Abstract
Background/Aims
Severe early childhood caries is a microbial infection that severely compromises the dentition of young children. The aim of this study was to characterize the microbiota of severe early childhood caries.
Methods
Dental plaque samples from 2- to 6-year-old children were analyzed using 16S rRNA gene cloning and sequencing, and by specific PCR amplification for Streptococcus mutans and Bifidobacteriaceae species.
Results
Children with severe caries (n = 39) had more dental plaque and gingival inflammation than caries-free children (n = 41). Analysis of phylotypes from operational taxonomic unit analysis of 16S rRNA clonal metalibraries from severe caries and caries-free children indicated that while libraries differed significantly (p < 0.0001), there was increased diversity than detected in this clonal analysis. Using the Human Oral Microbiome Database, 139 different taxa were identified. Within the limits of this study, caries-associated taxa included Granulicatella elegans (p < 0.01) and Veillonella sp. HOT-780 (p < 0.01). The species associated with caries-free children included Capnocytophaga gingivalis (p < 0.01), Abiotrophia defectiva (p < 0.01), Lachnospiraceae sp. HOT-100 (p < 0.05), Streptococcus sanguinis (p < 0.05) and Streptococcus cristatus (p < 0.05). By specific PCR, S. mutans (p < 0.005) and Bifidobacteriaceae spp. (p < 0.0001) were significantly associated with severe caries.
Conclusion
Clonal analysis of 80 children identified a diverse microbiota that differed between severe caries and caries-free children, but the association of S. mutans with caries was from specific PCR analysis, not from clonal analysis, of samples.
Key Words: Bifidobacteria, Clonal analysis, PCR, Severe early childhood caries, Streptococcus mutans
Introduction
Aggressive and progressing dental caries in young children is of high clinical significance because affected children can have both extensive caries in primary teeth and a high risk of spread of the caries to the permanent dentition [Li and Wang, 2002]. Early childhood caries (ECC), dental caries in the primary dentition, affects 28% of the US population, particularly disadvantaged ethnic and socioeconomic groups [Beltran-Aguilar et al., 2005]. Severe ECC can affect multiple teeth in children under 6 years of age [Drury et al., 1999], and these children frequently have painful abscesses and reduced ability to sleep or eat, which can lead to poor quality of life [Slade, 2001; Vargas et al., 2005; Clarke et al., 2006]. The extensive restorative therapy needed usually requires general anesthesia [Berkowitz, 2003].
The primary microbial etiology of ECC, determined mainly from selective cultural analysis, is Streptococcus mutans [Van Houte et al., 1982]. Bifidobacterium species have also been isolated using selective culture from childhood caries [Mantzourani et al., 2009b], and in older adults they were more strongly associated with caries than mutans streptococci [Mantzourani et al., 2009a]. Using 16S rRNA probes, Bifidobacterium species were also associated with ECC but were detected at lower levels than S. mutans [Aas et al., 2008]. Clonal analyses of environmental samples using broad-range primers have revealed a wide bacterial diversity, including previously uncultivated species [Hugenholtz et al., 1998]. Cloning and sequencing analysis of bacterial plaque samples from the oral cavity detected >700 phylotypes and species [Paster et al., 2001; Relman, 2002; Munson et al., 2004]. Oral species detected by clonal analysis of childhood caries included S. mutans and non-mutans Streptococcus species, Lactobacillus, Actinomyces, Bifidobacterium and Veillonella species, and uncultured taxa [Becker et al., 2002; Aas et al., 2008]. Both clonal [Becker et al., 2002; Aas et al., 2008] and cultural analysis of extensive childhood caries [Loesche et al., 1975], however, indicated that S. mutans formed only a low proportion of the total caries microbiota. These reports indicated that S. mutans was not the dominant species in dental caries, which suggested that other species, including uncultivated ones, could be important in the etiology of this infection.
The aim of this study was to characterize the microbiota associated with severe ECC by comparing the microbiota with that of caries-free children. A 16S rRNA gene broad-range PCR followed by clonal analysis was performed and data were analyzed by phylotypes generated by operational taxonomic unit (OTU) analysis and by comparison with the Human Oral Microbiome Database (HOMD) [Dewhirst et al., 2008]. A specific PCR was used to detect Streptococcus mutans and Bifidobacterium species. The study hypothesized that the microbiota would be different between severe ECC and caries-free children and that new caries-associated taxa would be candidates as caries pathogens.
Subjects and Methods
Clinical Methods
Eighty preschool children, 39 severe ECC and 41 caries-free, were recruited from 3 dental clinics in the Boston, Massachusetts, area (Boston Medical Center-Boston University, Floating Hospital-Tufts Medical Center, and Cambridge Health Alliance). Inclusion criteria were: the child was 2–6 years of age, was medically healthy, had not used antibiotics within the past 3 months, and the parent or guardian was willing to consent to the child's clinical examination and microbial sampling. The study design, protocol, and informed consent were approved by the institutional review boards of the institutions involved.
