Abstract
During early stage of embryonic development, the liver bud, arising from the foregut endoderm, is the beginning for the formation of future liver three-dimensional structure. While the gene expression profiles associated with this developmental stage have been well explored, the detailed cellular events are not as clear. Epithelial-mesenchymal transition (EMT) was thought to be essential for cell migration in the early vertebrate embryo but seldom demonstrated in human liver development. In this study, we tried to identify the cell populations with both stem cell and EMT features in the human liver bud. Our in situ studies show that the phenotype of EMT occurs at initiation of human liver development, accompanied by up-regulation of EMT associated genes. A human liver bud derived stem cell line (hLBSC) was established, which expressed not only genes specific to both mesenchymal cells and hepatic cells, but also Octamer-binding protein 4 (OCT4) and Nanog. Placed in appropriate media, hLBSC differentiated into hepatocytes, adipocytes, osteoblast-like cells and neuron-like cells in vitro. When transplanted into severe combined immunodeficiency mice pre-treated by carbon tetrachloride, hLBSC engrafted into the liver parenchyma and proliferated. These data suggest that there are cell populations with stem cell and EMT-like properties in the human liver bud, which may play an important role in the beginning of the spatial structure construction of the liver.
Keywords: epithelial-mesenchymal transition, liver bud, liver development, multipotent stem cells, fetal liver
1. Introduction
During early stage of embryonic development, the liver bud arises from the foregut endoderm by 8.5 days after coitus (dpc) in mice and by 3.5 gestational weeks (gw) in humans (Rossi JM et al., 2001; Zaret KS, 2001; Lemaigre FP, 2009). These endodermal cells eventually give rise to hepatoblasts, following specific signals from both the cardiac mesoderm and the cells of septum transversum mesenchyme, and accompanied by activation of specific transcription factors (Rossi JM et al., 2001; Zaret KS, 2001; Lemaigre FP, 2009). By coordination of proliferation, migration, intercellular adhesion and differentiation of these hepatoblasts, the liver expands to form the complicated three-dimensional structure. Although the molecular signals underlying liver genesis have been well described, the precise cellular morphogenic events have been seldom described during this stage.
Epithelial-mesenchymal transition (EMT) is thought to be essential for germ layer formation and cell migration in the early vertebrate embryo (Viebahn C, 1995; Davies JA,1996; Thiery JP et al., 2009) and has been demonstrated in vitro in both hepatocytes and progenitor cells in rodent and human neonatal (Pagan R et al., 1995, 1999) and fetal livers (Suzuki A et al., 2001; Chagraoui J et al., 2003; Dan YY et al.,2006; Inada M et al., 2008; Valdes F et al., 2002; Choi SS and Diehl AM, 2009). Since EMT could be induced in cultured cells, cell lines with EMT features may not truly represent the real cell status in vivo. Therefore Chagraoui J et al. (2003) tried to find cells expressing both cytokeratin (CK) 8 and alpha smooth muscle actin (αSMA) in biopsies from murine fetal liver. They observed EMT stromal cells in the murine fetal liver from 11.5dpc to 18.5dpc, with a time course parallel to that of hematopoiesis. Aside from this, there have been no other in vivo reports describing the involvement of EMT in liver development, especially in humans.
Do EMT involve in early human liver development? Lemaigre FP (2009) proposed that immediately after hepatic initiation, specific endoderm epithelium cells migrate through the basement membrane and invade the septum transversum to form the liver bud in a process putatively involving EMT. On this basis, the existence of stem cells with EMT features in early developmental stage of human fetal liver was postulated, but the cell populations involved in the EMT have never been definitively identified.
In this study, immunofluorescence was performed on paraffin imbedded human fetal livers between 5gw to 28gw to reveal co-expression of αSMA and CK8 only before 8gw, which suggest that EMT only occurs at initiation of liver development. Microarray analysis showed EMT-related genes were up-regulated in human liver bud around 5gw. To verify the presence of EMT-competent cells at 5gw, cells with EMT-like features were isolated from aborted human fetus of around 5gw. A distinct cell population with features of EMT and stem cell was established by culture medium optimization and clonal growth, which we named the human liver bud derived stem cells (hLBSC). In this report, we show the time course of EMT during early human liver development, and we report the isolation, long-term ex vivo culturing, and characterization of cell lines from human early liver bud with EMT-like properties and their competency in the repopulation of injured adult livers.
2. Materials and Methods
2.1. Human fetal livers
Human fetal livers were obtained from voluntary abortions performed in compliance with the Chinese legislation and in accordance with a protocol approved by the Institutional Ethics Review Board. The fetal age was established by standard clinical parameters and confirmed on anatomic criteria.
2.2. Isolation, culture, and clone of hLBSC line
Human fetal liver was peeled off thoroughly under an anatomical microscope and then washed by phosphate-buffered saline (PBS), pH 7.4. Single-cell suspensions were obtained by treatment with 0.25% Trypsin/EDTA (Gibcol BRL) for 5 minutes at 37°C and vigorous pipetting, then they were suspended in the optimized medium consisting of 50% IMDM (Gibcol BRL), 50% Ham F12 (Gibcol BRL), 2.5% FBS (Hyclone), 5 µg/ml insulin (Sigma St. Louis, MO), 20 ng/ml EGF (Sigma, St Louis, MO) and 1 mM GlutamaxTM (Gibcol BRL). The cells were plated at a 2×104 cells/cm2 in 12-well or 6-well plates (Gibcol BRL) pre-coated by 0.2% gelatin (Sigma St. Louis, MO). Once triangle-shaped cell colonies formed in 2 weeks in primary culture, the peri-clone cells were selectively detached from culture by scraping with cell scraper (Sarstedt, Newton, NC), and the colonies were treated by 0.05% Trypsin/EDTA (Gibcol BRL) and plated onto 6-well plates (Gibcol BRL) pre-coated by 0.2% gelatin.
