Abstract
Cone snail venoms have yielded pharmacologically-active natural products of exceptional scientific interest. However, cone snails are a small minority of venomous molluscan biodiversity, the vast majority being tiny venomous morphospecies in the family Turridae. A novel method called lumun-lumun opens access to these micromolluscs and their venoms. Old fishing nets are anchored to the sea bottom for a period of 1–6 months and marine biotas rich in small molluscs are established. In a single lumun-lumun community, we found a remarkable gastropod biodiversity (155 morphospecies). Venomous predators belonging to the superfamily Conoidea (36 morphospecies) were the largest group, the majority being micromolluscs in the family Turridae.
We carried out an initial analysis of the most abundant of the turrid morphospecies recovered, Clathurella (Lienardia) cincta (Dunker, 1871). In contrast to all cDNA clones characterized from cone snail venom ducts, one of the C. cincta clones identified encoded two different peptide precursors presumably translated from a single mRNA. The prospect of easily accessing so many different morphospecies of venomous marine snails raises intriguing toxinological possibilities: the 36 conoidean morphospecies in this one net alone have the potential to yield thousands of novel pharmacologically-active compounds.
Keywords: Marine Biodiversity, turrids, venom peptides
Introduction
Philippe Bouchet and co-workers, (e.g., Bouchet et al., 2002; 2009) comprehensively collected molluscs at several tropical marine sites and uncovered an unprecedented morphospecies richness in the tropical oceans. Several features of the Bouchet studies of tropical marine biodiversity are notable. The first is that a major fraction of all molluscan morphospecies catalogued are extremely small (which were therefore largely ignored by previous workers), with many of these small-shelled molluscan morphospecies not yet described. Furthermore, the most morphospecies-rich group found are venomous predatory snails of the family Turridae (sensu lato) (Bouchet et al., 2002); these are members of the superfamily Conoidea (“conoideans”), also comprising the cone snails (family Conidae) and auger snails (family Terebridae). It was something of a surprise that a carnivorous group could be one of the most morphospecies-rich lineages. In the Mediterranean and other temperate areas, herbivores such as the Rissoidae dominate molluscan biodiversity. An additional noteworthy feature of turrids is that although a number of turrid morphospecies have been described from shallower waters, the great majority are very small morphospecies from deeper water.
Environments such as tropical forests or shallow-water coral reefs are the habitats generally associated with the richness of the biodiversity of the earth. The discovery of an enormous biodiversity of small, venomous snails whose major adaptive radiation appears to be beyond the bathymetric range of normal coral reef habitats might not have been expected — what are the types of biological communities that generate this biodiversity, and why are there so many different morphospecies? Although such communities have neither been defined nor explored, they appear to make an important contribution to tropical marine biodiversity.
In this work, we describe how the diverse morphospecies that comprise such unexplored marine communities can be accessed through a highly specialized commercial activity developed by enterprising Philippine shell-gathering fisherfolk, called lumun-lumun. This is used to obtain microshells for hobbyists, primarily in Japan, who like to examine small natural objects (such as small molluscan shells) through a microscope. There is a continuous demand for such microshells, with sources of new specimens continuously being sought. It is for this commercial purpose that the lumun-lumun fishery was primarily developed in the Visayas.
The basic strategy that underlies lumun-lumun collection is to use old and defective fishing nets with a fine mesh; multiple nets are tied together, and dropped at the bottom typically at depths between 10–100 meters. One end of the conglomerate of nets is anchored down with a rock, with the other end attached to a rope that reaches a ledge in shallow water — the shallow end of the rope is tied to a rock a few meters below the low-tide line, which the fisherman can easily locate. The term lumun-lumun means “combine” or “mix together” in Cebuano, the language spoken in Cebu and Bohol, the islands in the Central Philippines where lumun-lumun is practiced. Thus, the word lumun-lumun conveys the general sense that a number of nets are bundled up tightly before being dropped into the water instead of being spread out in the water as nets usually are.
The bundled nets are generally set where there are currents rich in molluscan larvae, and usually left in place for 1–6 months before they are lifted. The fishermen have considerable empirical experience in which specific spots yield a productive lumun-lumun catch. The net contents are collected, and the molluscs are sieved into size classes, dried out and sold in packets to wholesalers who then distribute the material to retail shops serving the hobbyists. The development of lumun-lumun nets has therefore allowed collection of a spectrum of small molluscan shells, including many that had never previously been seen; these are the standard commercial product of the lumun-lumun fishery.
Thus the deployment of lumun-lumun provides an unprecedented access to the biological communities that settle in such nets; however, a systematic analysis of the contents of lumun-lumun nets has not previously been reported. In this report our goal was to examine the biodiversity to be found in one lumun-lumun net, and in particular, to assess how many different types of venomous molluscs may be present in one lumun-lumun community. This study establishes a base-line data set that will be the starting point for investigating these communities (e.g., how different is each such community? What does each of the predatory molluscan morphospecies prey on? Is there a morphospecies succession pattern?). Since there is a dearth of information regarding the apparently considerable biodiversity of small molluscs that flourish in tropical marine waters, lumun-lumun can be used as an experimental tool for defining and understanding this biodiversity.
We demonstrate in this study that lumun-lumun nets are a habitat for a diversity of venomous snails, and that the venom components of each conoidean micomollusc in a lumun-lumun net can be accessed by molecular genetics. Because each morphospecies has its own distinct complement of 50–200 biologically-active venom compounds, every lumun-lumun community has the potential to be a rich pharmacological and toxinological resource.
