Abstract
Acid ceramidase (aCDase) is one of several enzymes responsible for ceramide degradation within mammalian cells. As such, aCDase regulates the intracellular levels of the bioactive lipid ceramide. An inherited deficiency of aCDase activity results in Farber disease (FD), also called lipogranulomatosis, which is characterized by ceramide accumulation in the tissues of patients. Diagnosis of FD is confirmed by demonstration of a deficient aCDase activity and the subsequent storage of ceramide. Existing methods include extremely complex assays, many of them using radiolabeled compounds. Therefore, the aCDase assay and the in vitro enzymatic diagnosis of FD are still performed in only a very limited number of specialized laboratories. Here, the new fluorogenic substrate Rbm14-12 was synthesized and characterized as a new tool to determine aCDase activity. The resulting optimized assay was performed in 96-well plates, and different fibroblast and lymphoid cell lines derived from FD patients and controls were tested to measure aCDase activity. As a result, the activity in cells of FD patients was found to be very low or even null. This new fluorogenic method offers a very easy and rapid way for specific and accurate determination of aCDase activity and, consequently, for diagnosis of FD.
Keywords: acid ceramidase, Farber disease, diagnosis, fluorogenic assay
Farber disease (FD) is a rare inherited lipid storage disorder, also known as lipogranulomatosis, which is characterized by accumulation of ceramide in the cells and tissues of patients (1, 2). This accumulation is the consequence of a deficient intracellular activity of aCDase, a lysosomal hydrolase encoded by the ASAH1 gene. The main clinical features include painful and progressively deformed joints, subcutaneous nodules, a hoarse cry due to laryngeal involvement, and premature death. Hepatosplenomegaly and nervous system dysfunction may also occur (1, 2). Although the pathogenesis of FD is still unclear in terms of the molecular lesions caused by ceramide storage, the involvement of aCDase deficiency is unquestionable. Recent interest in aCDase also stems from the fact that this enzyme appears to modulate cell functions by controlling the levels of ceramide and sphingoid bases, which are both considered as putative bioactive molecules (3).
Diagnosis of FD must be biochemically confirmed by the demonstration of deficient activity of aCDase, which can then be further documented by characterization of the ASAH1 molecular defects. Although aCDase is a very well known enzyme, existing methods for determining its activity and for FD diagnosis still exhibit many disadvantages. The methods that have been used for FD diagnostic purposes can be classified into three groups: i) aCDase enzymatic assays (4–16), classically performed with radiolabeled substrates (4–13), which are rather water-insoluble and require at least one detergent (see Table 1) ; ii) loading tests, consisting of the addition of exogenous radiolabeled sphingolipids, e.g., ceramide (17, 18), sulfatide (19, 20), or sphingomyelin (21), on cultured living cells and the study of their metabolism; and iii) determination of accumulated ceramide, either by sophisticated chromatographic methods (22, 23) or through the use of the diacylglycerol kinase assay in the presence of γ[32P]ATP to measure the radiolabeled ceramide 1-phosphate produced (24). Some of the main drawbacks of these methods are the use of radiolabeled compounds, most of them being no longer commercially available; the laborious process of separation and identification of reaction products; the need for specific instruments such as a radiochromatoscanner, HPLC, GC-MS or phosphorimager; the possibility of ceramide breakdown by nonlysosomal ceramidases; the requirement of large quantities of biological material; and the complex incubation mixture in aCDase activity assays. All these disadvantages make both the aCDase assay and the diagnosis of FD restricted to very few expert laboratories.
TABLE 1.
