Abstract
Formalin-fixed paraffin-embedded (FFPE) tissues have been used to discover disease-associated protein changes using mass spectrometry. Protein post-translational modifications such as glycosylation are known to associate with disease development. In this study, we investigated whether FFPE tissues preserve such modifications and therefore can be used as specimen of choice to identify the disease-associated modifications. We isolated the glycopeptides from the tryptic digest of frozen and FFPE lung tissues using solid-phase extraction of glycopeptides and analyzed them using mass spectrometry. The glycopeptides identified from FFPE lung tissue were compared to the ones from frozen lung tissue regarding their relative abundance, unique glycosylation sites, and subcellular locations. The results from our study confirmed that glycosylation in FFPE tissues are preserved and FFPE tissues can be used for discovery of new disease associated changes in protein modifications. Furthermore, we demonstrated the feasibility of applying the strategy of glycopeptide isolation from tryptic peptides of FFPE tissue to other tissues such as liver and heart.
Keywords: N-glycosylation, FFPE tissue, protein post-translational modifications, quantitative analysis
INTRODUCTION
Surgical tissue has recently become a popular specimen of choice in the field of cancer biomarker discovery rather than serum 1 for two reasons. First, proteins in a given tissue are less complex and have a narrower dynamic range than those in serum. Second, like prostatic specific antigen (PSA), a tumor marker for prostate cancer, which has much higher concentrations in prostatic tissue than that in serum, proteins with cancer-biomarker potential may exist at higher concentrations and are readily identified in tissues than in serum. Hence, the idea of identifying potential cancer biomarkers in surgical tissues using quantitative proteomics and then targeting them in serum using antibody-based assays or mass spectrometry, so called tissue-targeted proteomic approach, make surgical tissues an important specimen for biomarker discovery 2, 3.
Two types of surgical tissues are available: fresh or frozen tissues and formalin-fixed paraffin-embedded (FFPE) tissues. Currently, the former are the main sources for tissue-targeted biomarker discovery. However, two issues with using fresh or frozen tissues are their limited availability in large qualities and variable quality due to lack of standardization of preparation and storage conditions. On the contrary, FFPE tissues, archived with pathological and clinical information processed by universal standard methodology, are readily available worldwide in hospitals and tissue banks. The concern of using FFPE tissues, however, is whether proteins in FFPE tissues, stabilized via cross-linking by the process of formalin-fix and paraffin-embed, are feasible for proteomic analysis.
The pioneering work of Hood et al has recently identified hundreds of proteins from soluble peptides extracted from FFPE tissues using mass spectrometry4–6. However, the feasibility of FFPE specimens for study of protein post-translational modifications such as glycosylation has not yet been examined. Protein glycosylation is known as the most abundant post-translational modification and ~50% of all proteins are estimated to be glycosylated 7, 8. Glycosylation is fundamental in normal cell functions such as cell adhesion, migration, and signal transduction 9–12, as well as in disease genesis and progression13, 14. Most glycosylated proteins are membrane associated or extracellular and readily released to body fluids and could serve as potential biomarkers in body fluids such as serum15, 16.
In this study, we investigated the feasibility of FFPE tissues for quantitative glycoproteomic analysis using the solid-phase extraction of N-liked glycopeptides (SPEG), a glycopeptide isolation technique that was reported recently by our group 17, 18. The FFPE tissues from mouse lung were proteolyzed to peptides and SPEG was then used to isolate glycopeptides followed by LC-MS/MS analysis. We compared quantitatively the glycopeptides from FFPE lung tissue with those isolated from frozen lung tissue and found that glycopeptides were identified in frozen tissues as well as FFPE tissues, although the relative abundances of the glycopeptides from FFPE tissue were reduced. To this end, our study demonstrated the feasibility of FFPE tissues for glycoproteomic analysis. This knowledge will facilitate the usage of FFPE tissues for study of protein glycosylation and may be useful to identify glycoproteins as potential biomarkers for disease diagnosis, progression and prognosis.
MATERIALS AND METHODS
Materials
Hydrazide resin and sodium periodate (Affi-Gel H2 Oxidizer) were from Bio-Rad (Hercules, CA). PNGase F was from New England Biolabs (Ipswich, MA). Sequencing grade trypsin was from Promega (Madison, WI). C18 columns (Sep-Pak Vac) were from Waters (Milford, MA). All other chemicals were from Sigma-Aldrich (Missouri, MO).
Tissues
Mouse tissues were collected and either snap frozen or fixed in 10% normal buffered formalin (NBF) for four hours. After fixation NBF tissues were transferred to 70% alcohol and processed to paraffin using a Shandon Hypercenter XP (GMI, Inc., Ramsey, Minnesota). For Shandon Hypercenter XP, a standard program of dehydration was carried out in ethanol 70%, 80%, 90%, 95%, 100%, 100% 1 hour each, followed by xylenes twice and 30 min each. Finally, the tissue was soaked in paraffin 30 min and repeated once and then hold in the second paraffin bath for at last 1 hour.