Methods
Lumun-lumun fieldwork
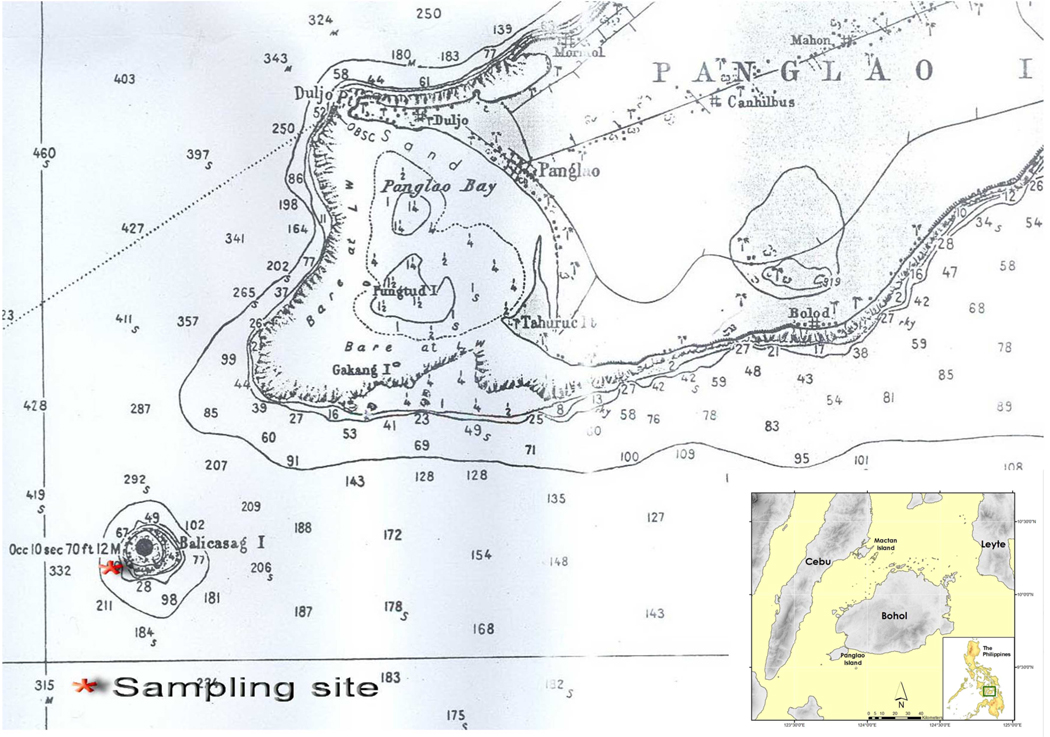
The lumun-lumun analyzed in this study were deployed and harvested off Balicasag Island, Bohol Province, in the Central Philippines; the sampling site is shown in Fig 1. A film crew recorded the lifting of this lumun-lumun (see Fig 2, taken from a video of the lifting of the actual lumun-lumun analyzed below). Two net bundles were laid down at a depth of 30–40 meters in July 2008, tied together to the same coral bench and lifted together on January 9th, 2009.
Figure 1.
Map showing Balicasag Island. The sampling site where the lumun-lumun net described in the text was deployed and harvested is indicated. Balicasag Island is located southwest of the larger island of Panglao. The location of Panglao relative to Bohol and other larger Philippine Islands is shown in the inset.
Figure 2.
The lumun-lumun net being lifted into the boat. Top panel: An underwater shot showing the very end of the net being harvested. On the left side is a diver observing lifting of the net, hanging on to the end of one of the outriggers attached to the boat. Lower panel: A lumun-lumun net shown in the boat. The hand winch used to lift the lumun-lumun net is near the top of the photograph. The photographs are from a video taken for the Howard Hughes Medical Institute and are reprinted here with their permission.
The lumun-lumun was set lifted on off the fore reef zone of Balicasag and the lifted net placed in a large plastic basin and brought to the shore, and then placed on a large polyethylene sheet (about 4m × 8m). The lumun-lumun net was shaken vigorously starting from one end to the other to remove the entangled animals. Additional specimens trapped in the net even after shaking were hand picked individually. All contents of the net were sorted by phyla and placed into the separate containers. The sorted live specimens were then preserved in 70% ethanol. The rest of lumun-lumun contents, comprising sediment and numerous micro-molluscs were soaked with an equivalent volume of 70% ethanol, placed into plastic containers, and the liquid carefully decanted and replaced with fresh 70% ethanol. This fraction was later sorted to separate the micro-molluscs from sediment and other debris. The molluscs were further sorted, first by family and subsequently into distinct morphospecies within each family using an Olympus SZ61 microscope.
Characterization of Clathurella cinta venom duct clones
A pool of Clathurella cincta venom ducts (>0.008g) was homogenized in 0.8 ml TRIzol reagent. The RNA was isolated by phase separation and precipitation according to the manufacturer’s standard protocol (TRIzol Total RNA Isolation - Life Technologies/Gibco BRL, Grand Island, NY).
PolyA mRNA was purified from total RNA using the Quaigen Oligotex spin-column protocol (Quiagen Inc., Valencia CA), according to the manufacturer’s standard protocol. First strand cDNA was prepared from C. cincta polyA mRNA using the Clontech SuperSMART PCR cDNA Synthesis Kit (Clontech Laboratories, Palo Atlo, CA) according to the manufacturer’s standard protocol. Second strand synthesis and amplification were performed by LD PCR, (Clontech SuperSMART PCR cDNA Synthesis, Clontech Laboratories, Palo Alto, CA) according to the manufacturer’s standard protocol, using an Applied Biosystems GeneAMpPCR System 9700 thermalcyler.