Comparison of enzyme assays for acid ceramidase
| Substrate | Detergent | Buffer | pH | Protein Amount (µg) | Incubation Time (h) | Detection Method | Km (µM) | Vmax(pmol/min/mg) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| [14C]C12, 16, 18 ceramide | Triton X-100 | Na acetate | 6 | 90–125 | 20 | Extraction+ TLC + imaging analyzer | – | – | 13 |
| [14C]C12, 18 ceramide | Triton X-100 + Tween20 + Na cholate | citrate/phosphate | 4.5 | 250–2500 | 5 | Extraction + TLC + radiochromatoscanner | 200 (for C12),1250 (for C18) | 330 (for C12),66 (for C18) | 6 |
| [3H]C16, 18 ceramide | Triton X-100 | Na acetate | 4.5 | 100–500 | 2 | Extraction +TLC + radiochromatoscanner | 110 | – | 10 |
| C12 ceramide | Triton X-100 + Tween20 + Na cholate + Nonidet P40 | Na acetate | 4.2 | 30 | 0.5 | Derivatization + HPLC | 149 | 2200 | 14 |
| NBD C12 ceramide | Triton X-100 | Na acetate | 4.5 | 50–100 | 0.5 | Extraction +TLC + fluorescence imager | 22.3 | 4.7 | 15 |
| BODIPY C12 ceramide | Triton X-100+ Na taurocholate | citrate/phosphate | 4.5 | 20 | 2 | HPLC | – | 28 | 16 |
| Rbm14-16 | Triton X-100 | Na acetate | 4.5 | 150 | 3 | microplate fluorescence reader | 113 | 3.6 | 25 |
| Rbm14-12 | none | Na acetate | 4.5 | 10–25 | 3 | microplate fluorescence reader | 25.9 | 334 | this paper |
Recently, our group published a versatile fluorimetric procedure to determine the activity of ceramidases by using a synthetic ceramide analog carrying a 2-oxo-2H-chromen-7-yloxy moiety in the CH3-terminal part of the sphingoid chain (25). This structure enables the release of umbelliferone after hydrolysis by ceramidase, periodate oxidation of the resulting aminodiol, and further β-elimination of the aldehyde oxidation product. The procedure provided an easy and useful assay that could be carried out in 96-well plate format for high-throughput screening of combinatorial libraries in the search of potential ceramidase inhibitors.
In order to improve the substrate specificity toward aCDase, we have synthesized several analogs of the original substrate that differ by their fatty acid chain length. From all the molecules tested as a substrate for aCDase, Rbm14-12, an analog that possesses a 12-carbon fatty acid chain length, proved to be more specific to determine aCDase activity. In this work, we have optimized the previous method using this new substrate. The high affinity and specificity for aCDase make this substrate very suitable for convenient determination of its activity and consequently, a very useful tool in FD diagnosis.
MATERIALS AND METHODS
Synthesis of substrates
The Rbm14 substrates were prepared by N-acylation of the coumarinic aminodiol under standard conditions using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, 1-hydroxybenzotriazole, and the fatty acid of choice. The long-chain base was prepared as reported (25) from Garner's aldehyde, which was transformed into tert-butyl (4S,2’S)-2,2-dimethyl-4-(oxiran-2-yl)oxazolidine-3-carboxylate as described (26). Treatment of the resulting epoxide with dimethylsulfonium methylide gave tert-butyl (4S,1’R)-4-(1-hydroxy-2-propenyl)-2,2-dimethyloxazolidine-3-carboxylate, which was hydroborated to afford tert-butyl (4S,1’R)-4-(1,3-dihydroxypropyl)-2,2-dimethyloxazolidine-3-carboxylate. Its primary mesyl ester, prepared by selective mesylation of the primary alcohol, was treated with the umbelliferone salt to give an adduct that afforded the corresponding aminodiol upon removal of the protective groups.
Details about synthesis and characterization of the compounds will be published elsewhere.
Cell lines
The cell lines used to characterize the substrate specificity of Rbm14-12 were FD1, a Farber disease fibroblast cell line with null aCDase activity, and FD1 AcCer10×, the same Farber cell line fully corrected due to overexpression of aCDase (27), The other Farber and control cell lines, whether skin fibroblasts or Epstein-Barr virus-transformed lymphoid cells, came from the Laboratoire de Biochimie Métabolique (Institut Fédératif de Biologie, CHU Purpan, Toulouse, France). Cells were cultured as previously described (24, 27).
Acid ceramidase enzyme assay
To measure aCDase activity, cultured cells were collected and washed twice with PBS. Cell pellets were resuspended in 100 µl of a 0.2 M sucrose solution and then they were sonicated. Cell homogenates were centrifuged at 15,000 g for 3 min. The supernatant was collected and used for protein quantification to work with equal amounts of protein. The enzymatic assay was carried out in 96-well plates. Briefly, each well contained a mixture of 74.5 µl of 25 mM sodium acetate buffer pH 4.5, 0.5 µl of a 4 mM Rbm14-12 substrate solution in ethanol (substrate final concentration 20 µM; ethanol final concentration 0.5%), and a fixed amount of protein (from 10 to 25 µg) in a volume of 25 µl of a 0.2 M sucrose solution. Negative control samples consisted in the same incubation mixture in the absence of protein extracts. The plate was incubated at 37°C for 3 h without agitation. Then, the enzymatic reaction was stopped by adding 50 µl of methanol and 100 µl of a 2.5 mg/ml NaIO4 fresh solution in 100 mM glycine/NaOH buffer pH 10.6 in each well. The plate was protected from light for 2 h and then the released fluorescence was quantified using a microplate fluorescence reader (λex 360 nm, λem 446 nm). The amount of umbelliferone released was calculated from the fluorescence intensity by using calibration curves with umbelliferone, in the range from 0 to 3000 pmol.