Dentists trained in study measurement criteria examined the children. The number of teeth and their caries status were recorded on a 0–4 scale by surface [Drury et al., 1999], dental plaque on a 0–4 scale, gingival inflammation on a 0–3 scale [Loe, 1967] and gingival bleeding as 0–1 (absent or present). The child's age, gender, race and ethnicity were recorded. Plaque samples were taken with sterile wooden toothpicks [Milgrom et al., 2000] from the buccal and interproximal surfaces of molars. In children with severe ECC, plaque from cavities was included. The samples were put in 100 μl of 50 mM Tris-EDTA buffer (pH 7.6) and stored frozen at −70°C [Paster et al., 2001].
Microbiological Methods
DNA was extracted from plaque samples and purified by chemical extraction using Master Pure kits (Epicenter, Madison, Wisc., USA) as previously described [Li et al., 2007]. The purified sample DNA was amplified with universal 16S rDNA primers so that clone libraries could be constructed. Cloning and sequencing were performed as previously described [Paster et al., 2001] with amplification of the 16S rRNA genes using universal 16S rRNA PCR primers (F24, 9–27 forward: 5′-GAG TTT GAT YMT GGC TCA G-3′ and F25, 1,525–1,541 reverse: 5′-AAG GAG GTG WTC CAR CC-3′). The PCR amplification product (1,500 bases) was examined using 1% agarose gel electrophoresis. The PCR product was extracted from the gel and purified using a Qiagen Gel Clean-Up Kit according to the manufacturer's specifications (Qiagen Inc., Valencia, Calif., USA).
Cloning of the PCR-amplified DNA was performed with a commercial kit (TOPO TA Cloning Kit, Invitrogen, San Diego, Calif., USA), according to the manufacturer's instructions. Transformation was performed in competent One Shot TOP 10 E. coli cells (Invitrogen, Calif., USA). The transformed cells for each sample were grown overnight at 37°C on Luria-Bertani agar plates supplemented with kanamycin (50 μg/ml). Ninety-six (first 16 subjects, 8 severe ECC, 8 caries-free) or 48 (rest of samples) transformed colonies were picked and suspended in 40 μl of 10 mM Tris buffer. One microliter of cell suspension was used as templates in a PCR with vector primers M13F and M13R (TOPO TA Cloning Kit, Invitrogen, Calif., USA). The amplicons were checked for correct size (approximately 1,500 bases) by 1% agarose gel electrophoresis. Clone amplicons were cleaned using exonuclease (Amersham Pharmacia Biotech Inc., Piscataway, N.J., USA) and shrimp alkaline phosphatase (Amersham). For 16S rRNA sequencing, clone amplicons were sequenced using an ABI prism cycle-sequencing kit on an ABI 3100 DNA Sequencer (Applied Biosystems, Foster City, Calif., USA). Sequencing used primer F15 (519–533 reverse: 5′-TTA CCG CGG CTG CTG-3′) as previously described [Paster et al., 2001].
Clone sequences were examined by BLAST analysis against a 16S rRNA reference sequence set at the HOMD [Dewhirst et al., 2008]. The HOMD (version 10) contains 619 species and phylotypes based on 98.5% similarity cutoffs of full 1,540 base 16S rRNA sequences. Each oral species or phylotype in the database has a Human Oral Taxon Number (HOT). The threshold for BLAST identification of 500 base partial sequences was 98% match (approximately 8 bases mismatch for partial sequences). Clones that did not match known oral taxa were examined with the Chimera Check program at the Ribosomal Database Project [Cole et al., 2009] and Mallard [Ashelford et al., 2006]. Non-chimera clones which did not match the HOMD references were clustered into provisional additional novel taxa. A description of the analysis of 35,000 oral 16S rRNA clone sequences to extend the HOMD reference set is in preparation. The phylogenetic trees for the species found in this study were created from the full-length reference sequences in HOMD using the neighbor-joining method [Saitou and Nei, 1987].
Specific 16S rRNA gene PCR assay was used to detect S. mutans and Bifidobacterium species. For S. mutans, S. mutans-specific reverse primer 5′-ACT CCA GAC TTT CCT GAC CG-3′ (position 587–608) and forward universal primer 5′-GAG TTT GAT YMT GGC TCA G-3′ (position 9–27) [Kanasi et al., 2010] was used. The samples were preheated (94°C, 5 min) followed by amplification under the following conditions: denaturation (94°C, 45 s), annealing (60°C, 45 s), elongation (72°C, 90 s plus an additional second per cycle for 32 cycles with a final extension (72°C, 15 min). Bifidobacterium species PCR was performed with 16S rRNA gene genus specific primers as previously described [Matsuki et al., 2004]. Amplicons were verified on 1% agarose gels. The Bifidobacterium primers, designed to detect intestinal bifidobacteria, produced appropriate size amplicons around 550 bp for Bifidobacterium subtile, Bifidobacterium dentium, Bifidobacterium longum, Scardovia inopinata and Parascardovia denticolens. The primers did not amplify strains of Rothia, Propionibacterium, Corynebacterium, Actinomyces, Streptococcus or Lactobacillus species. The reactions indicated that the Bifidobacterium primers detected species in the family Bifidobacteriaceae (Bifidobacterium, Scardovia, Parascardovia), but not other oral species, including in the phylum Actinobacteria.
Statistical Analyses
The subjects were divided into severe ECC and caries-free disease categories. Mean differences between severe ECC and caries-free children with respect to age, plaque, gingival and bleeding indices were evaluated by t test. Associations between disease category and gender and race-ethnicity were compared by χ2 test.