Tryptic digestion and SPEG isolation of N-linked glycopeptides
FFPE tissues block (~28mg) were placed in a 50~60°C oven for 30 min followed by incubation in a xylene bath for 5 min. This incubation step was then repeated twice. After that, excess liquid was shaken off from the tissues before they were soaked in each of the following solutions for 3 min by the order of fresh absolute ethanol, fresh 90% ethanol and fresh 80% ethanol. Then they were gently rinsed in running tap water for 30 s and placed in a PBS wash bath at room temperature for 30 min for further rehydration. After rehydration, the FFPE tissues were weighed to and ~50mg tissues were used for glycopeptide extraction in each experiment.
Rehydrated FFPE tissues or frozen tissues (~50mg from each tissue), sliced into 1~3mm in thickness, were vortexed in 100µl of 5mM phosphate buffer for 2–3 min and sonicated for 5 min in an ice-water bath to homogenize the tissues. After that, the tissue homogenate was incubated in 100µl of trifluoroethanol (TFE) at 60°C for 2 h followed by sonication for 2 min to denature proteins. Protein disulfide bonds were then reduced by a 30 min incubation of the tissues with 5mM tributylphosphine (TBP) at 60°C and alkylated by another 30 min incubation with 10mM iodoacetamide at RT in dark. Finally, the proteins were diluted 5-fold with 50mM NH4HCO3 (pH7.8) to reduce the TBP concentration to 1mM before digested using tryspin (enzyme to protein ratio of 1:50 w/w) overnight at 37°C with gently shaking. Proteins before and after tryptic digestion were analyzed by protein gel, and silver staining was used to determine whether the proteins from tissues were completely digested into peptides. The complete tryptic digestion was indicated by disappearance of protein bands with high molecular weight and appearance of peptide bands with low molecular weight (<10kDa). After trypsin digestion, samples were centrifugated at 13,000 rpm for 5 minutes to remove any particulate matter. Each sample contains ~ 4mg of peptides measured by the BCA assay. N-linked glycopeptides were then isolated from 2mg of peptides from the digests described above using SPEG 17. The enriched N-linked glycopeptides were concentrated by C18 columns and dried down and resuspended in 40µl of 0.4% acetic acid before LC-MS/MS analysis.
LC-MS/MS analysis
From 40µl of isolated N-linked glycopeptides, 3 µl (~1.5µg) were analyzed by ESI quadrupole-time-of-flight mass spectrometer (Q-TOF, Waters, Beverly, MA) using a linear acetonitrile gradient (from 5%–32% B over 100 min; A= 0.1% formic acid in water, B=0.1% formic acid in acetonitrile). Each sample was analyzed three times to decrease the errors induced by different MS runs in a label-free LC-MS method based on label-free peak intensity of the same peak among different LC-MS runs. Linear ion-trap mass spectrometer (LTQ, Thermo-Finnigan, San Jose, CA) was also used to increase the coverage of glycopeptides isolated from each sample and increase the number of identifications with an acetonitrile gradient from 5%–35% over 85 min at a flow rate of 0.15 ml/min.
Protein and glycosylation site identification
Raw MS data of LTQ and Q-TOF were converted into mzXML files by the MassWolf file converter 19. MS/MS spectra were searched with SEQUEST 20 against a mouse International Protein Index database (IPI, version3.13). The peptide mass tolerance is 2.0Da for Q-TOF and 3.0Da for LTQ. Other parameters of database searching are modified as follows: oxidized methionines (add methionine 16), a (PNGase F-catalyzed) conversion of Asn to Asp and cysteine modification (add cysteine 57). The output files were evaluated by INTERACT and ProteinProphet21, 22. The criterion of ProteinProphet analysis is the probability score ≥ 0.9 so that low probability protein identifications can be filtered out.
Number of peptide identifications represents total number of MS/MS spectra used to make the assignment of peptide sequences using SEQUEST20. The N-linked glycosylation site is the Asp contained within the consensus N-linked glycosylation motif.
Quantitative proteomic analysis
LC-MS method-MS spectra were aligned and quantitatively analyzed by SpecArray software suite 23. Briefly, SpecArray software suite uses LC-MS data generated by ESI-Q-TOF of peptides from each sample and sequentially performs the following tasks to determine relative abundance of peptides in different conditions: 1) Peptide peak picking: a list of peptide peaks was generated from each LC-ESI-MS run; 2) Peptide alignment: Peptides detected in individual LC-MS patterns were aligned based on peptide mass and retention time. The software tool accounted for shifts in the retention time in different LC-MS analyses during peptide alignment. 3) Peptide Array: For each peptide peak, an abundance ratio of matched peptides in different samples was determined for each peptide peak. An in-house developed software tool was then used to link the peptides identified from MS/MS spectra to their corresponding MS peaks by matching retention time, precursor mass, and charge states of the peptides.