The resulting PCR product was gel-purified and 600–1500 bp products recovered from agarose using the High Pure PCR Product Purification Kit (Roche Diagnostics, Indianapolis, IN) following the manufacturer’s suggested protocol for recovery of DNA from agarose. The eluted DNA was treated with End-It end-repair enzyme (Epicentre Biotechnologies, Madison WI), annealed to Novagen pSTBlue1 vector (EMD Biosciences, Gibbstown, NJ) using Fast-Link ligase (Epicentre Biotechnologies) and the resulting products electoporated into Invitrogen DH10B electrocompetent cells (Life Technologies Corporation, Carlsbad, CA), all following the manufacturers’standard protocols. The nucleic acid sequences of the resulting clones were determined according to the standard protocol for ABI automated sequencing.
Sequence analysis
Sequence data were analyzed for translation products using MacVector Version 10.6.0 sequence analysis software (MacVector, Inc., Cary, NC, info@macvector.com). Signal peptide cleavage sites were predicted by SignalP 3.0, a website-based computational method from the Center for Biological Sequence Analysis (http://www.cbs.dtu.dk/services/SignalP/).
Results
Contents of one lumun-lumun net
In its conventional commercial role, after being lifted from the sea bottom, lumun-lumun nets are brought to shore and the contents shaken out of the net, and only the molluscs are saved. The molluscan shells are then dried in the sun, sieved into size classes and sold to wholesalers; a photograph of the mixed molluscs harvested in this way is shown in Fig 3.
Figure 3.
The molluscan harvest from lumun-lumun. The two panels show two size classes sieved through different mesh sizes harvested from a lumun-lumun net. The top panel shows a finely sieved fraction, the bottom panel shows the coarsest fraction with larger shells. Most of the biodiversity arises from the more finely sieved fractions.
We analyzed all of the contents of a commercial lumun-lumun net set off Balicasag Island, Bohol, Philippines. A general overview of the contents of this lumun-lumun net is shown in Tables I and II.
Table I.
Summary of all of the contents of one lumum-lumun net.
| Phylum | Class | Number of Specimens |
Number of Morphospecies |
|---|---|---|---|
| Porifera | Calcarea | 2 | 2 |
| Demospongia | 20 | 16 | |
| Cnidaria | Anthozoa | 3 | 3 |
| Bryozoa | Gymnolaemata | 2 | 2 |
| Mollusca | Bivalvia | 158 | 68 |
| Gastropoda | 606 | 155 | |
| Annelida | Polychaeta | 40 | 10 |
| Arthropoda | Malacostraca | 258* | >20* |
| Echinodermata | Echinoidea | 6 | 3 |
| Ophioruidea | 25 | 7 | |
| Chordata | Osteichthyes | 7 | 19 |
Of the 258 crabs found, 109 were hermit crabs in small gastropod shells; the number of different hermit crab morphospecies could not be determined.
Table II.
Summary of Molluscs Sorted by Family
| A. Class Gastropoda | ||
|---|---|---|
| Family: | Specimens | Morphospecies |
| Fissurellidae | 1 | 1 |
| Turbinidae | 8 | 3 |
| Trochidae | 17 | 4 |
| Vitrinellidae | 1 | 1 |
| Cerithiidae | 115 | 8 |
| Vermetidae | 3 | 1 |
| Triphoridae | 50 | 22 |
| Turritellidae | 14 | 1 |
| Rissoidae | 3 | 2 |
| Strombidae | 3 | 2 |
| Lamellariidae | 1 | 1 |
| Cypraeidae | 21 | 5 |
| Eratoidae | 7 | 3 |
| Triviidae | 1 | 1 |
| Ovulidae | 1 | 1 |
| Personidae | 3 | 2 |
| Ranellidae | 27 | 3 |
| Buccinidae | 77 | 10 |
| Columbellidae | 74 | 12 |
| Fasciolariidae | 2 | 2 |
| Costellariidae | 10 | 9 |
| Mitridae | 5 | 5 |
| Cancellariidae | 3 | 1 |
| Marginellidae | 4 | 1 |
| Muricidae | 52 | 12 |
| Nassariidae | 1 | 1 |
| Scaphandridae | 14 | 5 |
| Terebridae | 1 | 1 |
| Turridae | 74 | 30 |
| Conidae | 13 | 5 |
| B. Class Bivalvia | ||
|---|---|---|
| Family: | Specimens | Morphospecies |
| Anomiidae | 2 | 2 |
| Arcidae | 17 | 11 |
| Cardiidae | 13 | 4 |
| Carditidae | 1 | 1 |
| Chamidae | 1 | 1 |
| Corbulidae | 2 | 2 |
| Glossidae | 1 | 1 |
| Limidae | 1 | 1 |
| Lucinidae | 1 | 1 |
| Mytilidae | 2 | 2 |
| Ostreidae | 7 | 6 |
| Pectinidae | 94 | 21 |
| Pinnidae | 1 | 1 |
| Plicatulidae | 2 | 2 |
| Psammobiidae | 1 | 1 |
| Pteriidae | 4 | 4 |
| Spondylidae | 1 | 1 |
| Tellinidae | 1 | 1 |
| Veneridae | 6 | 5 |
Animals from at least 8 different phyla were recovered. Analysis of the molluscs sorted to the family level revealed that a total of 49 different molluscan families were present, 30 in the class Gastropoda, and 19 in the class Bivalvia. There were 155 morphospecies of gastropods and 68 different morphospecies of bivalves, for a total of 223 different molluscan morphospecies.