To compare phylotypes from OTU analysis of 16S rRNA gene libraries, metalibraries were created by combining all sequences of severe ECC children, with a second metalibrary for sequences of caries-free children. These libraries were aligned to the 16S ribosomal SILVA reference database [Pruesse et al., 2007] and analyses performed using MOTHUR software [Schloss et al., 2009]. A distance matrix was created, and sequences were assigned to phylotypes based on OTU calculated from the genetic distance between the sequences. Estimates of phylotype richness were calculated using the abundance-based coverage estimator and the bias-corrected Chao1 [Chao, 1984; Schloss et al., 2009]. The number of observed and estimated phylotypes as a function of the number of clones sequenced was compared at evolutionary distances: 0.02, 0.05 and 0.10 [Hughes et al., 2001].
Good's coverage estimation [Good, 1953; Singleton et al., 2001] was calculated with MOTHUR [Schloss et al., 2009]. The ∫-LIBSHUFF algorithm was used to test whether the structures of 2 metalibraries were the same using the Cramér-von Mises test statistic with Bonferroni correction for multiple comparisons [Schloss et al., 2009] and a Monte Carlo test procedure. The numbers of sequences that were unique to each metalibrary and the probability that the observed differences between the metalibraries were due to chance were calculated. Coverage (percent of estimated diversity) was examined as a function of evolutionary distance. The coverage of severe ECC and caries-free libraries was compared.
To compare phylotypes with HOMD database, HOMD taxa as a percent of total clones identified by subject were averaged over subjects and compared between severe ECC and caries-free children by the McNemar test. Taxa detection frequencies from clonal analysis were calculated by child, then averaged across disease categories and compared by χ2 test. Detections of S. mutans and Bifidobacteriaceae in severe ECC and caries-free categories were compared by χ2 test. A significance threshold of ≤0.05 was used. The results were also adjusted for multiple comparisons using the false discovery rate (α = 0.05) [Benjamini and Hochberg, 1995]. Statistical analyses were performed with SPSS® software (SPSS Inc., Chicago, Ill., USA).
Results
The severe ECC and caries-free (control) groups were balanced by age (4.1 and 4.3 years, respectively) and gender (table 1). The racial classification of the children was: White (34%), Black (29%), Asian (25%) and other/mixed races (11%). Thirty percent of the children were Hispanic. Children with severe ECC exhibited mostly caries into the dentin (12% surfaces), followed by pulpal exposure (9% surfaces), white-spot lesions (7% surfaces) and enamel cavities (5% surfaces). Plaque, gingival and bleeding indexes were all significantly higher in severe ECC than caries-free children (p < 0.001). The sociodemographics did not differ between severe ECC and caries-free children [Palmer et al., 2010].
Table 1.
Demographic and clinical characteristics of the study population
| Caries-free (n = 41) | Caries (n = 39) | Caries-positive, % | p value | |
|---|---|---|---|---|
| Mean age ± SEM, years | 4.3 ± 0.2 | 4.1 ± 0.2 | 0.536a | |
| Gender | ||||
| Male | 26 | 23 | 47 | |
| Female | 15 | 16 | 52 | 0.684b |
| Race | ||||
| White | 18 | 9 | 33 | |
| Black | 11 | 13 | 54 | |
| Asian | 10 | 10 | 50 | |
| Other/mixed | 2 | 7 | 78 | 0.117b |
| Ethnicity | ||||
| Hispanic | 13 | 11 | 46 | |
| Non-Hispanic | 28 | 28 | 50 | 0.733b |
| Caries, percent surfaces affected | ||||
| Intact surface | 100 | 66 | ||
| White or opaque patches | 0 | 7 | ||
| Enamel cavity | 0 | 5 | ||
| Cavity into dentine | 0 | 12 | ||
| Pulpal exposure | 0 | 9 | ||
| Mean plaque index ± SEM | 0.65 ± 0.01 | 1.58 ± 0.01 | <0.001a | |
| Mean gingival index ± SEM | 0.26 ± 0.01 | 1.96 ± 0.02 | <0.001a | |
| Mean bleeding index ± SEM | 0.00 ± 0.00 | 0.57 ± 0.01 | <0.001a |
SEM = Standard error of the mean.
Student's t test.
χ2 test.
In the clonal analysis, out of 4,516 clones sequenced, 714 (15%) were not used because they were short, of poor quality or were chimeras. Of the remaining 3,802 clones, 1,846 were from severe ECC and 1,956 were from caries-free children. The observed and estimated diversity of phylotypes from OTU was similar for both metalibraries at 0.10 evolutionary distances (90% similarity). At 0.05 and 0.02 evolutionary distances (95 and 98% similarity, respectively) there were consistently higher values for the estimated than observed phylotypes. For the severe ECC library at 0.05 evolutionary distances there were 116 observed OTU and 147 estimated OTU; at 0.02 evolutionary distances 247 OTU were observed and 366 were estimated. For the caries-free library at 0.05 evolutionary distances there were 113 observed OTU and 133 estimated OTU; at 0.02 evolutionary distances 240 OTU were observed and 341 were estimated. Less than 50% of the observed and estimated phylotypes were shared between the severe ECC and caries-free libraries. The number of observed phylotypes at 98% sequence similarity was 92 for severe ECC, 84 for caries-free with 157 shared by severe ECC and caries-free, whereas estimated phylotypes were 134 for severe ECC, 117 for caries-free with 224 shared by severe ECC and caries-free libraries. Good's coverage estimation at 0.02 evolutional distances, which approximate species level, for severe ECC and caries-free libraries was 96.5 and 96.7%, respectively. At 0.05 evolutional distances, which approximate genus at a narrow cutoff, severe ECC and caries-free libraries had 98.6% coverage, and at 0.10 evolutional distances, which approximate genus at a broad cutoff, severe ECC and caries-free libraries had 99.2 and 99.4% coverage. There were significant differences by ∫-LIBSHUFF between severe ECC and caries-free metalibraries (p < 0.0001) with dCXY score: severe ECC – caries-free = 0.00017562; and caries-free – severe ECC = 0.00013673.