Subcellular location of identified proteins
Signal peptides were predicted using SignalP 2.0 24. Transmembrane (TM) regions were predicted using TMHMM (version 2.0)25. The TMHMM program predicts protein topology and the number of TM helices. Information from SignalP and TMHMM were combined to separate proteins into the categories: i) cell surface - proteins that contained predicted non-cleavable signal peptides and no predicted transmembrane segments; ii) secreted - proteins that contained predicted cleavable signal peptides and no predicted transmembrane segments; iii) transmembrane - proteins that contained predicted transmembrane segments and extracellular loops and intracellular loops; and iv) intracellular - proteins that contained neither predicted signal peptides nor predicted transmembrane regions. All protein sequences were taken from IPI mouse protein database (version 3.13).
Results and discussion
Glycoproteomic analysis of FFPE tissue
FFEP treatment cross-links proteins in tissues so that they are stabilized and locked in position for microscopic analysis. Given the harsh treatment conditions, it is reasonable to question the quality of proteins in FFPE tissues for proteomic analyses, especially the ones that aim to identify post-translational modification and sites. To this end, we investigated whether the process of formalin fixation and paraffin embedding would interfere with analyses of protein glycosylation by isolation of glycopeptides using SPEG and analyzing isolated glycopeptides by LC-MS and LC-MS/MS.
First, we compared the number of glycopeptide identifications from FFPE lung tissue with that from frozen lung tissue (Figure 1). When peptides were filtered using a probability score cut off of ≥ 0.9, 92% of the identified peptides (1243 out of 1348) from the frozen lung tissue and 85% of the peptides (485 out of 571) from the FFPE tissue were glycopeptides indicated by the inclusion of at least one of the consensus N-linked glycosylation motifs in the identified peptide sequences. In contrast, only 7% of tryptic peptides in the human proteome contain potential N-linked glycosylation motifs (N-X-T/S)26. This observation demonstrates that specific capture of glycopeptides by oligosaccharide tags using SPEG specifically enriches glycopeptides from FFPE tissue, evidenced by the fact that ~85% of the identified peptides are glycopeptides. Most important, it demonstrates that 1) protein glycosylation is preserved in FFPE tissues and 2) glycosylated proteins can be identified from them using mass spectrometry.
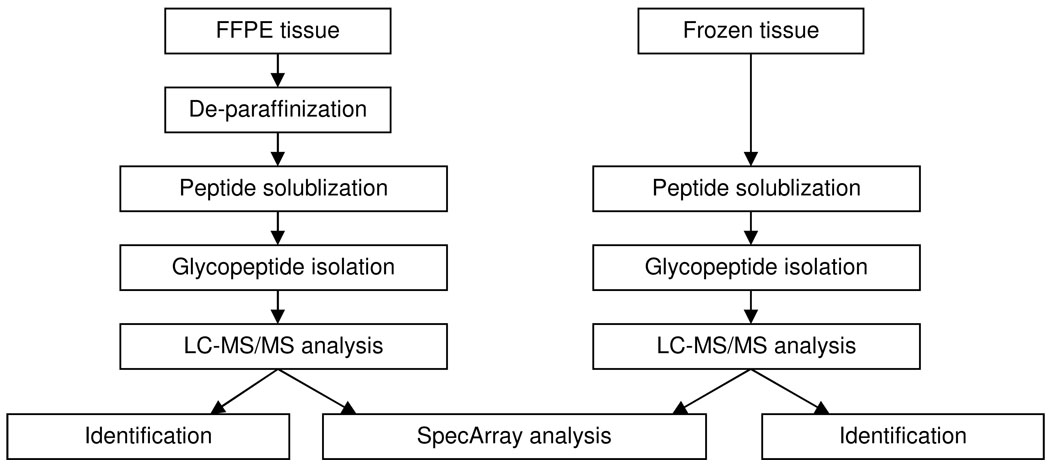
Figure 1.
Flow chart comparison of glycoprotein analyses from FFPE tissue and frozen tissue.