The census of molluscan families shown in Table II includes all shells found, whether living, occupied by hermit crabs, or empty (it was not always possible to determine definitively whether a shell was empty or had the animal retracted deep within the shell). A more detailed analysis of three of the families revealed quite different patterns of distribution of living, crabbed and empty shells. In the family Cerithiidae, with 8 morphospecies and 115 individuals, the 8 different morphospecies were all represented by living specimens. Of the 115 shells recovered, 87 were living, 28 had crabs and surprisingly, there were no empty shells recovered.
In contrast, in the family Cypraeidae, there were 5 morphospecies and 21 specimens. One of the morphospecies was only found as an empty shell (Cypraea punctata). There were a total of 14 living specimens and 7 empty specimen shells recovered without crabs. The contrast between these two families (Cerithiidae and Cypraeidae) suggests that both the crabbed and empty shells may be derived from specimens that while alive, were resident in the net. Hermit crabs efficiently occupy any dead certithiid shell, but apparently prefer not to occupy the empty cowrie shells (possibly because the narrow aperture of Cypraea makes them less optimal dwellings for hermit crabs). Although cowrie shells are found inhibited by hermit crabs, the lumun-lumun crabs presumably have a rich choice of alternatives.
This picture is not consistent across all of the taxa analyzed. In the case of the Buccinidae, there were 10 different morphospecies found, and only 3 were represented by living specimens. In one well-represented taxon, Engina mactanensis Cernohorsky, 1985, 19 specimens were recovered and all were with hermit crabs. Another morphospecies, Prodotia iostoma (Gray in Griffith and Pidgeon, 1834) was represented only by 7 specimens with crabs, with no living specimens and no empty shells. In contrast, a third buccinid morphospecies, Clivipollia pulchra (Reeve, 1846) had 2 living specimens, 4 empty shells and no crabbed specimens. These highly divergent patterns of the distribution between living, crabbed and empty shells in the various taxa (at both the family and morphospecies level) is noteworthy, and a variety of potential hypotheses to explain such results need to be tested. These data are suggestive that the dynamics of turnover of different families and morphospecies and/or patterns of predation may vary greatly within this community. However, for most taxa, the majority of specimens were obtained alive.
Conoidean gastropods in the lumun-lumun net: overview
We particularly focused on the conoidean gastropods present in the lumun-lumun net. There were a total of 36 morphospecies recovered that could be assigned to the superfamily Conoidea. The taxonomic status of different families in Conoidea has not been satisfactorily resolved: the traditional division into three families, Conidae, Turridae and Terebridae clearly needs revision. Because the morphological separation into these groups is relatively straightforward, we have retained the classical division into the three families for convenience, although we recognize that this does not accurately reflect true phylogenetic relationships. It has been pointed out that the family Turridae as traditionally defined is polyphyletic, and many of the morphospecies classically included in the family Turridae have been reassigned to other family groups in more recent taxonomic publications (Bouchet and Rocroi, 2005; Kohn, 1998; Puillandre et al., 2008; Taylor et al., 1993).
Of the 36 conoidean morphospecies found, there was one morphospecies of Terebridae, and 5 morphospecies and 13 individuals of Conus. We would not expect many Terebridae to be resident in a lumun-lumun net, since these are highly adapted sand-dwellers; the one Terebra specimen recovered appears to have been introduced into the net by a hermit crab. Of the five Conus morphospecies found, three could be identified: Conus viola Cernohorsky, 1977 (3 specimens); Conus circumcisus Born, 1778 (4 specimens) and one small juvenile tentatively assigned to Conus ichinoseana Kuroda, 1956. Conus circumcisus is a well-established fish-hunting Conus species. There are five additional juveniles representing two different species, neither of which could be definitively identified on the basis of shell morphology.
The great majority of the biodiversity of conoidean molluscs found in the net (30 species) were turrids, primarily belonging to the subfamily Clathurellinae (we have combined the traditional subfamilies Mangeliinae and Borsoninae into Clathurellinae); in the latest comprehensive taxonomic treatment of Philippine molluscs, this group has been treated as a family, Clathurellidae (Sysoev, 2008). Although representatives of other traditional subfamilies (i.e., Drilliinae, Crassispirinae, Raphitominae and Turrinae) were found (see Fig 4), together these are represented by only 11 morphospecies (versus the 19 in Clathurellinae). Consistent with the pattern noted by Bouchet et al. (2002), the number of specimens found for most of the morphospecies recovered was 1–2; for the turrids, the largest number of specimens for an individual morphospecies were the clathurelline Clathurella cincta (11 specimens), the turrine Turridrupa consobrina (8 specimens), and the crassispine Ceritoturris c.f. papillosa (6 specimens) (see Table III). The largest generic group in terms of morphospecies number (10) is the genus Clathurella (see Fig 5); we have treated Clathurella and Lienardia as synonymous. A significant fraction of the recovered morphospecies could not be definitively identified using standard taxonomic references, and it is likely that many of these are novel, undescribed morphospecies.
Figure 4.