2.3. RT-PCR analysis
RT-PCR was conducted as described (Li WL et al., 2006). Forward and reverse primers used for specific amplification can be found in Supplementary references, as listed in Supplementary Table 1.
2.4. Transduction of hLBSC
Transduction of hLBSC with the retroviral vector pLNCG-C1 was conducted as described previously (Tao XR et al., 2009) in order to establish the hLBSC-pLNCG-C1 cell line with stable EGFP expression for transplantation.
2.5. Immunofluorescence
hLBSC seeded onto chamber slides were permeabilized and fixed with methanol. Human fetus or fetal livers were fixed with 4% phosphate-buffered paraformaldehyde overnight at 4°C and embedded in OCT compound. Cells or tissues were incubated with the first antibody for 16 hours at 4°C, and then stained with the Dako Envision detection system (Dako, Calif). The first antibodies used included polyclonal rabbit anti-human ALB antibody (1:500, Dako), polyclonal rabbit anti-human α-1-anti-trypsin (α-1-AT) antibody (1:400, Dako), monoclonal mouse anti–human CK8, polyclonal rabbit anti-human αSMA antibody. Fluorescence was detected by Alexa Fluo 555 (1:200, Molecular probes/Invitrogen) or FITC-conjugated secondary antibodies (1:200, Molecular probes/Invitrogen). 40, 60-diamidino-2-phenylindole (DAPI) (Molecular Probes) was used to stain nuclei. The same reactions without primary antibody incubation were used as negative controls.
2.6. Microarray analysis
According to 4×44K Whole Genome Oligo Microarrays (one-color) Technical Manual (Agilent), total RNA was harvested from human liver bud around 5gw and adult human liver tissue, using TRIzol (Invitrogen) and the RNeasy kit (Qiagen) according to manufacturer’s instructions, including a DNase digestion step. After having passed RNA measurement on the Nanodrop ND-1000 and denaturing gel electrophoresis, the samples were amplified and labeled using the Agilent Quick Amp labeling kit and hybridized with Agilent whole genome oligo microarray in Agilent’s SureHyb Hybridization Chambers. After hybridization and washing, the processed slides were scanned with the Agilent DNA microarray scanner (part number G2505B) using settings recommended by Agilent Technologies. The resulting text files extracted from Agilent Feature Extraction Software (version 10.5.1.1) were imported into the Agilent GeneSpring GX software (version10.0) for further analysis.
2.7. Flow cytometry
For analysis of membrane antigens, hLBSC cell suspension was prepared by Trypsin/EDTA, and then 105~106 cells were stained sequentially with PerCP-conjugated anti-CD45 mAb, FITC-conjugated anti-CD34 mAb, FITC-conjugated anti-CD44 mAb, phycoerythrin-conjugated anti-CD29 mAb and relevant homotype control antibodies (all from PharMingen). Cells were also stained with 4 µg/ml propidium iodide in PBS before being passed through a FACS Calibur (Becton Dickinson, Mountain View, CA) flow cytometer. Gating was implemented based.
2.8. In vitro differentiation
To induce hepatocytic differentiation, the hLBSC cells were plated at a density of 2×104 cells/cm2 in medium A comprising 90% DMEM (Gibcol BRL), 10% FBS (Hyclone), 100 IU/ml penicillin G and 100 µg/ml streptomycin (Gibcol BRL). When the cells achieved about 60% confluence, 45 mg/ml SB was added. Ten days after SB supplement, expression of ALB, gamma glutamyl transpeptidase (GGT), dipeptidyl peptidase IV (DPPIV), CK19, tryptophan oxygenase (TO) and hepatocyte nuclear factor 4α (HNF4α) was detected by RT-PCR. Human albumin (Dako, Denmark) and human tyrosine aminotransferase (TAT) (Dako, Denmark) were analyzed by immunofluorescence. The PAS assay was used to visualize glycogen. For adipogenic differentiation, hLBSC were plated at a density of 5×104 cells/cm2 in the medium A as described previously. When the cells achieved about 100% confluence, 10% FBS (Hyclone) was substituted by 10% horse serum (Gibcol BRL). Two weeks later, Oil-red staining was performed and expression of PPAR-γ2 was detected by RT-PCR. To induce osteogenesis, hLBSC were plated at a density of 3×104 cells/cm2 in osteogenic medium containing DMEM-HG supplemented with 0.2 mM Vitamin C, 10−7 M Dexamethasone, and 10 mM β-glycerophosphate (all from Sigma). Fourteen days later the cells were subjected to alkaline phosphatase staining. And expression of collagen I was analyzed by RT-PCR. For neural differentiation, hLBSC were plated at a density of 3×104 cells/cm2 in the induction medium with 80% DMEM (Gibcol BRL), 20% FBS (Hyclone), 100 IU/ml penicillin G and 100µg/ml streptomycin (Gibcol BRL). When the cells achieved about 70% confluency, 20% FBS was removed and 1 mM BME was added. Four days after BME supplement, expression of nestin was detected by RT-PCR.