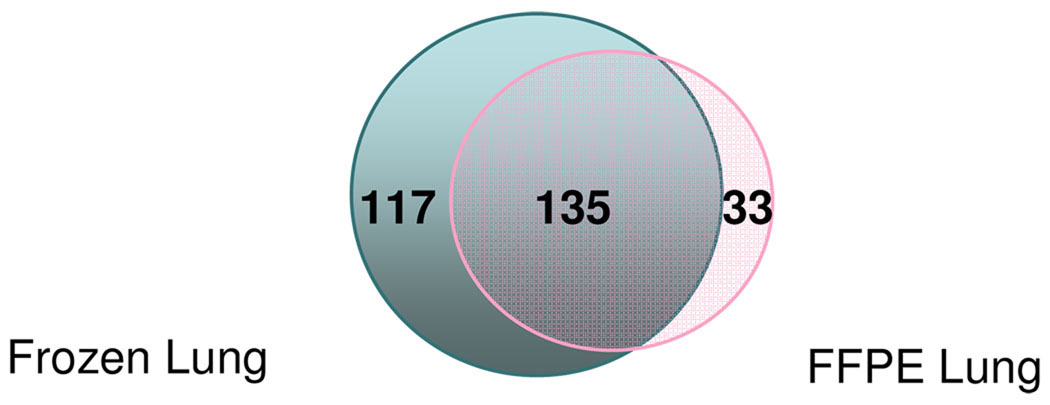
Second, we compared the number of unique glycosylation sites identified from FFPE and frozen lung tissues (Figure 2). 168 and 252 unique glycosylation sites were identified from the FFPE and frozen lung tissue, respectively. Although the number of unique glycosylation sites identified from the frozen lung tissue was greater than from the FFPE lung tissue, 80% of the glycosylation sites (135 out of 168) identified from the latter were present in the former (Figure 2). Some glycosylation sites are only identified from the FFPE lung tissue (33 out of 168). A larger number of glycosylation sites (117) were uniquely identified from frozen lung tissue but not from FFPE lung tissue.
Figure 2.
Venn diagram summary of unique glycosylation sites identified by LTQ: 168 in FFPE lung tissue, 252 in frozen lung tissue and 135 are in common.
To further investigate the reason attributed to the difference in peptide sequences identified in frozen lung or FFPE lung tissue, we compared the relative abundance of the glycopeptides identified from both tissues (Category I), only in the frozen tissue (Category II), and only in the FFPE tissue (Category III) in Table 1. The relative abundance of these glycopeptides were calculated by SpecArray using the relative MS intensity from Q-TOF 23 as were the peptide abundance ratio of frozen lung to FFPE lung (shown in Table 1). We also matched the identified glycopeptides from MS/MS spectra of the same Q-TOF run to its corresponding MS peak. A total of 47 identified N-linked glycopeptides were quantified in both specimens: 31 in Category I, 14 in Category II, and 2 in Category III. Among the 31 glycopeptides in Category I, 3 of them had higher abundance in FFPE tissue (ratios ≤0.5). This may be because the fixed tissues are substantially denser than frozen tissues caused by the shrinkage of fixed tissue. Palmer-Toy et al reported that more proteins were identified in the fixed/embedded tissue attributed to the same possible reason 6. Nevertheless, the majority of the peptides in this class (14 out of 31) had ratios ≥2, which indicates that the glycopeptide isolation is less efficient in FFPE tissue compared to frozen tissue. In category II, although 14 glycopeptides were only identified by MS/MS spectra in the frozen lungs, we were able to detect the corresponding MS peaks with low intensity in FFPE tissue and quantified the peptides using the MS intensity. Most of them were not identified due to the low abundance of these peptides and random sampling of mass spectrometer. The random sampling of mass spectrometer is that only a certain number of peptide peaks detected by mass spectrometry are randomly selected to be further analyzed by MS/MS, depending on the analyzing efficiency of the instrument. A very similar observation was made in Category III, in which the glycopeptides were only identified in FFPE tissue (Table 1). Based on these quantitative data, the difference of identification could be mainly attributed to the low concentration of peptides and limited detection ability of the mass spectrometer, rather than a loss of the peptide. This specific difference could be compensated by increasing the number of MS analyses to achieve complete and reproducible coverage of the proteome. In addition, sample pre-fractionation by multiple dimensions prior to mass spectrometry, such as gas-phase fractionation of the m/z dimension or capillary isoelectric focusing based multidimensional separation coupled with nano-RPLC (reversed-phase liquid chromatography), have also been reported27, 28.
Table 1.
Glycoproteins identified in FFPE lung and frozen lung, with their corresponding peak intensity detected by MS.