Comparison of phylotypes with HOMD indicated that 139 HOMD taxa were identified, with an additional 35 provisional taxa. From severe ECC children >98% clones were identified at the >98% sequence similarity level to HOMD groups and 1.78% to additional provisional groups, whereas from caries-free children 97.02% clones were identified to HOMD groups and 2.28% to provisional groups. Of the 139 HOMD taxa identified, 88 (63%) were detected in both severe ECC and caries-free children, 22 (16%) were detected only in severe ECC children, and 29 (21%) were detected only in caries-free children. The numbers of taxa detected in severe ECC (mean ± SEM = 14.8 ± 0.73) and caries-free children (mean ± SEM = 16.6 ± 0.77) (p = 0.09, t test) did not differ.
Unidentified clones in provisional phylotypes were closely related to recognized species, and in descending order of detection frequencies were identified to the genera or higher taxonomic level: Capnocytophaga, Veillonella, Actinomyces, Kingella, Lachnospiraceae, Peptococcus, Selenomonas, Streptococcus, Aggregatibacter, Gemella, Granulicatella, Lautropia, Neisseria and Synergistetes. There were no differences in mean percentages or detection frequencies in the provisional phylotypes between severe ECC and caries-free children. These potentially novel taxa are being evaluated for inclusion into HOMD.
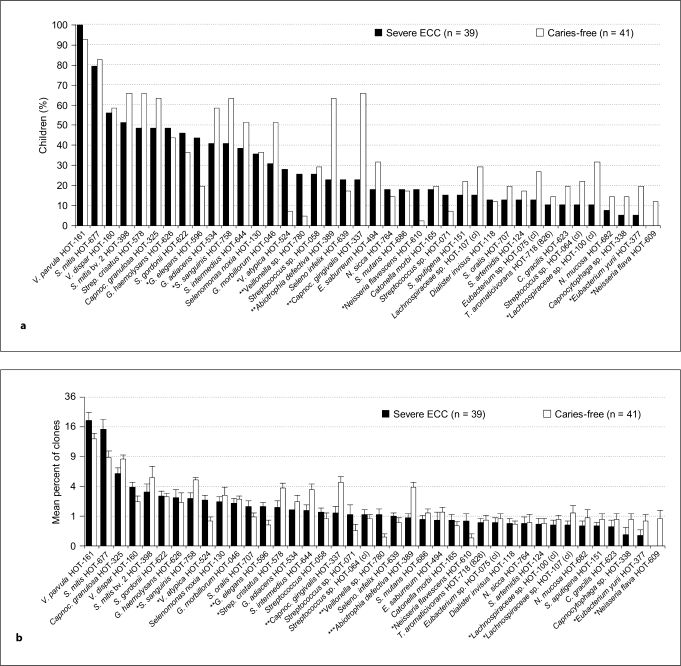
Clones identified to HOMD species and phylotype groups are shown in phylogenetic trees (fig. 1), which include the numbers of children and the taxa detected. The most frequently detected taxa (fig. 2), Veillonella, Streptococcus and Capnocytophaga, were detected in >50% of the children, particularly Veillonella parvula, Streptococcus mitis 1 and 2, and Capnocytophaga granulosa. Among the Proteobacteria, Bacteroidetes and Actinobacteria (fig. 1a) most clones were Neisseria and Capnocytophaga. Neisseria flavescens and Campylobacter showae were detected more frequently in severe ECC children, while Capnocytophaga gingivalis and Neisseria flavia occurred more frequently and in higher proportions in caries-free children (fig. 1a, 2a, b). Clostridia species and phylotypes within Firmicutes (fig. 1b) included 6 species of Selenomonas, 11 closely related Selenomonas phylotypes, 3 species of Veillonella and 2 Veillonella phylotypes. Veillonella atypica and Veillonella sp. HOT-780 (fig. 1b) were detected more frequently in severe ECC, whereas Lachnospiraceae sp. HOT-100 and Eubacterium yurii were found more frequently in caries-free children (fig. 1b, 2a, b). The phylogenetic tree of the Bacillus branch of Firmicutes included 19 species and 10 phylotypes in Streptococcus, 4 species of Lactobacillus, 2 Granulicatella and 2 Gemella species (fig. 2b). Granulicatella elegans and Streptococcus salivarius were detected more frequently in severe ECC children, whereas Abiotrophia defectiva and Streptococcus sanguinis (fig. 1c), also Streptococcus cristatus in mean levels (fig. 2b), were associated with caries-free children. Associations for A. defectiva and C. gingivalis remained significant after adjustment for multiple comparisons [Benjamini and Hochberg, 1995].