Lysosomal beta-galactosidase assay
The acid β-galactosidase activity was determined on the same cell lysates used for the measure of aCDase activity. This assay is based on a published fluorimetric method (28) with some modifications to adapt it to the 96-well plate format. Briefly, each well contained 20 µl of a 0.5 M sodium acetate buffer pH 4.5, 20 µl of a 20 mM EDTA solution, 40 µl of a 3 mM solution of 4-methylumbelliferyl-β-D-galactopyranoside (final concentration of 1.2 mM), 10 µl of lysate, and 10 µl of ultra pure water. After incubation at 37°C for 30 min, the reaction was stopped by the addition of 100 µl of a 100 mM glycine/NaOH buffer pH 10.6. The fluorescence released was quantified using a microplate fluorescence reader (λex 360 nm, λem 446 nm) as described above.
Effect of protein amount and incubation time
To determine the time-dependence of Rbm14-12 hydrolysis, the enzyme assay was carried out at different incubation times, ranging from 0.5 to 6 h. To determine the detection limit of the assay, we used variable amounts of protein extract, from 2.5 to 180 µg.
pH-dependence of substrate hydrolysis
The pH-dependence of Rbm14-12 hydrolysis was determined by performing the assay with protein extracts from three different cell lines with different degrees of aCDase activity: FD1 and FD1 AcCer10× fibroblasts, and normal lymphoid cells. Reactions were carried out in different buffers without detergents: glycine-HCl buffer (pH 2.5 to 3.0), sodium acetate buffer (pH 3.5 to 5.5), sodium phosphate buffer (pH 6.0 to 8.1) and glycine-NaOH buffer (pH 8.9 to 9.8).
Determination of kinetic constants
Kinetic constants were calculated from Lineweaver-Burk representations of at least five aCDase activity assays, using different concentrations of Rbm14-12 (from 2.5 to 80 µM), and in the presence of protein amounts from 10 to 25 µg.
RESULTS
Substrate specificity of acid ceramidase
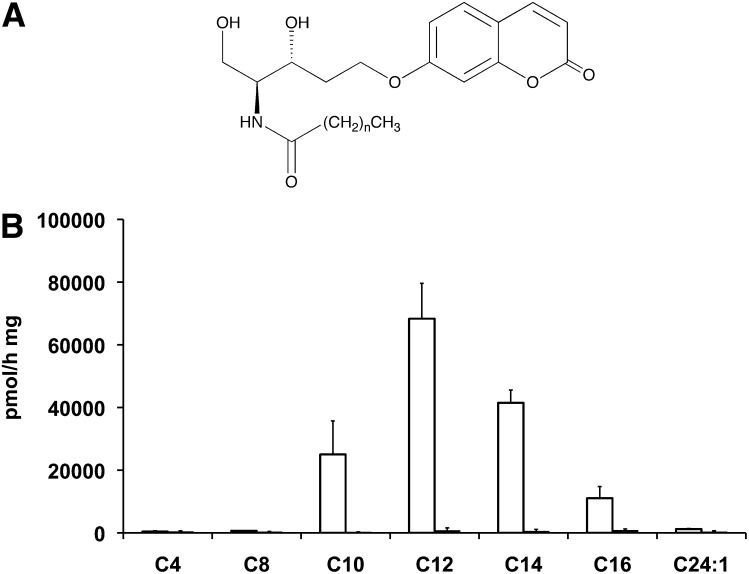
We tested the synthesized substrates on FD1 and FD1 AcCer10× cell lysates in order to check whether these substrates are hydrolyzed by and specific for aCDase. Under the same incubation conditions, we found that among the seven substrates tested, only four, i.e., Rbm14-10, 12, 14 and 16 were hydrolyzed by lysates of cells overexpressing AcCer (Fig. 1). These results agree with the previously reported fatty acid chain specificity of aCDase. In addition, the lack of detectable substrate hydrolysis by FD lysates indicates the specificity of these substrates for aCDase. The aCDase specific activity obtained using the already published compound Rbm14-16 showed that, among all the molecules tested, this analog exhibited the lowest affinity for aCDase, whereas Rbm14-12 showed the highest hydrolysis rates, up to 7 times those of the C16 homolog. Taking into account these results, the enzymatic assay for aCDase was optimized using Rbm14-12 as a novel fluorogenic substrate.