Representatives of each of the subfamilies of turrids that were collected in the Balicasag net. Top left, Crassispirinae (Crassispira cerithina). Top right, Raphitominae (Tritonoturris amabilis). Center, Turrinae (Turridrupa consobrina). Bottom left, Drillinae (Clavus bilineatus). Bottom right, Clathurellinae (Mitromorpha stepheni). As discussed in the text, a broad consensus on the phylogeny for the “turrids” has not been reached. We have defined Clathurellinae to include the genera indicated by Sysoev (2008), except for the Raphitominae.
Table III.
Detailed list of Turrid Morphospecies
| Subfamily: | Morphospecies: | # of specimens |
|---|---|---|
| Drillinae | ||
| Clavus bilineatus (Reeve, 1845) | 3 | |
| Clavus protentus (Hervier, 1896) | 1 | |
| Crassispirinae | ||
| Crassispira certhina (Anton, 1838) | 5 | |
| Graciliclava costata (Hedley, 1922) | 1 | |
| Carinapex minutissima (Garrett, 1873) | 2 | |
| Ceritoturris c.f. papillosa (Garrett, 1873) | 6 | |
| Anacithara sp. | 2 | |
| Turrinae | ||
| Turridrupa consobrina (Powell, 1967) | 8 | |
| Turridrupa c.f. jubata (Reeve,1843) | 2 | |
| Clathurellinae | ||
| Clathurellinae (similar to Clathurella) | 2 | |
| Macteola sp. | 1 | |
| Etrema polydesma (Hedley, 1922) | 2 | |
| Etrema c.f. scalarina (Deshayes, 1863) | 5 | |
| Clathurella c.f. rubida (Hinds, 1843) | 2 | |
| Clathurella crassicostata (Pease, 1860) | 1 | |
| Clathurella cincta (Dunker, 1871) | 11 | |
| Clathurella roseotincta (Montrouzier, 1872) | 1 | |
| Clathurella sp. 1 | 1 | |
| Clathurella sp. 2 | 1 | |
| Clathurella sp. 3 | 1 | |
| Clathurella sp. 4 | 1 | |
| Clathurella sp. 5 | 1 | |
| Clathurella sp. 6 | 2 | |
| Hemilienardia hersilia (Hedley, 1922) | 2 | |
| Hemilienardia idiomorpha (Hervier, 1897) | 1 | |
| Heterocythara sp. | 3 | |
| Mitromorpha sp. | 1 | |
| Mitromorpha c.f. stepheni (Melvill & Standen, 1897) | 1 | |
| Raphitominae | ||
| Tritonoturris c.f. amabilis (Hinds, 1843) | 3 | |
| Pseudodaphnella c.f. tincta (Reeve, 1846) | 1 |
Figure 5.
Shells of morphospecies of Clathurella that were recovered in the Balicasag net. A detailed morphospecies list is shown in Table 3. Of the 10 different morphospecies recovered, only 4 could be identified using standard taxonomic references: Top row, left to right: C. c.f. rubida C. sp. 1 and C. cincta. Middle row: C. crassicostata C. sp. 6, C. sp. 4 and C. sp. 2. Bottom row: C. sp. 3, C roseotincta, and C. sp. 5. Some of these morphospecies (e.g. sp. 6) are only provisionally assigned to Clathurella. We have treated Clathurella and Lienardia as synonyms.
Toxinological analysis of Clathurella cincta
An initial analysis of the venom duct of the Clathurella morphospecies for which the largest number of specimens were recovered, Clathurella cincta, was carried out. Since all specimens were under 5mm in length, dissecting the venom ducts was difficult, and a very small amount of messenger RNA was recovered from each individual. The RNA from C. cincta specimens recovered from several lumun-lumun nets was pooled to accumulate sufficient mRNA. Standard protocols were used in the analysis (see Methods); a variety of technical difficulties were encountered, requiring a much larger number of clones to be analyzed to obtain putative toxin sequences.
Compared to most Conus peptides, the predicted Clathurella venom polypeptides are larger and less disulfide rich. Shown in Table IV are the three predicted polypeptide sequences resulting from our analysis; one of these is predicted to have an odd number of cys residues. This is rare in Conus, although not unprecedented. Conus peptides with odd number of cysteine residues have recently been shown to function as a dimer (Quinton et al., 2009; Walker et al., 2009). This raises the possibility that the peptide from Clathurella acts as a dimer under physiological conditions, with the two subunits cross-linked through a disulfide bond.
Table IV.
Sequence of an Open Reading Frame from Clathurella cincta with two Venom Peptide Precursors
 |
| C. Predicted mature peptides after processing. |
|
The complete nucleotide sequence (A) and a translated sequence (B) from a clone in the Clathurella cincta venom duct cDNA library is shown. The two open reading frames are indicated by the yellow and green color, respectively. The arrows indicate where signal sequences are predicted to be cleaved; the double arrow indicates a canonical propeptide cleavage site. The first reading frame from the 5' end of the message has a typical prepropeptide organization; the predicted gene product of this open reading frame is very unconventional in having 3 vicinal cys residues (and no other cys residues in the predicted mature polypeptide). For the second downstream open reading frame, cleavage at the end of the predicted signal sequence would release a polypeptide with 4 cys residues, the gene product predicted to be found in venom after posttranslational modification. There is no propeptide region in this precursor sequence. The predicted mature toxin regions from this clone and another clone from the C. cincta library are shown in C.
One of the clones was unique relative to conopeptide encoding clones from Conus venom ducts: a single messenger RNA apparently encodes two different venom peptides. The relevant open reading frame is shown in Table IV. Both of the predicted translation products yield mature gene products that are 43 amino acids in length, one with two disulfide linkages, and a most unusual peptide with three cys residues all vicinal to each other. This sequence was independently obtained from two clones, although one clone had an incomplete sequence.