Fig. 1.
Phylogenetic trees of species and phylotypes identified from clones compared with the HOMD. The trees were created from full 16S rRNA reference sequences although it is recognized that use of 500-bp data from study clones could have created a different, but less accurate, treeing topography. Taxa in trees include identifications at >0.1 mean percent positive taxa by subject. A total of 139 taxa were identified. Numbers of children with taxa detected are in the table to the right of each tree. Species or phylotypes that differed in detection frequencies are noted at ∗ p ≤ 0.05, ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001, by χ2 analysis, or Fishers exact test. a Phylogenetic tree of 51 taxa in Proteobacteria, Bacteroidetes and Actinobacteria detected in severe caries and caries-free children, with positions of Clostridia and Bacillus as phylogenetic references. The tree includes 28 taxa of Proteobacteria, 15 taxa of Bacteroidetes, 1 Treponema species, 5 Actinobacteria and 2 Leptotrichia taxa. b Phylogenetic tree of 49 taxa in the Clostridia branch of Firmicutes, which includes the Gram-negative Veillonella and Selenomonas genera, detected in severe ECC and caries-free children. This group comprises several named and unnamed taxa, including the Centipeda species. c Phylogenetic tree of 38 taxa in the Bacillus branch of Firmicutes detected in severe caries and caries-free children. The tree includes 29 Streptococcus species, comprising 10 unnamed species, 2 Gemella, 2 Granulicatella and 4 Lactobacillus species.
Fig. 2.
The most frequently detected species from severe caries and caries-free children. The scale represents the square root of data to enhance the visualization of taxa less frequently detected or in lower proportions. a Detection frequencies of taxa by child in severe caries and caries-free children. Taxa are ordered in decreasing order of detection in severe ECC children. Taxa that differed in detection frequencies are noted at ∗ p ≤ 0.05, ∗∗ p ≤ 0.01 by χ2 analysis. b Mean proportions of taxa detected in severe caries and caries-free children. Taxa are given in decreasing order of mean clone levels in severe ECC children. Taxa that differed in detection frequencies are noted at ∗ p ≤ 0.05, ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001 by Mann Whitney U test. Error bars = standard error of the mean.
S. mutans was detected by clonal analysis in 20% of the severe ECC and caries-free children, at around 1% clones, with no significant differences between severe ECC and caries-free children (fig. 1c, 2b). Streptococcus sobrinus was detected in 1 caries child. No Bifidobacteriaceae (Bifidobacterium, Scardovia or Parascardovia) species were detected in the clonal analysis.
S. mutans and Bifidobacteriaceae Species Detection by Specific PCR
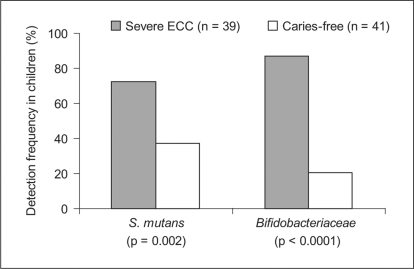
S. mutans was detected in 73% of the severe ECC children and 38% of the caries-free children (p = 0.002, χ2 test, fig. 3). S. mutans detection was significantly associated with plaque (p = 0.004), gingival inflammation (p = 0.003) and bleeding (p = 0.003) (Kruskal Wallis test) (data not shown). Bifidobacteriaceae species were detected in 87% of the severe ECC children and 21% of the caries-free children (p < 0.0001, χ2 test, fig. 3) Bifidobacteriaceae detection was significantly associated with plaque (p < 0.0001), gingival inflammation (p < 0.0001) and bleeding (p < 0.0001) (Kruskal Wallis test) (data not shown).
Fig. 3.
Detection frequencies by child of S. mutans and Bifidobacteriaceae species assayed by species-specific PCR. Streptococcus mutans (p < 0.002) and Bifidobacteriaceae species (p < 0.0001) were strongly associated with severe early childhood caries.
Discussion
The study aimed to use 16S rRNA gene broad-range PCR followed by cloning and sequencing, a noncultural approach to detect the diversity of cultured and uncultured taxa in an environment, to describe the microbiota of severe ECC. The study examined 80 children to overcome variation between individuals [Diaz et al., 2006; Aas et al., 2008], and to detect significant microbial differences between severe-ECC and caries-free populations. Statistical tools were developed [Schloss et al., 2009] to analyze extensive sequence-based data from metagenomic studies of the Sargasso Sea, which had 2 million sequencing reads [Venter et al., 2004]. We applied these OTU analyses to evaluate observed and estimated population diversity and coverage and also differences between clone libraries from severe ECC and caries-free children. Significant differences were detected between severe ECC caries-free children using the clone sequence data by OTU analysis and phylotype classification using HOMD. Association of selected target caries pathogens with severe ECC, however, required specific 16S rRNA PCR analysis of samples.
DNA sequences from the clonal libraries were grouped into OTU phylotypes at different levels of evolutionary distance to determine the numbers of observed phylotypes. The estimated phylotypes, which indicate the total diversity of the population [Hughes et al., 2001; Schloss and Handelsman, 2005] were greater than observed at each threshold, suggesting that there is increased diversity in the populations than reported from this clonal analysis. Coverage of the populations, however, was quite high at 0.02 evolutionary distances, suggesting that clinically relevant comparisons between health and disease could be made. The differences in childhood caries and caries-free populations previously reported from clonal analyses [Becker et al., 2002; Aas et al., 2008] was confirmed in the OTU phylotypes with over half of the phylotypes detected only in either severe ECC or caries-free populations, at differences of evolutionary distance <0.02 or 98% similarity.