2.9. In vivo transplantation
pLNCG-C1 transfected hLBSC (2.4×106) were suspended in PBS and then injected intrasplenically into SCID mice with acute liver damage in the Balb/c background (n = 7). Acute liver damage in SCID mice was induced by intraperitoneal injection of 5 ml/kg WT CCL4 dissolved in olive oil (1: 5 v/v) one day before transplantation. The mice were sacrificed 3 weeks after implantation. Liver cryostat sections in 10 µm thickness were observed under a fluorescence microscope to detect the transplanted cells. Quantification of EGFP-positive cells in liver was performed in cryostat sections (10 random ×100 visual fields were selected to get mean value for every mouse). The same Liver cryostat sections were stained by Hematoxylin and eosin (H&E) to show the locus of transplanted cells. Expression of human α-1-AT was determined by immunofluorescence on 4µm liver sections fixed in 4% phosphate-buffered paraformaldehyde overnight at 4°C and embedded in OCT compound.
3. Results
3.1. Presence of cells in EMT in early gestational stage of human fetal liver
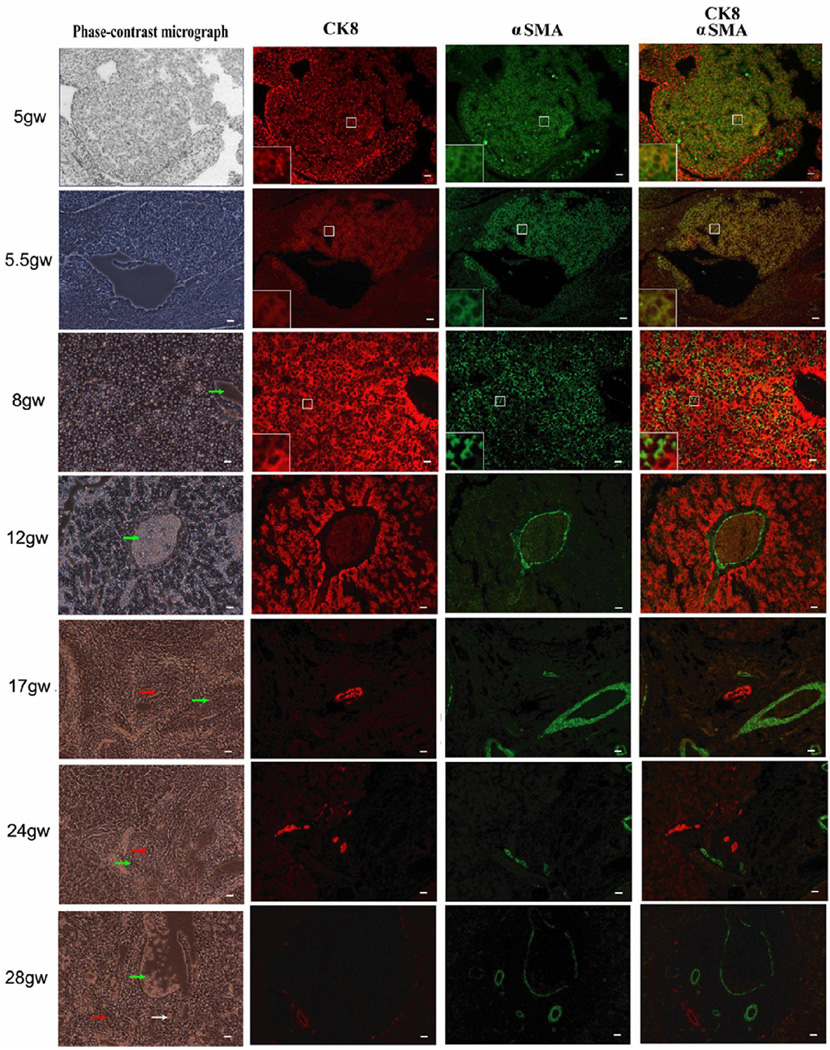
To assess the occurrence of EMT during human fetal liver development, multi-immunostaining with R555-conjugated anti-CK8 and FITC-conjugated anti-αSMA was performed on paraffin imbedded human fetal livers between 5gw to 28gw. As shown in Fig. 1, co-expression of CK8 and αSMA was observed in most non-hematopoietic cells of human fetal liver at early gestational stages (5gw and 5.5gw), in only a few cells at 8gw, but in none of the cells at later gestational stages (12gw, 17gw, 24gw and 28gw). By 12gw, expression of CK8 and αSMA was segregated. CK8 was positive in hepatocytes constituting the lobules at 12gw and only in bile ducts at later development times. In contrast, the αSMA-positive signal, probably originating from smooth muscle cells, was observed in most nonparenchymal cells at 8gw and in the wall of interlobular veins and arteries after 12gw. The results, based on co-expression of CK8 and αSMA, strongly indicate that EMT only occurs at early stages of human liver development.
Fig. 1. Immunofluorescent study of human fetal livers from 5gw to 28gw.
Multi-immunostaining with R555-conjugated anti–CK8 (red), FITC-conjugated anti-αSMA (green) and DAPI (blue). Co-expression of CK8 and αSMA is observed in most non-hematopoietic cells of human fetal liver at early gestational stages (5gw and 5.5gw), in only a few cells at 8gw, but in none of the cells at later gestational stages (12gw, 17gw, 24gw and 28gw). By 12gw, expression of CK8 and αSMA was segregated. Some cells in centre box were magnified as shown in left-down box. Arrows in green, white and red represent interlobular veins, interlobular arteries and bile ducts respectively. Scale bar = 20 µm.