| IPI | protein name | Peptide | cellular locatin |
P* | tryptic Ends |
FFPE lung | FrozenLung | ratio # |
|---|---|---|---|---|---|---|---|---|
| Category I proteins identified in both FFPE lung and frozen lung | ||||||||
| IPI00108041 | Stromal interaction molecule 1 | R.LAVTN*TTM#TGTVLK.M | TM | 1 | 2 | 36.7±22.9 | 93.0±5.8 | 2.5 |
| IPI00111013 | Cathepsin D | K.YYHGELSYLN*VTR.K | S | 1 | 2 | 93.7±0.1 | 149.8±10.3 | 1.6 |
| IPI00113539 | Fibronectin | R.DQCIVDDITYNVN*DTFHK.R | S | 1 | 2 | 29.0±2.1 | 163.5±57.8 | 5.7 |
| IPI00114958 | Splice Isoform HMW of Kininogen-1 | K.HSIEHFNN*NTDHSHLF.T | S | 1 | 2 | 236.0±33.6 | 447.6±78.6 | 1.9 |
| IPI00116913 | Laminin alpha-5 chain | R.LLLGGLPVSGTFHN*FSGCIS.N | S | 1 | 1 | 5.8±0.2 | 15.2±0.7 | 2.6 |
| IPI00117857 | Alpha-1-antitrypsin 1–6 | R.ELVHQSN*TSNIFF.S | S | 0.99 | 2 | 53.6±6.5 | 40.9±4.1 | 0.8 |
| IPI00117957 | Asporin | R.ITDIEN*GTFANIPR.V | S | 1 | 2 | 92.8±25.7 | 26.0±8.7 | 0.3 |
| IPI00121362 | F11r protein | R.AFM#N*SSFTIDPK.S | TM | 1 | 2 | 13.7±6.3 | 63.7±19.7 | 4.7 |
| IPI00122231 | Advanced glycosylation end product-specific receptor | A.QILPN*GSLLLPATGIVDEGTFR.C | TM | 1 | 1 | 32.5±13.3 | 62.7±15.7 | 1.9 |
| IPI00122973 | Splice Isoform 1 of Intercellular adhesion molecule 1 | K.FLFKN*QTLELH.V | TM | 0.97 | 1 | 97.3±34.2 | 45.7±11.3 | 0.5 |
| IPI00123223 | Murinoglobulin-1 | V.M#AQGSIIQTGN*HTHQVEPGEAPVK.G | A | 1 | 2 | 27.2±3.4 | 66.7±2.2 | 2.5 |
| IPI00123831 | SDR1 protein | K.ENGVFEEISN*SSGR.F | TM | 0.99 | 2 | 61.8±5.2 | 59.0±9.0 | 1.0 |
| IPI00125058 | Splice Isoform B of Laminin alpha-3 chain | Q.LLPLGNISDNVDR.I | S | 0.97 | 2 | 50.8±33.8 | 190.7±24.2 | 3.8 |
| IPI00125813 | Dipeptidyl peptidase 4 | K.LDFIVLN*ETR.F | TM | 0.98 | 2 | 13.5±5.7 | 7.1±4.9 | 0.5 |
| IPI00126194 | Alpha-2-macroglobulin | I.SIDTSN*FTAPFLR.V | S | 1 | 2 | 8.4±0.7 | 47.6±5.1 | 5.7 |
| IPI00127352 | AMBP protein | K.KEDSCQLN*YSEGPCLGM#QER.Y | S | 1 | 2 | 53.7±14.8 | 145.6±12.0 | 2.7 |
| IPI00128249 | Alpha-2-HS-glycoprotein | H.ALDPTPLAN*CSVR.Q | S | 1 | 2 | 574.7±76.3 | 1592.0±242.4 | 2.8 |
| IPI00128484 | Hemopexin | R.VAEVEN*GTKPDSDVPEHC.L | S | 0.91 | 1 | 75.9±32.3 | 102.4±50.7 | 1.4 |
| IPI00131830 | Serine protease inhibitor A3K | K.NLINDYVSNQTQGM#IK.E | S | 1 | 2 | 234.0±28.6 | 851.5±242.9 | 3.6 |
| IPI00133751 | Splice Isoform 1 of Microfibrilassociated glycoprotein 4 | R.VDLEDFEN*NTAYAK.Y | S | 0.99 | 2 | 401.5±38.4 | 356.0±29.7 | 0.9 |
| IPI00134585 | Glutamyl aminopeptidase | K.IALN*LTM#YLK.S | TM | 1 | 2 | 5.9±1.3 | 6.2±2.0 | 1.1 |
| IPI00139788 | Serotransferrin | K.N*STLCDLCIGPLK.C | S | 1 | 2 | 37.5±2.2 | 74.2±2.0 | 2.0 |
| IPI00169815 | Procollagen, type VI, alpha 2 | R.GTFTDCALAN*M#TQQIR.Q | S | 0.99 | 2 | 42.9±3.8 | 72.0±4.4 | 1.7 |
| IPI00169896 | Splice Isoform 2 of Choline transporter-like protein 2 | K.TCNPETFPLRN*ESLQCPTAR.C | TM | 1 | 2 | 43.5±16.0 | 45.5±2.6 | 1.1 |
| IPI00273822 | 16 days embryo lung cDNA | K.IGTYEVLN*GSR.L | S | 1 | 2 | 25.6±6.0 | 49.7±6.2 | 1.9 |
| IPI00322447 | RA175 | R.FQLLN*FSSSELK.V | TM | 1 | 2 | 12.3±2.1 | 13.5±2.2 | 1.1 |
| IPI00331214 | Platelet glycoprotein IV | K.VISNN*CTSYGVLDIGK.C | TM | 0.98 | 2 | 49.0±15.6 | 224.9±12.0 | 4.6 |
| IPI00345112 | IntegrIn alpha 8 | R.M#VVCDLGNPM#VTGTN*FSLGLR.F | TM | 1 | 2 | 21.9±1.7 | 15.7±2.0 | 0.7 |