The three toxin sequences obtained in this initial analysis demonstrate that it should be possible to identify putative venom polypeptide toxins, generally conforming to the standard Conus paradigm but with some divergent features. The predicted open reading frames elucidated in this initial analysis are unusual compared to most Conus peptides, and some of the rules long established for Conus (“one toxin encoded by one mRNA”) may not be applicable to Clathurella venom peptides. However, given the extremely small size of most lumun-lumun morphospecies, technical innovations will be required to make the elucidation of venom polypeptide sequences more facile in the future. Using standard protocols, it took considerable effort to obtain the few sequences presented here, but the results of this initial analysis provides proof-of-principle that venom peptides can be accessed from Conoidean microspecies in a lumun-lumun net.
Discussion
Net contents versus the dynamic lumun-lumun community
The contents of the lumun-lumun net analyzed include at least 8 different phyla of animals; a notable feature was the preponderance and great diversity of the molluscs found (over 220 morphospecies). Compared to all other groups, molluscs dominated the biodiversity recovered, both with respect to morphospecies diversity and total number of individuals. An issue immediately raised is whether the collection method is biased towards preferential recovery of molluscs. Several factors likely lead to preferential recovery of molluscs in a lumun-lumun net (relative to their actual proportion in the dynamic community).
A greater loss of individuals in some taxa seems likely. As a net lies at the bottom, there is undoubtedly a larger diversity of fish that swim in and out than are recovered. The fish trapped in the net as it is being lifted represent likely only a small a fraction of the total spectrum of fish that participate in the community. Another potential artifact is that as the net is being pulled for harvesting, a higher proportion of individuals in other taxa may escape compared to the relatively slow moving or completely sessile molluscs. The arthropods may be under represented in the harvest: crabs and isopods may crawl out and shrimps could swim out (approximately as many crabs and hermit crabs were recovered as shrimp; no isopods were found in this net, although they are present in other lumun-lumun nets) polychaete worms enriched at substrate levels could be left behind in the substrate as the net is dragged. These lead to a potential underestimate in number of individuals and morphospecies in these taxa. Living molluscs in the net might remain attached to the net, leading to higher recovery.
Additionally, remains of soft-bodied animals would disappear, while the remains of molluscs that have fallen victim to predators could be retained in the net as empty shells. Thus, the contents of the net may reflect the historical record of molluscan morphospecies in the community, but for taxa like fish and polychaetes, only a fraction of live animals present at the precise instant in time that the net is lifted are detected. Thus, the results shown in the Tables are necessarily a distortion of the actual working community, and must be understood as such. It should also be noted that the fishermen do not place the lumun-lumun nets randomly; they clearly have experience regarding which localities are particularly productive, so this is a source of sampling bias. Despite these limitations, it serves as a rare, if imperfect, window to an entire marine ecosystem, about which little is presently known.
Autonomous Reef Monitoring Structures (ARMS), developed by CReefs scientist to attract colonizing non coral invertebrates, is another technique reported to have potential to discover new morphospecies (www.coml.org). ARMS was deployed at a backreef site, a lagoon patch reef site, and fore reef sites, from 1 to 14 m in French Frigate Shoals in the Northwestern Hawaiian Islands. After a year CReefs scientists reported the mollusks as the most abundant group. Lumun-lumun in this study deployed for about six months, yielded more than 300 morphospecies, mollusks being the most abundant taxa. Lumun-lumun composed of old fishing nets served as artificial substrate and afforded habitat structural complexity. Relatively many more morphospecies were recorded than those obtained by other methods, like the tangle nets laid in the sub tidal and suction sampler and brushing basket special equipment using SCUBA in the inter tidal areas (Bouchet, 2009). Apparently, lumun-lumun is more efficient in collecting micro mollusks that settle or are sampled in the net compared to the other collection methods mentioned.
Molluscan biodiversity in one lumun-lumun community
By any measure, the variety of different molluscs found in the net is remarkable (49 different families, over 220 different morphospecies). As might be expected, there is a greater diversity of gastropods than bivalves, but the number of morphospecies of bivalves is very high (68). The largest bivalve groups are the scallops, family Pectinidae (94 individuals, 21 morphospecies) and the ark shells, family Arcidae (17 specimens, 11 morphospecies). Among gastropods, the largest group based on the total number of shells recovered was the cerithids (115 shells, vs. a total of 74 turrid and 50 triphorid shells). The most biodiverse gastropod families were the triphorids, family Triphoridae (21 morphospecies), and the turrids, family Turridae (30 morphospecies).
A significant fraction of the 30 morphospecies recovered in the family Turridae, such as the majority of the Clathurella morphospecies illustrated in Fig 5, are apparently undescribed. Thus, a significant fraction of the lumun-lumun biodiversity will need to be formally described in a taxonomically suitable manner. One obvious biological question is what the different Turrid morphospecies prey on. It would be desirable to determine phylogenetic relationships between morphospecies, identify their prey, and evaluate whether there is a correspondence between the phylogeny of predators and of the prey. The living specimens can be analyzed using standard genetic markers (including the COI barcode) to determine phylogenetic relationships. In principle, those live specimens that ingested prey shortly before the net was harvested are potential candidates for a PCR analysis of gut contents to identify prey. This approach has been carried out successfully on some larger turrids (M. Astilla, unpublished results); “ecological barcoding” of feeding has also been carried out on coralliophids and their cnidarian hosts (Oliverio et al., 2009).