To understand the molecular events in early human liver development, microarray analysis was performed to compare mRNA expression between human liver bud around 5gw and adult human liver tissue. As shown in Supplementary Table 2, those genes related with EMT and up-regulated more than 2-fold in the liver bud, relative to adult liver, were mapped to pathways extracted from KEGG (Mlecnik B, 2005) and BIOCARTA (http://www.biocarta.com/genes/allPathways.asp), which may interact with Twist and Snail transcription factors that play important roles in the process of EMT (Kalluri R and Weinberg RA, 2009; Zeisberg M and Neilson EG, 2009; Kalluri R, 2009; Acloque H et al., 2009) and early liver development (Rossi JM et al., 2001; Zaret KS, 2001; Lemaigre FP, 2009).
3.2. Establishment of hLBSC lines from the human Liver bud
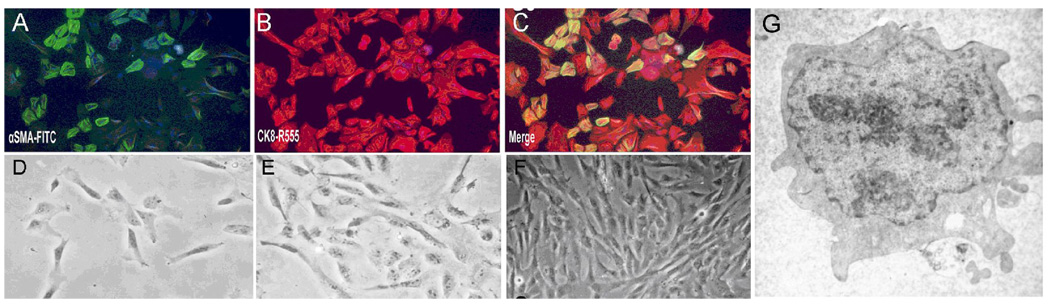
To isolate EMT cells from early human fetal liver, we used fresh liver tissues from aborted human fetus at around 5gw. The initial cell population in primary culture was a mixture of large fibroblast-like and epithelial-like cells, and suspended hematopoietic cells. By the 3rd day in culture, 60% of the cells were double positive for αSMA and CK8 (Fig. 2A–C), among which we can see a few very small cells in triangle shape. Within two weeks in culture, only the small triangle-shaped cells survived, and these cells proliferated rapidly and grew confluent in 3~4 days thereafter (Fig. 2D, E). The confluent cell colonies were picked and purified by limited serial dilution and further passaged in culture for up to 25 continuous passages. After repeated subpassaging, the cell line which we eventually named hLBSC had small fibroblastic shaped morphology, a high cytoplasm to nucleus ratio, and few organelles in the cytoplasm (Fig. 2F, G). Eleven hLBSC colonies were derived respectively from eleven fetal livers (Supplementary Table 3), all of which exhibited homogenous morphology and vigorous growth characteristics. hLBSC clones derived from the liver FL031204-2 was used for further analysis. The growth curve of hLBSC at the fifth passage is shown in Supplementary Fig. 1, with a population doubling time of 28.1 hours according to the formula TD = T×log2/log (N/N0). These cells had normal karyotype. After transplanted subcutaneously into SCID mice (n=4), hLBSC at passage 16 did not develop any visible neoplasma within 14 weeks of observation.
Fig. 2. Establishment of human liver bud derived stem cell (hLBSC).
(A–C) bi-immunostaining of human fetal liver cells at the 3rd day of primary culture with FITC-conjugated anti-αSMA (green, A) and R555-conjugated anti–CK8 (red, B), Merged image (C) showed 60% cells were double positive for both αSMA and CK8 (yellow), among which we can see a few very small cells in triangle shape. Within two weeks in culture, only the small triangle-shaped cells survived (D), and these cells proliferated rapidly and grew confluent in 3~4 days thereafter (E). After repeated subpassaging, the cell line which we eventually named hLBSC had small fibrous shaped morphology(F), a high cytoplasm to nucleus ratio, and few organelles in the cytoplasm (G).