To identify the clones, sequences were identified by BLAST analysis against the HOMD reference set of 619 species and phylotypes. A wide diversity of bacterial taxa (species and phylotypes) was detected including more species/phylotypes of Streptococcus, Capnocytophaga, Selenomonas and Neisseria than previously reported in children [Becker et al., 2002; Aas et al., 2008], reflecting, in part, the large number of clones sequenced and identification to HOMD taxa.
The microbiota of children with severe ECC was associated with V. atypica and Veillonella sp. HOT-780 compared with caries-free children. In other studies Veillonella species were related to ECC by culture [Marchant et al., 2001] and DNA probes [Becker et al., 2002]. Veillonella species also dominated in early and established dental cavities in primary and secondary teeth of children by clonal and 16S rRNA probe analyses [Aas et al., 2008]. Physiologically Veillonella species use lactate as a carbon energy source, and their detection in dental caries reflects the lactic acid rich environment derived from bacterial metabolic end products, particularly S. mutans. In biofilm experiments, Veillonella enhanced growth of S. mutans, suggesting a close symbiotic relationship between these species [Egland et al., 2004; Chalmers et al., 2008]. Granulicatella elegans was detected in higher mean proportions from severe-ECC than caries-free children. This species, previously recognized as a nutritionally variant Streptococcus, has been detected in children with caries [Aas et al., 2008], but clinical significance of this species has not been reported. Neisseria flavescens was not reported from other clonal analyses of plaque samples of young children but was detected in refractory periodontitis [Paster et al., 2001]. The non-mutans streptococci, S. mitis and Streptococcus sp. HOT-071 in the S. mitis/S. oralis group, were detected more frequently from severe-ECC than caries-free children while not significantly, and have also been associated with childhood caries by 16S rRNA probes [Becker et al., 2002]. Associations of certain species with severe-ECC may also reflect the increased levels of gingival inflammation in these children with abundant dental plaque and frequently jagged teeth or root stumps irritating the gingival margin.
The microbiota of caries-free children was associated with higher proportions of C. gingivalis, Abiotrophia defectiva, S. cristatus, S. sanguinis and Lachnospiraceae sp. HOT-100 clones than severe ECC children. C. gingivalis is frequently detected in subgingival plaque and was found in young children but not as frequently as C. granulosa [Corby et al., 2005], as observed in the current study. Abiotrophia defectiva, another nutritionally variant Streptococcus, has been detected in endocarditis and bacteremia and generally requires 16S rRNA sequence data for identification [Senn et al., 2006]. A. defectiva was previously detected at higher levels in caries-free than caries children [Becker et al., 2002; Corby et al., 2005]. S. cristatus and S. sanguinis are health-related species. S. cristatus was associated with caries-free children in primary and secondary teeth [Aas et al., 2008]. S. sanguinis was previously associated with caries-free children [Becker et al., 2002; Corby et al., 2005] and had an inverse relationship with S. mutans with respect to childhood caries; children colonized with S. sanguinis before S. mutans showed delayed S. mutans infection and detection of dental caries [Caufield et al., 2000]. Lachnospiraceae sp. HOT-100 is an uncultivated phylotype which appears from our data associated with caries-free children. The role of these species in dental health should be examined further.
Despite an extensive literature relating S. mutans to ECC, lack of S. mutans association by clonal analysis was previously reported in childhood caries, advanced carious lesions and root caries [Chhour et al., 2005; Aas et al., 2008; Preza et al., 2008]. While bias in the clonal analyses could have resulted from PCR and primer bias [Ishii and Fukui, 2001; Munson et al., 2004] low proportions of S. mutans in caries were also reported in a cultural analysis of children with extensive caries [Loesche et al., 1975]. Our findings are thus consistent with literature studies. Streptococcus sobrinus was detected in only 1 child with severe ECC in the current study, consistent with lower S. sobrinus detection levels than S. mutans and lack of association with childhood caries by 16S rRNA probes [Becker et al., 2002].
Plaque samples were also analyzed by specific PCR to detect S. mutans and Bifidobacteriaceae. S. mutans detection by PCR was significantly associated with severe ECC. Recent studies linking S. mutans with severe ECC have been performed by culture [Marchant et al., 2001], PCR [Seki et al., 2006] and by 16S rRNA probes [Becker et al., 2002; Corby et al., 2005; Aas et al., 2008]. The discrepancy between clonal counts and selective detection (PCR, selective isolation, DNA probes) is likely related to these different approaches used for species detection, with selective detection having the ability to identify species in low proportions of the microbiota. These data are thus consistent with the observation that carious sites may harbor widely diverse bacteria, and S. mutans, while important in disease, may be present only in low proportions of carious sites.
The strong association of species in Bifidobacteriaceae represents new findings by this PCR detection method, although bifidobacteria have been detected and previously associated with childhood caries [Becker et al., 2002; Aas et al., 2008] and initial caries [Van Houte et al., 1996; Svensater et al., 2003]. These studies, however, did not report associations at the detection frequency and significance of the current study. Future clonal analyses need to be expanded to detect species in Bifidobacteriaceae.