Since the conoidean morphospecies recovered are mostly believed to prey on polychaete worms, a molecular analysis of worms in the net (10 morphospecies) should be carried out so predators of the polychaetes can be paired to its prey. Little is known about specific prey of most turrid taxa, even at the generic level, and therefore there may be some surprises (turrids not eating polychaete worms). The family Turridae is only one of the 49 families of molluscs found. Many of the other families recovered (e.g., Muricidae) are also carnivorous, and therefore a parallel investigation of the prey of these groups can be carried out to define biotic interactions in the lumun-lumun community.
From conoidean biodiversity to pharmacological diversity
The discovery that ∼25% of the recovered gastropod biodiversity (36 different morphospecies) in the lumun-lumun net are venomous conoidean snails has implications for the discovery of novel pharmacologically-active natural products. Prior work on the superfamily Conoidea, (and in particular, on one small segment of this biodiverse group, the fish-hunting cone snails in the genus Conus) has demonstrated the pharmacological potential of compounds produced by these snails (for a review, see Olivera and Teichert, 2007). Each conoidean venom has between 50–200 different pharmacologically active compounds, and one has become an FDA-approved drug for intractable pain (Prialt) with at least 5 others from cone snail venoms reaching human clinical trials (Miljanich, 2004; Terlau and Olivera, 2004). Initial toxinological work done on other conoidean groups indicates that other conoideans have venoms of comparable complexity to cone snails (Heralde et al., 2008; Imperial et al., 2007; Imperial et al., 2003; López-Vera et al., 2004; Watkins et al., 2006)
Since conoideans use venom to capture prey, defend against predators and deter potential competitors (Olivera, 2002), their venom is chemically complex, with 50–200 different compounds in the venom of each morphospecies. Because the genes encoding venom components rapidly diverge from each other; each morphospecies of venomous conoidean has its own particular complement of venom compounds, with essentially no overlap at all between two different morphospecies, (Olivera, 2006). Within a single morphospecies however, the venom components are the same from one individual to the next, except for allelic variation. Thus, the discovery of 36 morphospecies of conoideans in a single lumun-lumun net implies that there are probably far more new compounds in the venoms of morphospecies in this one net than have been elucidated in all decades that conoidean venoms have been investigated. The potential for discovering novel toxinological mechanisms from the contents of a single lumun-lumun net is therefore very high. We have taken the first step in accessing such sequences (see Table IV). Two of the venom peptides isolated have a structural framework similar to that of the J-superfamily of Conus (Imperial et al., 2006), members of which have been shown to target K+ channels. However, these peptides do not share any extensive sequence homology with the Conus superfamily.
It was not so long ago that to access compounds from the venoms of animals as small as the conoidean microspecies in a lumun-lumun_net would have been inconceivable. However, a number of technical developments, conceptual insights and the use of molecular genetics/genomics to fuel discovery (Olivera and Teichert, 2007) now make even miniscule amounts of tissue sufficient, in principle, for chemical and pharmacological discovery. In theory, we should be able to access the potentially novel pharmacological agonists that have been evolved over millions of years by the diverse conoidean morphospecies found in a lumun-lumun net. As has been reviewed elsewhere (Terlau and Olivera, 2004), it is highly likely that such venom compounds will target specific ion channels, receptors and transporters in nervous systems, and therein lies their pharmacological potential.
The ability to easily access novel conoidean biodiversity using lumun-lumun nets should therefore open the door to a vast treasure trove of new biologically-active natural products. In practice, a number of technical difficulties need to be overcome before easy access to the natural products in the venoms of conoidean microspecies can be obtained in a facile manner. Because the morphospecies are so small, dissecting their venom ducts quickly is challenging, and the lag in dissection probably results in significant RNA degradation. Furthermore, venom ducts of some turrid morphospecies are disproportionately small, and thus even under optimal conditions, a tiny amount of RNA is recovered from each specimen. When only small amounts of mRNA are available, the analysis of cDNA clones becomes technically challenging using standard protocols. Thus, in order to obtain cDNA sequences to access the venom peptides efficiently, a number of methodological innovations will be required. Nevertheless, we have demonstrated here that we can indeed obtain venom peptide sequences from the tiny conoidean morphospecies in a lumun-lumun net, and that these peptides exhibit intriguing differences from Conus venom peptides.
Acknowledgements
The work described in this report was supported by NIH Grant GM48677 (to BMO) and by a PharmaSeas grant (to GPC and PMA) from the Department of Science and Technology of the National Government of the Republic of the Philippines. This work was also supported by the Howard Hughes Medical Institute (through a Professor’s award to BMO). The top photograph in Figure 2 was kindly provided by an HHMI team that came to film in the Philippines (Satoshi Amagai, Dennis Liu, and Blake Porch). A large complement of students, assistants and aides participated in the field trip to Bohol; we would particularly like to acknowledge the contributions of Maria Diarey Tianero, Tatiana Steyker, Rowena Antemano in organizing activities during the field trip, and of Meljune Chicote in sorting specimens from the net. We thank Francis Fletcher Freire for providing a map, and Belinda Longakit for providing her expertise in sponge taxonomy.
Footnotes
Conflict of Interest Statement: The authors declare that there are no conflicts of interest.