3.3. hLBSC show both epithelial and mesenchymal features
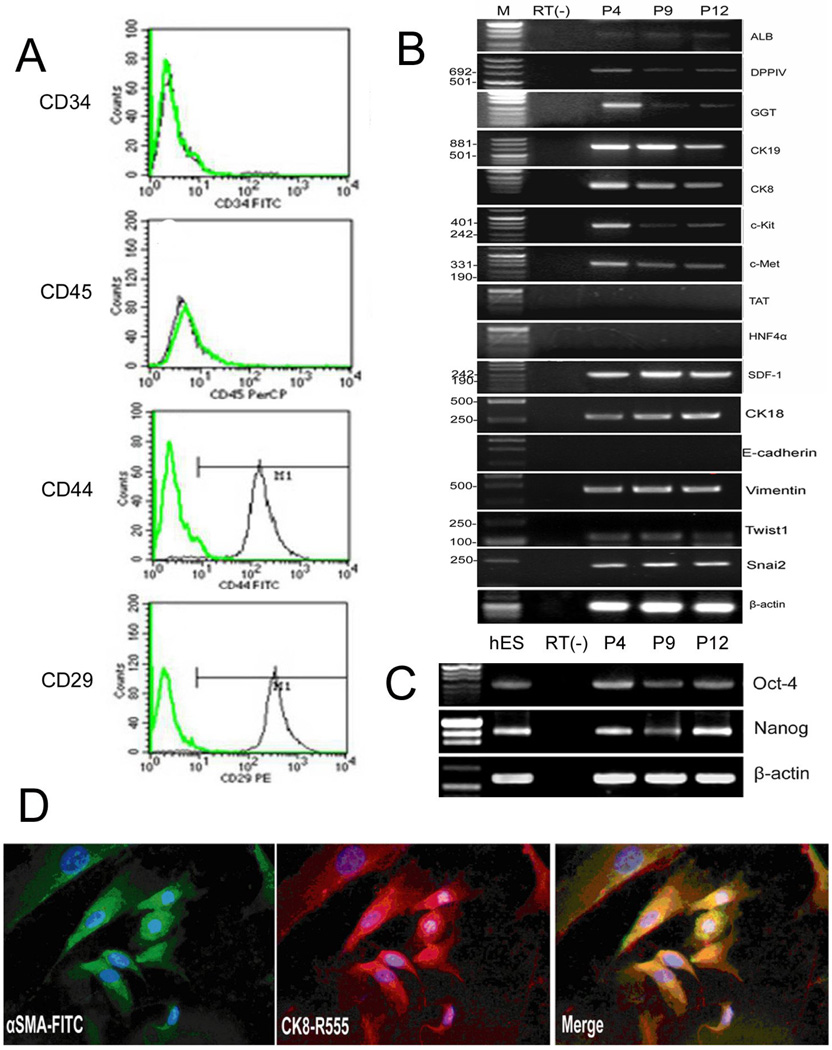
To characterize the phenotype of hLBSC, we used RT-PCR, immunocytochemistry, double immunofluorescence, and fluorescence-activated cell sorting (FACS) to detect the gene expression profile of hLBSC. First, no expression of hematopoietic markers CD34 and CD45 was detected by FACS (Fig. 3A). RT-PCR of cells in 4th, 9th, 12th passages showed that they expressed hepatocytic markers ALB, DPPIV, biliary markers CK19, GGT, miscellaneous markers CK8, CK18, hepatocyte growth factor receptor (c-Met), and oval cell related marker stem cell factor receptor (c-Kit), but no E-cadherin, TAT or HNF4α (Fig. 3B). Over 99% of the cells expressed the mesenchymal markers, CD44 and CD29 (Fig. 3A). Vimentin, Twist1, Snai2, Stromal cell-derived factor-1 (SDF-1) was also expressed by the hLBSCs (Fig. 3B). The patterns of gene expression strongly implicate the co-expression of mesenchymal and epithelial markers within the same cells, which was confirmed by double immunofluorescence. hLBSCs stained positively for αSMA and for CK8, mesenchymal and epithelial markers, respectively. αSMA was localized in microfilaments forming stress fibers, while CK-8 formed perinuclear intermediate filaments (Fig. 3D). Moreover, after hepatic induction, there was an increase in expression of CK-8 and a decrease in expression of αSMA, as the Supplementary Fig. 2 showed. This phenotype is reminiscent of an EMT as described by others (Pagan R et al., 1995; Chagraoui J et al., 2003; Kiassov AP et al., 1995). Simultaneously, RT-PCR detected expression of stem cell transcription factors octamer-binding protein 4 (Oct-4) and Nanog in 4th, 9th, and 12th passage of the hLBSC cells (Fig. 3C), further suggesting that hLBSC might be a primitive stem cell population in human liver bud.
Fig. 3. Gene expression profile of hLBSC.
RT-PCR analysis show that hLBSC express most markers characteristic for hepatic progenitor cells, such as ALB, DPPIV, GGT, CK19, CK8, CK18, c-Kit, and c-Met, but no expression of E-cadherin, TAT and HNF4α (B). hLBSC also display some characteristics of mesenchymal cells, such as CD44, CD29 (A), vimentin, Twist1, Snai2 and SDF-1 (B), together with expression of Oct4 and nanog (C). But no hematopoietic markers CD45 and CD34 (A) were detected by FACS. Double immunofluorescence using the anti-αSMA polyclonal antibody and anti–CK8 monoclonal antibody reveal co-expression of CK8 (red) and aSMA (green) in hLBSC, with DAPI (blue) nuclear counterstaining (D). Plots in A show isotype control IgG-staining profile (green line) versus specific antibody staining profile (black line). P4, P9, P12 represents hLBSC in 4, 9, 12 passages (B, C). M: pUC Mix Marker or D2000. RT (−): mRNA sample without reverse transcription.