Fig. 1.
A: General chemical structure of Rbm14 compounds, (e.g., for Rbm14-12, n = 10). B: Comparison of the hydrolysis rates of the seven substrate analogs. The ceramide analogs (all at 40 µM) with the indicated side chain length were incubated for 3 h in the presence of FD1 AcCer10× (empty bars) or FD1 (solid bars) cell lysates (20 µg protein) in the absence of detergents. The activities measured in FD1 cells (solid bars) are too low to be seen. Results are the means of three independent experiments.
Enzyme assay optimization
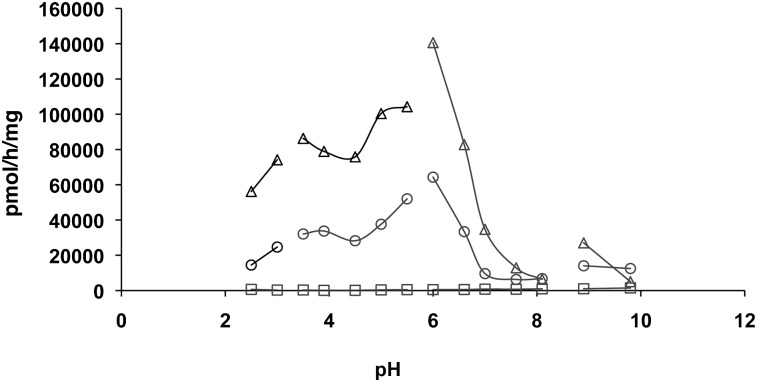
We investigated the pH dependence of Rbm14-12 hydrolysis in FD1 AcCer10×, but also in FD1 fibroblasts and normal lymphoid cells to further test the substrate specificity for aCDase. Using the FD1 cell line, we found no hydrolysis of the substrate at any pH tested, whereas the FD1 AcCer 10× cells hydrolyzed Rbm14-12 at acidic pH with optimal activity around pH 6 (Fig. 2). Normal lymphoid cells showed the same pattern as FD1 AcCer10× fibroblasts but exhibited lower activity levels. Although enzyme activity peaked at pH 6, comparable results, albeit with lower fluorescence intensity values, were obtained at pH 4.5. Therefore, all subsequent assays were performed at pH 4.5 as this is close to the physiological pH in lysosomes and because any possible interference by nonlysosomal ceramidases needs to be avoided for specific determination of aCDase activity and FD diagnosis.
Fig. 2.
pH-dependence of Rbm14-12 hydrolysis by human cells. The ceramide analog (at 20 µM) was incubated for 3 h with lysates (20 µg protein) from three cell lines exhibiting different degrees of aCDase activity: FD1 AcCer10× fibroblasts (△), normal lymphoid cells (○), and FD1 fibroblasts (□). The buffers were: glycine-HCl (pH 2.5 to 3), sodium acetate (pH 3.5 to 5.5), sodium phosphate (pH 6 to 8.1), and glycine-NaOH (pH 8.9 to 9.8). Results are representative of at least four independent experiments.
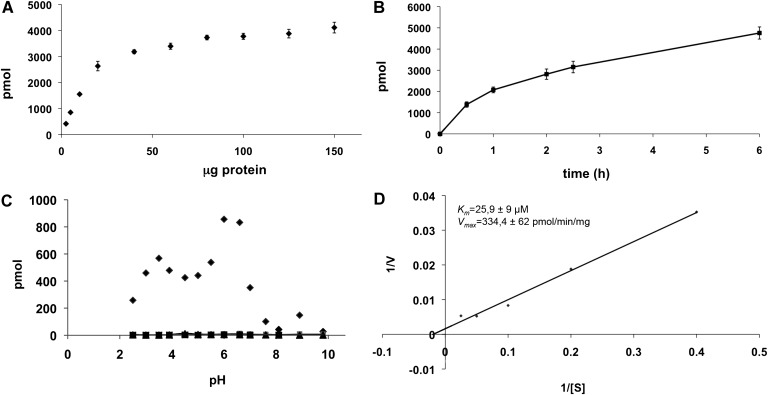
Hydrolysis of Rbm14-12 was dependent on protein concentration. Incubation of the substrate in the presence of different amounts of FD1 AcCer 10× protein extracts, ranging from 2.5 to 180 µg, showed a linear response up to 25 µg of protein, reaching a plateau at 50 µg (Fig. 3A). Therefore, a 10 to 25 µg quantity was chosen as an optimal protein amount in subsequent aCDase assays.