| IPI00406434 | Mini-agrin | K.NELM#LN*SSLM#R.I | S | 1 | 2 | 93.6±29.7 | 228.5±7.1 | 2.4 |
| IPI00466371 | 16 days neonate cerebellum cDNA, | R.GELQSEN*SSLTLSSSNR.K | TM | 0.98 | 2 | 63.4±9.0 | 110.4±19.8 | 1.7 |
| IPI00467944 | 61 kDa protein | L.DLGGASTQITFVPQN*STIESPENSLQFR.L | TM | 1 | 1 | 12.8±1.0 | 56.3±1.4 | 4.4 |
| Categroy II proteins identified only in frozen lung | ||||||||
| IPI00113797 | Napsin A | W.FN*LTGQDYVIK.I | S | 0.96 | 1 | 333.5±32.5 | 191.0±24.0 | 0.6 |
| IPI00113824 | Basement membrane-specific heparan sulfate proteoglycan core protein | R.SLTQGSLIVGNLAPVN*GTSQGK.F | A | 1 | 2 | 4.1±0.1 | 40.0±1.2 | 9.8 |
| IPI00113869 | Splice Isoform 2 of Basigin | W.FKTSDTGEEEAITN*STEANGK.Y | A | 0.99 | 2 | 30.2±29.6 | 19.9±4.7 | 0.7 |
| IPI00115516 | EMILIN-1 | R.LEDRFN*STLGPSEEQEK.N | S | 0.99 | 2 | 33.8±5.3 | 110.4±14.1 | 3.3 |
| IPI00116105 | Corticosteroid-binding globulin | M.M#VQSGN*ISYFR.D | S | 1 | 2 | 161.9±25.4 | 90.0±8.2 | 0.6 |
| IPI00117093 | Laminin beta-3 chain | R.QTACTPGDCPGELCPQDN*GTACGSHCR.G | A | 1 | 2 | 35.2±2.3 | 38.8±6.2 | 1.1 |
| IPI00123957 | Cd97 protein | R.DFNPATVN*YTIQK.L | TM | 1 | 2 | 38.9±27.9 | 111.7±9.5 | 2.9 |
| IPI00126050 | Bone marrow macrophage cDNA, RIKEN full-length enriched library | K.EVM#NLLQPLN*VTK.V | S | 0.94 | 2 | 167.8±22.3 | 877.8±119.4 | 5.2 |
| IPI00132474 | Integrin beta-1 | K.SAVGTLSGN*SSNVI.Q | TM | 0.96 | 1 | 67.5±22.8 | 403.2±99.5 | 6.0 |
| IPI00272381 | 2 days neonate sympathetic ganglion cDNA | K.DM#SDGFISN*LTIQR.Q | S | 1 | 2 | 45.5±2.8 | N/A | |
| IPI00272690 | Angiotensin-converting enzyme, somatic isoform | K.TFDVSN*FQN*SSIKR.I | TM | 1 | 2 | 177.1±64.9 | 177.5±15.5 | 1.0 |
| IPI00420955 | Sortilin 1 | K.DITNLIN*NTFIR.T | TM | 0.92 | 2 | 6.2±0.1 | 9.7±1.9 | 1.6 |
| IPI00469123 | Melanoma cell adhesion molecule | N.SLGSN*TTTIVLK.L | TM | 0.98 | 1 | 169.5±21.3 | 105.2±13.6 | 0.6 |
| IPI00649550 | Protein | E.KQELSEYN*ATAIK.S | TM | 1 | 1 | 29.7±4.7 | 76.7±18.5 | 2.6 |
| Category III proteins identified only in FFPE lung | ||||||||
| IPI00330833 | Complement C5 | F.VLNLPSN*VTVLK.F | S | 0.99 | 2 | 21.3±3.5 | 0.0 | |
| IPI00417180 | UDP glycosyltransferase 1 family, polypeptide A10 | K.EFEAYVN*ASGEHGI.V | TM | 1 | 1 | 529.5±77.6 | 324.1±22.0 | 0.6 |
probability score of peptides by ProteinProphet
ratio of peptide intensity from frozen lung vs. FFPE lung
Subcellular location of glycoproteins
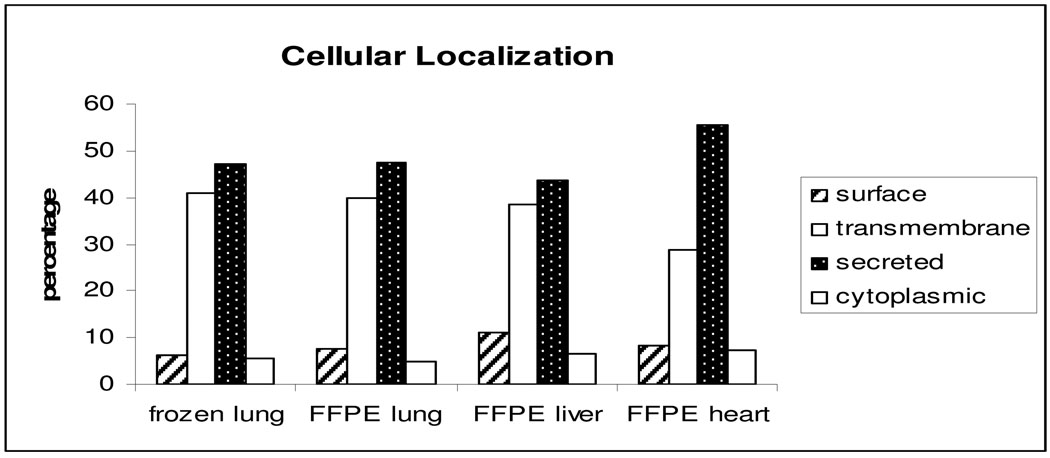
We compared the subcellular location of glycoproteins identified from the frozen and FFPE lung tissues as another measure of the quality of FFPE tissues for glycoprotein analysis. Glycoprotein subcellular distribution from the frozen and FFPE lung tissues were identical: 7.7% and 6.4% cell surface proteins, 39.9% and 40.6% transmembrane proteins, 47.6% and 47.4% secreted proteins, and 4.8% and 5.6% cytoplasmic proteins were from FFPE and frozen lung, respectively (Figure 3). The classification of the identified proteins showed identical subcellular distributions. Since glycoproteins are most likely transmembrane, cell surface, or secreted proteins, the subcellular distribution of the glycoproteins enriched from both FFPE lung and frozen lung is different from global proteome analysis5.