3.4. hLBSC have hepatocytic, adipogenic, osteogenic and neural differentiation potential
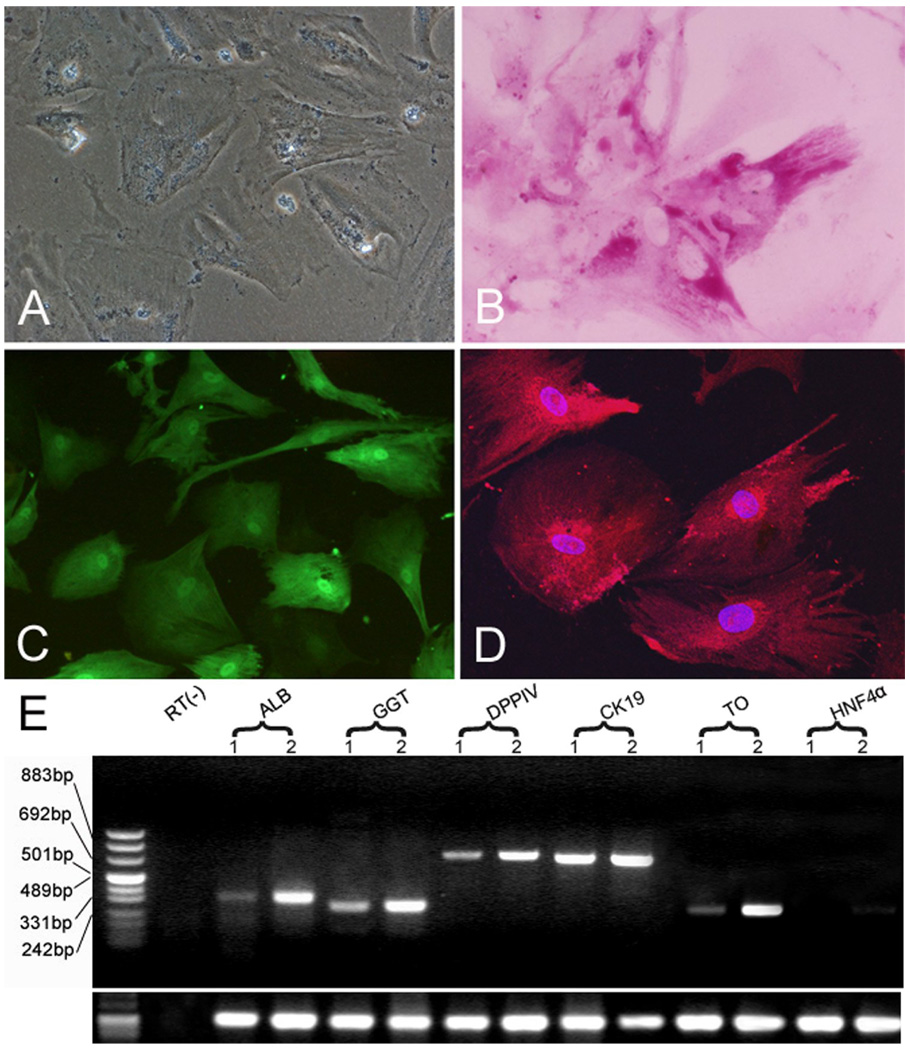
To investigate the multi-lineage differentiation potential of hLBSC, we performed several in vitro induction protocols on the cells. Sodium butyrate (SB) had been used to induce hepatic differentiation of mouse embryonic hepatoblasts (Rogler LE, 1997). When hLBSC was exposed to 45mg/ml SB, cell growth was significantly arrested. Morphologically, cells flattened and became larger with significantly more cytoplasm within 24 hours. Binuclear cells were observed in 2 days after SB treatment, and by 10 days, binuclear cells comprised 10–15% of the total cell population (Fig. 4A). Enhanced expression of ALB, GGT, DPPIV, CK19, TO and HNF4α was detected by RT-PCR (Fig. 4E). Abundant glycogen stores were seen in the cytoplasm of most binuclear cells by Periodic Acid-Schiff (PAS) staining (Fig. 4B). Immunocytofluorescene showed enhanced expression of TAT and ALB (Fig. 4C, D). Taken together, these results reveal that hLBSC can give rise to hepatocytes when cultured in vitro in the presence of the appropriate inducing agents. Enhanced expression of bile ductular markers CK19 and GGT was also detected (Fig. 4E), suggesting that hLBSC may have the ability to differentiate into cholangiocytes.
Fig. 4. Hepatic differentiation of hLBSC.
Binuclear cells were observed in 2 days after SB treatment, and by 10 days, binuclear cells comprised of 10–15% of the total cell population (A). Abundant glycogen stores were seen in the cytoplasm of most binuclear cells by PAS staining (B). Immunocytofluorescene showed expression of TAT and ALB in the hLBSC treated by SB (C, D). Enhanced expression of ALB, GGT DPPIV, CK19, TO and HNF4α was detected by RT-PCR (E). Lane 1: samples without SB treatment; Lane 2: samples with SB treatment. RT (−): mRNA sample without reverse transcription.
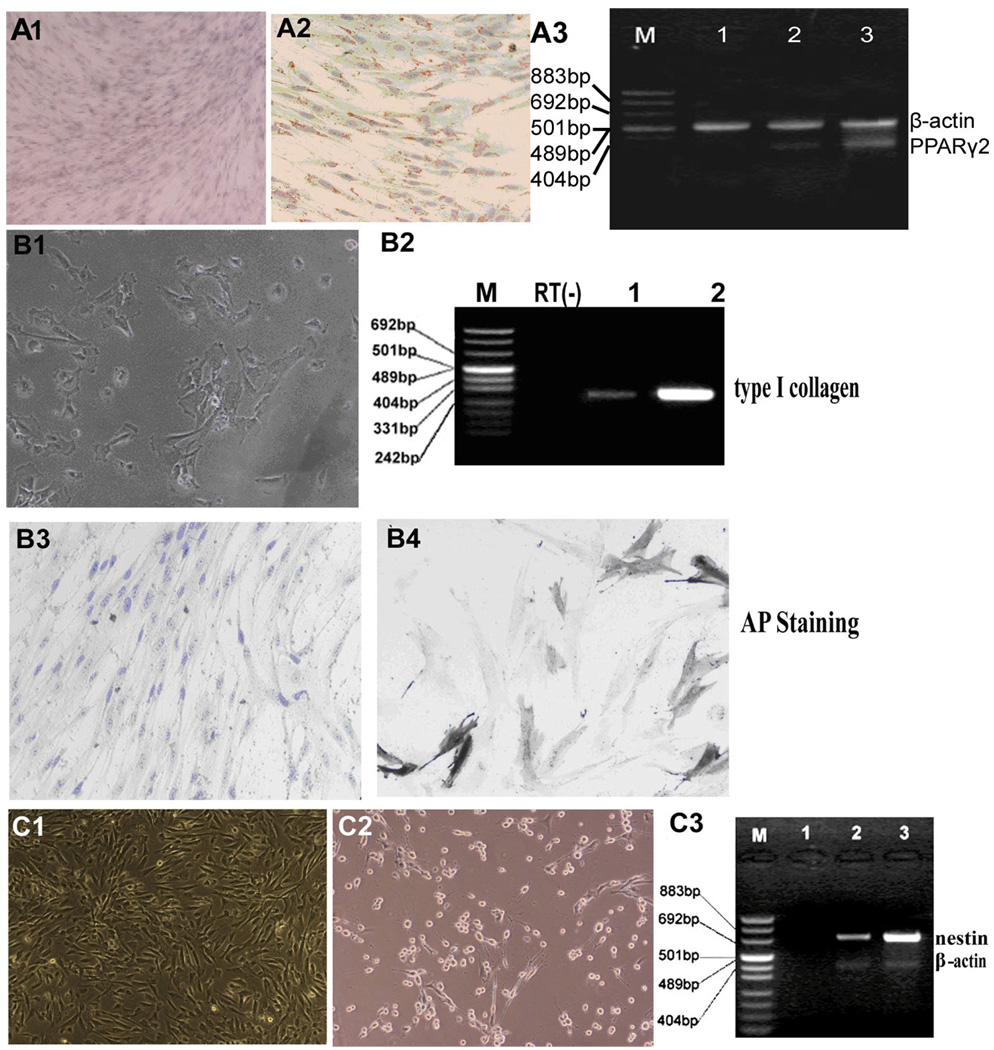
Since hLBSC express some mesenchymal markers, we tested the ability of hLBSC to differentiate into adipocytes using established conditions for inducing bone marrow mesenchymal stem cells to adipocytes (Reyes M et al., 2001). In adipogenic induction medium, hLBSC adapted a long spindle shape. Cytoplasmic lipid droplets of different sizes and with a high refraction ratio were seen in 10% cells within 2 days, and in 60% of the cells by 10 days. By 14 days, 90% cells had abundant droplets which stained positively by Oil-red-O, a fat specific dye (Fig. 5, A2). Peroxisome Proliferators-Activated Receptor (PPAR)-γ2, a marker specific for adipocytes, was detected by RT-PCR (Fig. 5, A3), confirming the adipocytic differentiation potential of hLBSC.