Fig. 3.
Characteristics of Rbm14-12 hydrolysis. A: Effect of protein concentration on Rbm14-12 hydrolysis. The substrate (at 20 µM) was incubated for 3 h in the presence of different amounts of FD1 AcCer10× fibroblast cell lysates. Results are representative of two independent experiments. B: Time-dependence of Rbm14-12 hydrolysis. The substrate (at 20 µM) was incubated for the indicated time periods in the presence of FD1 AcCer10× cell lysate (20 µg of protein). Data are representative of two independent experiments. C: Effect of detergents on Rbm14-12 hydrolysis. The substrate (at 20 µM) was incubated for 3 h at different pH in the presence or absence (♦) of 0.5% sodium cholate (▪), 0.05% Tween-20 (▴) or 0.1% Triton X-100 (X). Results represent the mean of two independent experiments performed in duplicate. D: Kinetics of Rbm14-12 hydrolysis. The substrate was incubated for 3 h at different concentrations in the presence of FD1 AcCer10× cell lysate (10 to 25 µg of protein). Lineweaver-Burk representation and the resulting parameters are shown. Results are the means of five independent experiments.
When studying the optimal time of incubation, we found linearity up to 6 h (Fig. 3B). An incubation time of 3 h was selected in aCDase assays as high values of fluorescence intensity with low amounts of protein were already obtained at this time point.
We also tested the influence of detergents on Rbm14-12 hydrolysis. Indeed, previously reported enzyme assays for aCDase include the use of at least one detergent and often a mixture of different detergents (6). The effect of Triton X-100, sodium cholate, or CHAPS in the incubation buffer was tested at different pH. In the presence of detergents, we could not measure any release of fluorescence (Fig. 3C), suggesting that the above detergents decreased fluorescence intensity or, more likely, have an inhibitory effect on Rbm14-12 hydrolysis. This was unexpected because the previously published compound for aCDase activity determination, Rbm14-16, worked in the presence of 0.25% (w/v) of TritonX-100. Later experiments demonstrated that the hydrolysis of this substrate was 20-times higher in the absence of Triton X-100 and that reduction of substrate hydrolysis was dose-dependent (activity was reduced by 20% already at a 0.001% final concentration of Triton X-100; data not shown). The same pattern was observed for Rbm14-12, suggesting that this effect of Triton X-100 is related to the nature of their sphingoid base. The possibility that Triton X-100 inhibited the oxidation or β-elimination processes was ruled out, because its addition at different concentrations after the enzymatic reaction did not affect the production of fluorescence (data not shown). A possible explanation of this inhibition of hydrolysis is that Triton X-100 affects the accessibility of these artificial substrates (e.g., by engulfing the substrate into micelles and preventing interaction with the catalytic center of the enzyme), which display a reduced partition coefficient as compared with natural ceramides (predicted logP values of 5.8 and 13 for Rbm14-12 and C16:0-ceramide, respectively). Thus, all assays were then performed in sodium acetate buffer 25 mM without addition of detergents.
Determination of kinetic constants
When the substrate concentration was varied at pH 4.5 under the above-mentioned conditions, maximum aCDase activity was observed above 40 µmol/L of substrate with an apparent Km of 25.9 ± 9 µM, and a Vmax of 334 ± 62 pmol/min/mg (Fig. 3D).
Acid ceramidase activity in Farber disease cell lines
The optimized assay was then used to determine aCDase activity in a series of FD cell lines, to assess its usefulness for diagnosing FD. Five lines of FD fibroblasts and five lines of FD lymphoid cells were tested. In both cases, the aCDase activity in FD cell lines was very low or undetectable, whereas in control cells specific aCDase activities ranged from 4 to 9 nmol/h/mg in fibroblasts and from 8 to 15 nmol/h/mg in lymphoid cells (Table 2). As a control for integrity of lysosomal hydrolases in the FD cell lines tested, the activity of acid β-galactosidase was determined. Despite interindividual variations, β-galactosidase activity was measurable in all samples. Calculating the aCDase to β-galactosidase activity ratio clearly demonstrated the aCDase deficiency in all FD cell lines.
TABLE 2.