Figure 3.
Distribution summary of N-linked glycoproteins identified by LTQ in tissues from frozen lung, FFPE lung, FFPE liver, and FFPE heart, according to predicted cellular location.
Efficiency of trypsin digestion
It is commonly thought that crosslinking caused by formalin-fixation might interfere with trypsin digestion. To investigate whether the formalin fixation could impact the efficiency and specificity of trypsin digestion, the information of tryptic ends of the identified peptides from FFPE and frozen lung is shown in Table 2. The percentage of peptides identified from FFPE lung with arginyl and lysyl termini was 51.63% and 48.37%, respectively, compared to these in frozen lung, 50.43% and 49.57%. The prevalence of these two residues within proteins is considered roughly equivalent5. Furthermore, 63.10% and 64.68% peptides with one tryptic end, 36.9% and 35.3% peptides with two tryptic ends, and 16.07% and 17.06% peptides with missed cleavage sites were identified in FFPE lung and frozen lung, respectively. All the data suggest the formalin fixation did not make dramatic differences in the efficiency and specificity of trypsin from frozen tissue, in accordance with the previous report5.
Table 2.
Comparison of tryptic ends and missed cleavage sites in FFPE lung and fresh frozen lung.
| % of 1 trypt | % of 2 tryptic end | % of -R | % of -K | missed cleavage site % | |
|---|---|---|---|---|---|
| FFPE lung | 63.10 | 36.9 | 51.63 | 48.37 | 16.07 |
| Frozen lung | 64.68 | 35.32 | 50.43 | 49.57 | 17.06 |
Investigation of other FFPE tissues
In addition to lung, we also investigated whether SPEG method could also be applied to other FFPE tissues such as heart and liver. Using the same approach for lung tissues, we identified 166 and 135 unique glycosylation sites from FFPE heart and liver, as compared to 168 glycosylation sites from lung. This indicated the feasibility of SPEG method to other FFPE tissues. We also compared glycoprotein subcellular location among FFPE lung, heart and liver tissues to further determine the specificity of the SPEG method, which indicated that the extracellular proteins can be significantly enriched in FFPE heart and liver, similar to FFPE lung (Figure 3).
Conclusion
In this study we were able to identify glycopeptides with glycosylation sites from FFPE lung and compared the identified glycopeptides with that from frozen lung using the SPEG and mass spectrometry. The data indicated that FFPE tissue can be used for glycoproteomic analysis, although the abundance of glycopeptides from FFPE tissue was reduced. Such differences in intensity were also reported in a previous study by qualitative analysis of protein extraction from FFPE 4. The intensity differences affected the identification of glycosylation sites to a certain degree. Furthermore, the protein cellular distributions from FFPE tissue, based on prediction from sequence, shows extreme similarity with frozen tissue. In short, the glycosylation of proteins is preserved in FFPE tissues. This may imply that other post-translational modification, such as phosphorylation and sulphation, may be also preserved in FFPE tissues and such modified proteins can be analyzed using specific enrichment followed by analysis using mass spectrometry. Furthermore, the SPEG method can also be successfully applied to other FFPE tissues, such as liver and heart. This study may facilitate the usage of FFPE tissues for analyses of proteins with diverse glycosylation forms for biomarker discovery.
Acknowledgment
This work was supported with federal funds from the National Cancer Institute, National Institutes of Health, by Grants R21-CA-114852 and this work was partially supported by the HERA Foundation.