Fig. 5. Multipotent differentiation of hLBSC.
(A1-3) Adipogenic differentiation of hLBSC. By 14 days in adipose induction media, 90% cells had abundant droplets which stained positively by Oil-red-O, a fat specific dye (A2), while hLBSC without adipocytic induction were negatively stained (A1). And expression of PPAR-γ2 (351bp) was detected at 0 days (lane 1), 10 days (lane 2), and 14 days (lane 3) by RT-PCR (A3), with β-actin (516bp) as control. M: pUC Mix Marker. (B1-4) Osteogenic differentiation of hLBSC. Under osteogenic induction conditions, most of the cells acquired cuboidal or polygonal shape, accompanied by the appearance of calcification spots with high refraction ratio (B1). Thirty percent induced cells were positive for AP staining (B4), with untreated hLBSC as control (B3). Expression of type I collagen (299bp) was detected by RT-PCR (B2). M: pUC Mix Marker; Lane 1: control samples; Lane 2: samples with osteogenic induction. RT (−): mRNA sample without reverse transcription. (C1-3) Neural differentiation of hLBSC. In the presence of β-mercaptoethanol, hLBSC underwent morphological transformation with appearance of long processes (C2), with untreated hLBSC as control (C1). Enhanced expression of nestin (718bp) was detected by RT-PCR (C3). M: pUC Mix Marker; Lane 1: mRNA sample without reverse transcription. Lane 2: control samples without BME treatment; Lane 2: samples with BME treatment.
When hLBSC were exposed to osteogenic induction conditions, most of the cells acquired cuboidal or polygonal shape, accompanied by the appearance of calcification spots with high refraction ratio (Fig. 5, B1). Thirty percent cells were positive for alkaline phosphotase staining (Fig. 5, B4). Expression of collagen I was detected (Fig. 5, B2), further indicating the osteogenic differentiation potential of hLBSC.
In the presence of β-mercaptoethanol (Hung SC et al., 2002), hLBSC underwent morphological transformation and acquired the appearance of long processes (Fig. 5, C2). The morphological changes, accompanied by enhanced expression of nestin, detectable by RT-PCR (Fig. 5, C3), suggested the possibility of neural differentiation. Together, these results showed that hLBSC had multipotential differentiation capability into different embryonic layers derived cells.