Acid ceramidase activity in normal and Farber disease cell lines
| FIBROBLASTS | LYMPHOID CELLS | ||||||
|---|---|---|---|---|---|---|---|
| aCDase (pmol/h/mg) | lysosomal β Gal (nmol/h/mg) | aCDase/lysosomal β Gal | aCDase (pmol/h/mg) | lysosomal β Gal (nmol/h/mg) | aCDase/lysosomal β Gal | ||
| CTRL1 | 5816 ± 553 | 1546 ± 38 | 3.761 | CTRL5 | 8396 ± 1675 | 260 ± 25 | 32.32 |
| CTRL2 | 3785 ± 404 | 2114 ± 370 | 1.790 | CTRL6 | 14367 ± 2781 | 516 ± 42 | 27.86 |
| CTRL3 | 5628 ± 1337 | 767 ± 53 | 7.330 | CTRL7 | 15625 ± 4725 | 468 ± 114 | 33.36 |
| CTRL4 | 9388 ± 2731 | 2827 ± 333 | 3.321 | CTRL8 | 8416 ± 2892 | 420 ± 59 | 20.06 |
| FD1 | −32 ± 76 | 2726 ± 1139 | −0.012 | FD6 | 235 ± 153 | 480 ± 105 | 0.49 |
| FD2 | −1 ± 115 | 2145 ± 489 | −0.001 | FD7 | 241 ± 211 | 395 ± 233 | 0.61 |
| FD3 | 45 ± 125 | 2634 ± 162 | 0.017 | FD8 | 539 ± 369 | 605 ± 305 | 0.94 |
| FD4 | 55 ± 74 | 1815 ± 695 | 0.031 | FD9 | 707 ± 438 | 664 ± 121 | 0.66 |
| FD5 | −18 ± 13 | 846 ± 311 | −0.022 | FD10 | 200 ± 76 | 268 ± 181 | 0.74 |
aCDase activity was determined using Rbm14-12 at 20 µM after incubation for 3 h with lysates (20 µg protein) from fibroblasts or lymphoid cells derived from control individuals (CTRL) and patients affected with FD. As a control enzyme, β-galactosidase activity was determined. Results are the average of three independent experiments.
DISCUSSION
Here we present a novel method to determine aCDase activity that proved useful for FD diagnosis. The rationale behind this assay has already been reported by our group (25). Briefly, it is based on the use of a fluorogenic substrate having the same stereochemistry as that of natural ceramide and bearing a coumarin group in the sphingoid base moiety. After enzymatic cleavage of the amide bond by ceramidase and subsequent oxidation by external NaIO4, the product undergoes a β-elimination at alkaline pH to eventually release the fluorescent product umbelliferone.
The novel fluorogenic substrate Rbm14-12 exhibits a higher affinity for aCDase as compared with the reported homolog with an N-palmitoyl chain. This was exemplified by a 7-fold higher intensity of fluorescence under the same assay conditions. The kinetic parameters values are improved with respect to the previously reported assay: whereas Km and Vmax values for Rbm14-12 were 25.9 µM, and of 334 pmol/min/mg, respectively (giving a Vmax / Km ratio of 12.9), a Km of 113 µM and a Vmax of 3.6 pmol/min/mg (Vmax / Km ratio of 0.032) were reported for Rbm14-16, indicating that the efficiency of hydrolysis of the new compound under these conditions is about 400-fold higher than that of the previous reported one. In addition, the specificity of Rbm14-12 toward aCDase was demonstrated in comparative assays between protein samples derived from several control and FD cell lines. These properties make this molecule an excellent tool for accurate measurement of aCDase activity and for FD diagnosis.
As compared with the previously described methods for diagnosing FD through aCDase assay, the present enzyme assay exhibits many advantages (see also Table 1): i) this new substrate is not radiolabeled and, consequently, does not require any specific instrument for detection of radioactivity nor authorization; ii) our substrate is a water-soluble analog of ceramide, does not need the use of detergents for solubilisation, and does not require halogenated solvents for extraction (thus reducing the quantity of contaminants and limiting hazard); iii) the substrate is stable under storage (e.g., at least 6 months when stored in solution at −20°C); iv) the assay does not require separation of the product from the substrate and thus avoids complex and tedious procedures (such as TLC, HPLC, or GC); v) detection of the product is based on the measurement of fluorescence intensity using common instruments; vi) the assay with Rbm14-12 is very sensitive, exhibiting a detection limit of about 50 pmoles, and requires very minute amounts of cell lysates (about 10–20 µg protein); vii) measurement of ceramidase activity can be applied to various biological materials (such as human blood lymphocytes, cell culture medium, cultured skin fibroblasts, and other cultured cell lines); viii) the assay is fast (the overall duration of the assay does not exceed 5.5 h) and can be miniaturized for high-throughput screening and simultaneous multiple measurements; ix) the assay is specific for aCDase, and allows the assessment of the hydrolytic activity only, but not the reverse reaction (i.e., formation of ceramide), catalyzed by aCDase; and finally, x) when performed at pH 4.5, this assay is well suited for diagnosing inherited aCDase deficiency (FD) using clinical samples as nonlysosomal ceramidases do not interfere.
