 1999. Molecular cloning and expression of lipid transfer inhibitor protein reveals its identity with apolipoprotein F. J. Biol. Chem. 274: 1814–1820. [DOI] [PubMed] [Google Scholar]
- 36.Morton R. E., Greene D. J. 1994. Regulation of lipid transfer between lipoproteins by an endogenous plasma protein: selective inhibition among lipoprotein classes. J. Lipid Res. 35: 836–847. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.