Discrepancies in the current study data compared with literature reports included lack of detection using clonal analysis of Bifidobacteria and several other species in the Actinobacteria phylum (Actinomyces, Corynebacterium, Rothia) and Lactobacillus, especially since selected Lactobacillus species were recently associated with ECC [Kanasi et al., 2010]. Possible reasons for not detecting certain species include DNA extraction and bacterial lysis methods, and PCR and primer bias [Ishii and Fukui, 2001; Munson et al., 2004]. While differences in populations detected varied with different primers in dental caries [Munson et al., 2004], a recent report clarified improved primers to detect Actinobacteria [Frank et al., 2008]. Only 1 spirochete species, Treponema denticola, was detected, thus several primer sets would be needed for the recovery of all major taxa in the microbiota of dental caries.
In summary, the present study compared the microbiota of severe ECC with caries-free children by clonal analysis. Phylotypes based on OTU analyses indicated that there was more diversity than detected in this clonal analysis and that severe ECC and caries-free populations differed. Significant differences were also detected in HOMD taxa between severe ECC and caries-free children. V. atypica, Veillonella sp. HOT-780, N. flavescens and G. elegans were associated with severe ECC, and Streptococcus sp. HOT-071 was detected more frequently from and in higher proportions with severe ECC but not significantly as in other studies. By clonal analysis S. mutans was detected infrequently and detection did not differ between severe ECC and caries-free populations, but S. mutans and Bifidobacteriaceae species were significantly associated with severe ECC using specific PCR assays. We conclude that candidates as new caries pathogens from this study were G. elegans and species within Bifidobacteriaceae. Other caries-associated species including Neisseria and Veillonella are not acidogenic and thus not caries pathogens but could play a key role in supporting the biofilm of this infection.
Disclosure Statement
There are no conflicts of interest.
Acknowledgements
We acknowledge Mohammed Dahlan, Rachel Montgomery, Benita Demirza, Caroline Young, David Okuji and Sam Merabi for subject recruitment and clinical measurements and Sara Barbuto for assistance with clone sequencing. We thank Patrick Schloss, Department of Microbiology & Immunology, University of Michigan, Ann Arbor, Mich., USA, for reviewing the manuscript and providing helpful feedback. This work was supported by Public Health Service Grants the DE-014264 (A.T.), DE-015847 (A.T.), DE-016937 (F.D.), DE-007151 (E.K.), DE-007327 (N.C. and E.K.) from NIDCR and a Medical Faculty/PhD training grant, Umeå University, Sweden (E.K.).
References
- Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16s rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, Boches SK, Dewhirst FE, Griffen AL. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran-Aguilar ED, Barker LK, Canto MT, Dye BA, Gooch BF, Griffin SO, Hyman J, Jaramillo F, Kingman A, Nowjack-Raymer R, Selwitz RH, Wu T. Surveillance for dental caries, dental sealants, tooth retention, edentulism, and enamel fluorosis – United States, 1988–1994 and 1999–2002. MMWR Surveill Summ. 2005;54:1–43. [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series BMethodol. 1995;57:289–300. [Google Scholar]
- Berkowitz RJ. Acquisition and transmission of mutans streptococci. J Calif Dent Assoc. 2003;31:135–138. [PubMed] [Google Scholar]
- Caufield PW, Dasanayake AP, Li Y, Pan Y, Hsu J, Hardin JM. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect Immun. 2000;68:4018–4023. doi: 10.1128/iai.68.7.4018-4023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers NI, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J Bacteriol. 2008;190:8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Non-parametric estimation of the number of classes in a population. Scand J Stat. 1984;4:265–270. [Google Scholar]
- Chhour KL, Nadkarni MA, Byun R, Martin FE, Jacques NA, Hunter N. Molecular analysis of microbial diversity in advanced caries. J Clin Microbiol. 2005;43:843–849. doi: 10.1128/JCM.43.2.843-849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Locker D, Berall G, Pencharz P, Kenny DJ, Judd P. Malnourishment in a population of young children with severe early childhood caries. Pediatr Dent. 2006;28:254–259. [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby PM, Lyons-Weiler J, Bretz WA, Hart TC, Aas JA, Boumenna T, Goss J, Corby AL, Junior HM, Weyant RJ, Paster BJ. Microbial risk indicators of early childhood caries. J Clin Microbiol. 2005;43:5753–5759. doi: 10.1128/JCM.43.11.5753-5759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Wade WG, Yu WH: The human oral microbiome database. 2008. http://www.homd.org [DOI] [PMC free article] [PubMed]
- Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ, Jr, Kolenbrander PE. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury TF, Horowitz AM, Ismail AI, Maertens MP, Rozier RG, Selwitz RH. Diagnosing and reporting early childhood caries for research purposes: a report of a workshop sponsored by the national institute of dental and craniofacial research, the health resources and services administration, and the health care financing administration. J Public Health Dent. 1999;59:192–197. doi: 10.1111/j.1752-7325.1999.tb03268.x. [DOI] [PubMed] [Google Scholar]