Acknowledgments
The authors thank Dr. J. A. Medin for FD1 AcCer10X cells, M. A. Gauthié for FD cell culture, and S. Carpentier for ceramide determinations.
Footnotes
Abbreviations:
- aCDase
- acid ceramidase
- FD
- Farber disease
This work was supported by INSERM, Université Paul Sabatier, the VML foundation, the Spanish Ministry of Science and Innovation (SAF2008-00706), and Generalitat de Catalunya (2009-SGR-1072). Dr. C. Bedia was supported by grants from Obra Social La Caixa and AGAUR [(2007-BP-A00129), Generalitat de Catalunya].
REFERENCES
- 1.Moser H. W., Moser A. B., Chen W. W., Schram A. W. 1989. Ceramidase deficiency: Farber lipogranulomatosis. The metabolic basis of inherited disease. Scriver C. R., Beaudet A. L., Sly W. S., Valle D., McGraw-Hill, New York: 1645–1654. [Google Scholar]
- 2.Levade T., Sandhoff K., Schulze H., Medin J. A. 2009. Acid ceramidase deficiency: Farber lipogranulomatosis. In Scriver's OMMBID (Online Metabolic and Molecular Bases of Inherited Disease), Valle D., et al., New York: McGraw-Hill; http://www.ommbid.com [Google Scholar]
- 3.Mao C., Obeid L. M. 2008. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim. Biophys. Acta. 1781: 424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nilsson A. 1969. The presence of sphingomyelin- and ceramide-cleaving enzymes in the small intestinal tract. Biochim. Biophys. Acta. 176: 339–347. [DOI] [PubMed] [Google Scholar]
- 5.Sugita M., Willians M., Dulaney J. T., Moser H. W. 1975. Ceramidase and ceramide synthesis in human kidney and cerebellum. Description of a new alkaline ceramidase. Biochim. Biophys. Acta. 398: 125–131. [DOI] [PubMed] [Google Scholar]
- 6.Momoi T., Ben-Yoseph Y., Nadler H. L. 1982. Substrate-specificities of acid and alkaline ceramidases in fibroblasts from patients with Farber disease and controls. Biochem. J. 205: 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spence M. W., Beed S., Cook H. W. 1986. Acid and alkaline ceramidases of rat tissues. Biochem. Cell Biol. 64: 400–404. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Yoseph Y., Gagne R., Parvathy M. R., Mitchell D. A., Momoi T. 1989. Leukocyte and plasma N-laurylsphingosine deacylase (ceramidase) in Farber disease. Clin. Genet. 36: 38–42. [DOI] [PubMed] [Google Scholar]
- 9.Fusch C., Huenges R., Moser H. W., Sewell A. C., Roggendorf W., Kustermann-Kuhn B., Poulos A., Carey W. F., Harzer K. 1989. A case of combined Farber and Sandhoff disease. Eur. J. Pediatr. 148: 558–562. [DOI] [PubMed] [Google Scholar]
- 10.Wertz P. W., Downing D. T. 1990. Ceramidase activity in porcine epidermis. FEBS Lett. 268: 110–112. [DOI] [PubMed] [Google Scholar]
- 11.Azuma N., O'Brien J. S., Moser H. W., Kishimoto Y. 1994. Stimulation of acid ceramidase activity by saposin D. Arch. Biochem. Biophys. 311: 354–357. [DOI] [PubMed] [Google Scholar]
- 12.Chatelut M., Harzer K., Christomanou H., Feunteun J., Pieraggi M. T., Paton B. C., Kishimoto Y., O'Brien J. S., Basile J. P., Thiers J. C., et al. 1997. Model SV40-transformed fibroblast lines for metabolic studies of human prosaposin and acid ceramidase deficiencies. Clin. Chim. Acta. 262: 61–76. [DOI] [PubMed] [Google Scholar]
- 13.Mitsutake S., Tani M., Okino N., Mori K., Ichinose S., Omori A., Iida H., Nakamura T., Ito M. 2001. Purification, characterization, molecular cloning, and subcellular distribution of neutral ceramidase of rat kidney. J. Biol. Chem. 276: 26249–26259. [DOI] [PubMed] [Google Scholar]