REFERENCE
- 1.Kondo T. Tissue proteomics for cancer biomarker development: laser microdissection and 2D-DIGE. BMB Rep. 2008;41(9):626–634. doi: 10.5483/bmbrep.2008.41.9.626. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Liu AY, Loriaux P, Wollscheid B, Zhou Y, Watts JD, Aebersold R. Mass spectrometric detection of tissue proteins in plasma. Mol Cell Proteomics. 2007;6(1):64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Tian Y, Kelly-Spratt KS, Kemp CJ, Zhang H. Indentification of glycoproteins from mouse skin tumors and plasma. Clin Proteom. 2008 doi: 10.1007/s12014-008-9014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KS. Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab Invest. 2005;85(11):1405–1415. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 5.Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4(11):1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Palmer-Toy DE, Krastins B, Sarracino DA, Nadol JB, Jr, Merchant SN. Efficient method for the proteomic analysis of fixed and embedded tissues. J Proteome Res. 2005;4(6):2404–2411. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- 7.Gu J, Taniguchi N. Regulation of integrin functions by N-glycans. Glycoconj J. 2004;21(1–2):9–15. doi: 10.1023/B:GLYC.0000043741.47559.30. [DOI] [PubMed] [Google Scholar]
- 8.Wong CH. Protein glycosylation: new challenges and opportunities. J Org Chem. 2005;70(11):4219–4225. doi: 10.1021/jo050278f. [DOI] [PubMed] [Google Scholar]
- 9.Isaji T, Gu J, Nishiuchi R, Zhao Y, Takahashi M, Miyoshi E, Honke K, Sekiguchi K, Taniguchi N. Introduction of bisecting GlcNAc into integrin alpha5beta1 reduces ligand binding and down-regulates cell adhesion and cell migration. J Biol Chem. 2004;279(19):19747–19754. doi: 10.1074/jbc.M311627200. [DOI] [PubMed] [Google Scholar]
- 10.Ono M, Handa K, Withers DA, Hakomori S. Glycosylation effect on membrane domain (GEM) involved in cell adhesion and motility: a preliminary note on functional alpha3, alpha5-CD82 glycosylation complex in ldlD 14 cells. Biochem Biophys Res Commun. 2000;279(3):744–750. doi: 10.1006/bbrc.2000.4030. [DOI] [PubMed] [Google Scholar]
- 11.Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406(6794):411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 12.Zheng M, Fang H, Hakomori S. Functional role of N-glycosylation in alpha 5 beta 1 integrin receptor. De-N-glycosylation induces dissociation or altered association of alpha 5 and beta 1 subunits and concomitant loss of fibronectin binding activity. J Biol Chem. 1994;269(16):12325–12331. [PubMed] [Google Scholar]
- 13.Axford JS, Cunnane G, Fitzgerald O, Bland JM, Bresnihan B, Frears ER. Rheumatic disease differentiation using immunoglobulin G sugar printing by high density electrophoresis. J Rheumatol. 2003;30(12):2540–2546. [PubMed] [Google Scholar]
- 14.Holland M, Yagi H, Takahashi N, Kato K, Savage CO, Goodall DM, Jefferis R. Differential glycosylation of polyclonal IgG, IgG-Fc and IgG-Fab isolated from the sera of patients with ANCA-associated systemic vasculitis. Biochim Biophys Acta. 2006;1760(4):669–677. doi: 10.1016/j.bbagen.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr Opin Chem Biol. 2004;8(6):617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2(7):521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 17.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2(2):334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 19.Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22(11):1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 20.Eng JMAL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:13. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 21.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 22.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 23.Li XJ, Yi EC, Kemp CJ, Zhang H, Aebersold R. A software suite for the generation and comparison of peptide arrays from sets of data collected by liquid chromatography-mass spectrometry. Mol Cell Proteomics. 2005;4(9):1328–1340. doi: 10.1074/mcp.M500141-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997;8(5–6):581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 25.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Loriaux P, Eng J, Campbell D, Keller A, Moss P, Bonneau R, Zhang N, Zhou Y, Wollscheid B, Cooke K, Yi EC, Lee H, Peskind ER, Zhang J, Smith RD, Aebersold R. UniPep--a database for human N-linked glycosites: a resource for biomarker discovery. Genome Biol. 2006;7(8):R73. doi: 10.1186/gb-2006-7-8-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blonder J, Rodriguez-Galan MC, Lucas DA, Young HA, Issaq HJ, Veenstra TD, Conrads TP. Proteomic investigation of natural killer cell microsomes using gas-phase fractionation by mass spectrometry. Biochim Biophys Acta. 2004;1698(1):87–95. doi: 10.1016/j.bbapap.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Guo T, Wang W, Rudnick PA, Song T, Li J, Zhuang Z, Weil RJ, DeVoe DL, Lee CS, Balgley BM. Proteome analysis of microdissected formalin-fixed and paraffin-embedded tissue specimens. J Histochem Cytochem. 2007;55(7):763–772. doi: 10.1369/jhc.7A7177.2007. [DOI] [PubMed] [Google Scholar]