- Egland PG, Palmer RJ., Jr Kolenbrander PE: Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci USA. 2004;101:16917–16922. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16s rRNA genes. Appl Environ Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Fukui M. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl Environ Microbiol. 2001;67:3753–3755. doi: 10.1128/AEM.67.8.3753-3755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanasi E, Johansson I, Lu SC, Kressin NR, Nunn ME, Kent R, Jr, Tanner AC. Microbial risk markers for childhood caries in pediatricians' offices. J Dent Res. 2010;89:378–383. doi: 10.1177/0022034509360010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ge Y, Saxena D, Caufield PW. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J Clin Microbiol. 2007;45:81–87. doi: 10.1128/JCM.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang W. Predicting caries in permanent teeth from caries in primary teeth: an eight-year cohort study. J Dent Res. 2002;81:561–566. doi: 10.1177/154405910208100812. [DOI] [PubMed] [Google Scholar]
- Loe H. The gingival index, the plaque index and the retention index systems. J Periodontol. 1967;38(suppl):610–616. doi: 10.1902/jop.1967.38.6.610. [DOI] [PubMed] [Google Scholar]
- Loesche WJ, Rowan J, Straffon LH, Loos PJ. Association of Streptococcus mutans with human dental decay. Infect Immun. 1975;11:1252–1260. doi: 10.1128/iai.11.6.1252-1260.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantzourani M, Fenlon M, Beighton D. Association between Bifidobacteriaceae and the clinical severity of root caries lesions. Oral Microbiol Immunol. 2009a;24:32–37. doi: 10.1111/j.1399-302X.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- Mantzourani M, Gilbert SC, Sulong HN, Sheehy EC, Tank S, Fenlon M, Beighton D. The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res. 2009b;43:308–313. doi: 10.1159/000222659. [DOI] [PubMed] [Google Scholar]
- Marchant S, Brailsford SR, Twomey AC, Roberts GJ, Beighton D. The predominant microflora of nursing caries lesions. Caries Res. 2001;35:397–406. doi: 10.1159/000047482. [DOI] [PubMed] [Google Scholar]
- Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R. Quantitative PCR with 16s rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. 2004;70:167–173. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgrom P, Riedy CA, Weinstein P, Tanner AC, Manibusan L, Bruss J. Dental caries and its relationship to bacterial infection, hypoplasia, diet, and oral hygiene in 6- to 36-month-old children. Community Dent Oral Epidemiol. 2000;28:295–306. doi: 10.1034/j.1600-0528.2000.280408.x. [DOI] [PubMed] [Google Scholar]
- Munson MA, Banerjee A, Watson TF, Wade WG. Molecular analysis of the microflora associated with dental caries. J Clin Microbiol. 2004;42:3023–3029. doi: 10.1128/JCM.42.7.3023-3029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CA, Kent R, Loo CY, Hughes CV, Stutius E, Pradhan N, Dahlan M, Kanasi E, Vasquez S, Tanner A. Diet and caries-associated bacteria in severe early childhood caries. J Dent Res. 2010 doi: 10.1177/0022034510376543. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preza D, Olsen I, Aas JA, Willumsen T, Grinde B, Paster BJ. Bacterial profiles of root caries in elderly patients. J Clin Microbiol. 2008;46:2015–2021. doi: 10.1128/JCM.02411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with arb. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. New technologies, human-microbe interactions, and the search for previously unrecognized pathogens. J Infect Dis. 2002;186(suppl 2):S254–S258. doi: 10.1086/344935. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Yamashita Y, Shibata Y, Torigoe H, Tsuda H, Maeno M. Effect of mixed mutans streptococci colonization on caries development. Oral Microbiol Immunol. 2006;21:47–52. doi: 10.1111/j.1399-302X.2005.00253.x. [DOI] [PubMed] [Google Scholar]
- Senn L, Entenza JM, Greub G, Jaton K, Wenger A, Bille J, Calandra T, Prod'hom G. Bloodstream and endovascular infections due to Abiotrophia defectiva and Granulicatella species. BMC Infect Dis. 2006;6:9. doi: 10.1186/1471-2334-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Furlong MA, Rathbun SL, Whitman WB. Quantitative comparisons of 16s rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol. 2001;67:4374–4376. doi: 10.1128/AEM.67.9.4374-4376.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade GD. Epidemiology of dental pain and dental caries among children and adolescents. Community Dent Health. 2001;18:219–227. [PubMed] [Google Scholar]
- Svensater G, Borgstrom M, Bowden GH, Edwardsson S. The acid-tolerant microbiota associated with plaque from initial caries and healthy tooth surfaces. Caries Res. 2003;37:395–403. doi: 10.1159/000073390. [DOI] [PubMed] [Google Scholar]
- Van Houte J, Gibbs G, Butera C. Oral flora of children with ‘nursing bottle caries’. J Dent Res. 1982;61:382–385. doi: 10.1177/00220345820610020201. [DOI] [PubMed] [Google Scholar]
- Van Houte J, Lopman J, Kent R. The final ph of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J Dent Res. 1996;75:1008–1014. doi: 10.1177/00220345960750040201. [DOI] [PubMed] [Google Scholar]
- Vargas CM, Macek MD, Goodman HS, Wagner ML. Dental pain in Maryland school children. J Public Health Dent. 2005;65:3–6. doi: 10.1111/j.1752-7325.2005.tb02780.x. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]