- 14.Bernardo K., Hurwitz R., Zenk T., Desnick R. J., Ferlinz K., Schuchman E. H., Sandhoff K. 1995. Purification, characterization, and biosynthesis of human acid ceramidase. J. Biol. Chem. 270: 11098–11102. [DOI] [PubMed] [Google Scholar]
- 15.Tani M., Okino N., Mitsutake S., Ito M. 1999. Specific and sensitive assay for alkaline and neutral ceramidases involving C12-NBD-ceramide. J. Biochem. 125: 746–749. [DOI] [PubMed] [Google Scholar]
- 16.He X., Li C. M., Park J. H., Dagan A., Gatt S., Schuchman E. H. 1999. A fluorescence-based high-performance liquid chromatographic assay to determine acid ceramidase activity. Anal. Biochem. 274: 264–269. [DOI] [PubMed] [Google Scholar]
- 17.Chen W. W., Moser A. B., Moser H. W. 1981. Role of lysosomal acid ceramidase in the metabolism of ceramide in human skin fibroblasts. Arch. Biochem. Biophys. 208: 444–455. [DOI] [PubMed] [Google Scholar]
- 18.Sutrina S. L., Chen W. W. 1982. Metabolism of ceramide-containing endocytotic vesicles in human diploid fibroblasts. J. Biol. Chem. 257: 3039–3044. [PubMed] [Google Scholar]
- 19.Kudoh T., Wenger D. A. 1982. Diagnosis of metachromatic leukodystrophy, Krabbe disease, and Farber disease after uptake of fatty acid-labeled cerebroside sulfate into cultured skin fibroblasts. J. Clin. Invest. 70: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inui K., Furukawa M., Nishimoto J., Okada S., Yabuuchi H. 1987. Metabolism of cerebroside sulphate and subcellular distribution of its metabolites in cultured skin fibroblasts derived from controls, metachromatic leukodystrophy, globoid cell leukodystrophy and Farber disease. J. Inherit. Metab. Dis. 10: 293–296. [DOI] [PubMed] [Google Scholar]
- 21.Levade T., Tempesta M. C., Moser H. W., Fensom A. H., Harzer K., Moser A. B., Salvayre R. 1995. Sulfatide and sphingomyelin loading of living cells as tools for the study of ceramide turnover by lysosomal ceramidase–implications for the diagnosis of Farber disease. Biochem. Mol. Med. 54: 117–125. [DOI] [PubMed] [Google Scholar]
- 22.Krivit W., Hammarstrom S. 1972. Identification and quantitation of free ceramides in human platelets. J. Lipid Res. 13: 525–530. [PubMed] [Google Scholar]
- 23.Sugita M., Iwamori M., Evans J., McCluer R. H., Dulaney J. T., Moser H. W. 1974. High performance liquid chromatography of ceramides: application to analysis in human tissues and demonstration of ceramide excess in Farber's disease. J. Lipid Res. 15: 223–226. [PubMed] [Google Scholar]
- 24.Chatelut M., Feunteun J., Harzer K., Fensom A. H., Basile J. P., Salvayre R., Levade T. 1996. A simple method for screening for Farber disease on cultured skin fibroblasts. Clin. Chim. Acta. 245: 61–71. [DOI] [PubMed] [Google Scholar]
- 25.Bedia C., Casas J., Garcia V., Levade T., Fabrias G. 2007. Synthesis of a novel ceramide analogue and its use in a high-throughput fluorogenic assay for ceramidases. ChemBioChem. 8: 642–648. [DOI] [PubMed] [Google Scholar]
- 26.Moore W. J., Luzzio F. A. 1995. Synthethic studies directed toward the liposidomycins: preparation and reactions of serine-derived epoxides. Tetrahedron Lett. 36: 6599–6602. [Google Scholar]
- 27.Medin J. A., Takenaka T., Carpentier S., Garcia V., Basile J. P., Segui B., Andrieu-Abadie N., Auge N., Salvayre R., Levade T. 1999. Retrovirus-mediated correction of the metabolic defect in cultured Farber disease cells. Hum. Gene Ther. 10: 1321–1329. [DOI] [PubMed] [Google Scholar]
- 28.Wenger D. A., Williams C. 1991. Screening for lysosomal disorders. Techniques in Diagnostic Biochemical Genetics: A Laboratory Manual. Hommes F. A., editor Wiley-Liss, Inc, New York: 587–617. [Google Scholar]