References
- Bouchet P, Lozouet P, Maestrati P, Heros V. Assessing the magnitude of species richness in tropical marine environments: high numbers of molluscs at a New Caledonia site. Biological Journal of the Linnean Society. 2002;75:421–436. [Google Scholar]
- Bouchet P, Lozouet P, Sysoev AV. Deep-Sea Research, Fred Grassle Festschrift. 2009 [Google Scholar]
- Bouchet P, Ng P, Largo D, Panglao 2004-Investigations of the marine species richness in the Philippines. The Raffles Bulletin of Zoology. 2009 Supplement No. 20:1–19. [Google Scholar]
- Bouchet P, Rocroi JP. Malacologia - International Journal of Malacology. ConchBooks; 2005. Malacologia: International Journal of Malacology, Classification and Nomenclator of Gastropod Families. [Google Scholar]
- Heralde FM, 3rd, Imperial J, Bandyopadhyay PK, Olivera BM, Concepcion GP, Santos AD. A rapidly diverging superfamily of peptide toxins in venomous Gemmula species. Toxicon. 2008;51(5):890–897. doi: 10.1016/j.toxicon.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperial JS, Kantor Y, Watkins M, Heralde FM, 3rd, Stevenson B, Chen P, Hansson K, Stenflo J, Ownby JP, Bouchet P, Olivera BM. Venomous auger snail Hastula (Impages) hectica (Linnaeus, 1758): molecular phylogeny, foregut anatomy and comparative toxinology. Journal of Experimenta Zoology Part B: Molecular and Developmental Evolution. 2007;308(6):744–756. doi: 10.1002/jez.b.21195. [DOI] [PubMed] [Google Scholar]
- Imperial JS, Watkins M, Chen P, Hillyard DR, Cruz LJ, Olivera BM. The augertoxins: biochemical characterization of venom components from the toxoglossate gastropod Terebra subulata. Toxicon. 2003;41:391–398. doi: 10.1016/s0041-0101(03)00169-7. [DOI] [PubMed] [Google Scholar]
- Kohn AJ. Superfamily Conoidea., in: Mollusca: Southern Synthesis. In: Beesley PL, Ross GJB, Wells A, editors. Fauna of Australia. Melbourne: CSIRO Publishing; 1998. pp. 846–854. [Google Scholar]
- López-Vera E, Heimer de la Cotera EP, Maillo M, Riesgo-Escovar JR, Olivera BM, Aguilar MB. A novel structural class of toxins: the methionine-rich peptides from the venoms of turrid marine snails (Mollusca, Conoidea) Toxicon. 2004;43:365–374. doi: 10.1016/j.toxicon.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Miljanich GP. Ziconotide: neuronal calcium channel blocker for treating severe chronic pain. Current Medicinal Chemistry. 2004;11:3029–3040. doi: 10.2174/0929867043363884. [DOI] [PubMed] [Google Scholar]
- Olivera BM. Conus venom peptides: reflections from the biology of clades and species. Annual Review of Ecology, Evolution and Systematics. 2002;33:25–42. [Google Scholar]
- Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. Journal of Biological Chemistry. 2006;281(42):31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: a model for concerted pharmacological discovery. Molecular Interventions. 2007;7(5):251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- Oliverio M, Barco A, Modica MV, Richter A, Mariottini P. Ecological barcoding of corallivory by ITS2 sequences: hosts of coralliophiline gastropods detected by the cnidarian DNA in their stomach. Molecular Ecology Resources. 2009;9:94–103. doi: 10.1111/j.1755-0998.2008.02388.x. [DOI] [PubMed] [Google Scholar]
- Poppe GT. Philippine Marine Mollusks, in: (Gastropoda - Part 2) In: Poppe GT, editor. ConchBooks; 2008. p. 848. [Google Scholar]
- Puillandre N, Samadi S, Boisselier MC, Sysoev AV, Kantor YI, Cruaud C, Couloux A, Bouchet P. Starting to unravel the toxoglossan knot: molecular phylogeny of the "turrids" (Neogastropoda: Conoidea) Molecular Phylogenetic Evolution. 2008;47(3):1122–1134. doi: 10.1016/j.ympev.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Quinton L, Gilles N, De Pauw E. TxXIIIA, an atypical homodimeric conotoxin found in the Conus Textile venom. Journal of Proteomics. 2009;72(2):219–226. doi: 10.1016/j.jprot.2009.01.021. [DOI] [PubMed] [Google Scholar]
- Sysoev AV. Turridae - (Clathurellidae), in: Philippine Marine Mollusks - Vol. II. In: Poppe GT, editor. Hackenheim, Germany: ConchBooks; 2008. pp. 732–755. [Google Scholar]
- Taylor JD, Kantor Y, Sysoev AV. Foregut anatomy, feeding mechanisms, relationships and classifiction of the Conoidea (=Toxoglossa) (Gastropoda) Bulletin, Natural History Museum, London (2001) 1993;(59):125–170. [Google Scholar]
- Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiological Reviews. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- Walker CS, Jensen S, Ellison M, Matta JA, Lee WY, Imperial JS, Duclos N, Brockie PJ, Madsen DM, Isaac JTR, Olivera BM. A Novel Conus Snail Polypeptide Causes Excitotoxicity by Blocking Desensitization of AMPA Receptors. Current Biology. 2009;19:1–9. doi: 10.1016/j.cub.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins M, Hillyard DR, Olivera BM. Genes expressed in a turrid venom duct: divergence and similarity to conotoxins. Journal of Molecular Evolution. 2006;62(3):247–256. doi: 10.1007/s00239-005-0010-x. [DOI] [PubMed] [Google Scholar]