3.5. hLBSC can repopulate injured mouse liver in vivo
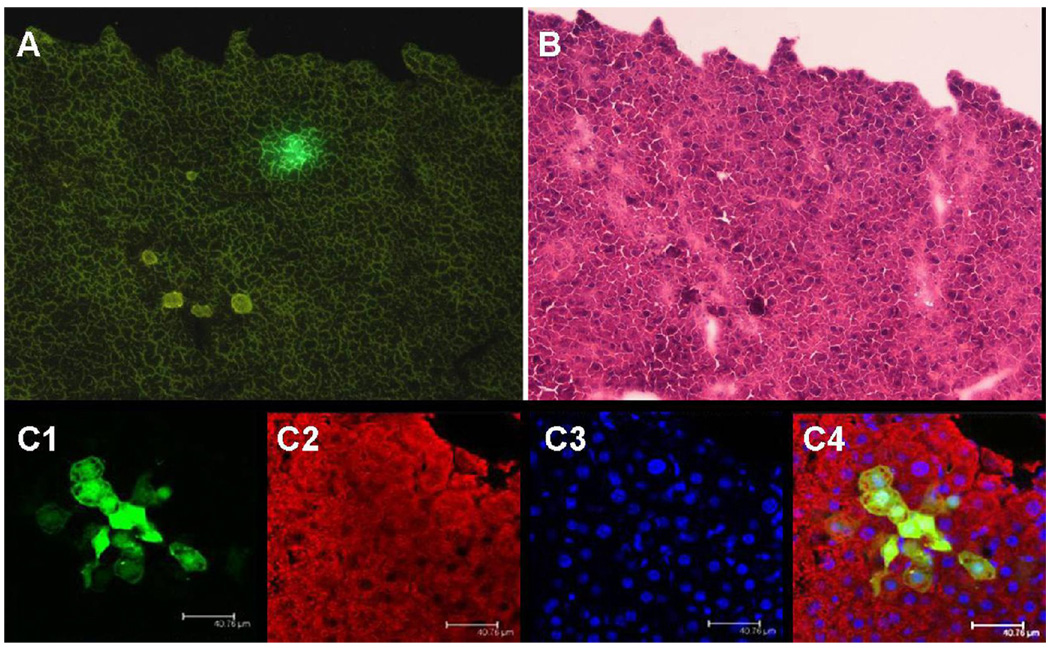
To assess the differentiation ability of hLBSC cells in vivo, we prepared recipient animals by using the CCl4 protocol frequently used to elicit acute liver injury in recipients for cell transplantations (Suzuki A et al., 2002; Yamamoto H et al., 2003). To do this, a hLBSC subline, the hLBSC-pLNCG-C1 cell line with stable EGFP expression was established. Then the EGFP-expressing hLBSCs were transplanted intrasplenicly into SCID mice pre-treated by CCL4. The mice were sacrificed 3 weeks (n = 7) after implantation to monitor the outcome of the transplantation. EGFP-positive cell clusters, indicating engrafted hLBSC proliferated and differentiated into hepatocyte lineage. The EGFP-positive cells comprised 2.7±1.2 (n=7) percent of the total cells in the transplanted liver (Fig. 6A). HE staining confirmed that they were clusters of hepatocytes in the hepatic cord (Fig. 6B). The overlay image of EGFP and the same section stained by anti-α-1-AT antibody clearly shows that all EGFP-positive donor cells were also α-1-AT-positive hepatocytes (Fig. 6C). Taken together, these observations demonstrate the ability of hLBSC to engraft and differentiate into hepatocyte lineage in vivo after transplantation into recipient animals.
Fig. 6. In vivo transplantation of hLBSC into CCl4-treated SCID mice.
hLBSC- pLNCG C1 cell line with stable EGFP expression was established. Then the EGFP expressing cells were transplanted introsplenicly into SCID mice pre-treated by CCL4. Three weeks after implantation, EGFP-positive cell clusters were detected in the recipient mice (A). H&E staining confirmed that they were clusters of hepatocytes in the hepatic cord (B). The overlay image of EGFP (green, C1) and the same section stained by anti-α-1-AT antibody (red, C2) clearly shows that all EGFP-positive donor cells were also α-1 AT-positive hepatocytes (yellow, C4), with DAPI nuclear counterstaining (blue, C3). (Relative magnification A, B: 100×; C: 200×)
Discussion
In this report, we document the occurrence of EMT in early human liver development, mainly by cells co-expressing CK8 and αSMA, markers for epithelial and mesenchymal cells, respectively (Chagraoui J et al., 2003; Zeisberg M and Neilson EG, 2009). EMT occurred predominantly at early gestational stages at 5 gw, declined at 8 gw and were largely absent by 12 gw. There was up-regulation of EMT related genes at 5gw, suggesting the involvement of cells in EMT during early liver development. We have also isolated and stably cultured a multipotent stem cell population (hLBSC) with characteristics of EMT from human liver bud. hLBSC have a high self-renew capacity, and show hepatocytic, adipogenic, osteogenic and neural differentiation potential. What’s more, hLBSC can contribute to the regeneration of the liver parenchyma in SCID mice.
The human liver arises from the foregut endoderm after 3.5 weeks of gestation and develops rapidly. The liver begins hematopoiesis at the 6th week and functions as the main hematopoietic organ from the 15th week through mid-gestation (Timens W and Kamps WA, 1997). The fetal livers that we used for the isolation of hLBSC were around 5 weeks of gestation, which contained a large percentage of stem cells and a relatively small percentage of hematopoietic cells. To eliminate the possibility of artifacts of culture or transformation, hLBSC were isolated and clonally propagated repeatedly from different fetal livers, and no differences were found with respect to growth pattern and cellular morphology. There was no tumor formation in the mice transplanted with hLBSC. These data strongly suggested that hLBSC was a normal cell population existing in the human liver bud rather than an artifact of culture or transformation.
hLBSC differed from previously described HSPC or mesenchymal stem cells (MSC), based on their origin, morphology, gene expression profile and differentiation potential (Campagnoli C et al., 2001; Malhi H et al., 2002; Suzuki A and Nakauchi H, 2002). The hLBSC that we have isolated were different from previously reported HSPC in that the hLBSC simultaneously expressed some mesenchymal markers and stem cell transcription factor Oct4 and Nanog, which has been considered a hallmark of embryonic stem cells (ESC), and play an important role in maintaining pluripotency of ESC (Sato N et al., 2004; Boyer LA et al, 2005). In contrast, HSPC did not express most of mesenchymal markers, such as CD44, CD29 and SDF-1, but expressed hepatic and/or bile ductular markers. What set hLBSC apart from MSC is that hLBSC express some hepatic markers, such as ALB, GGT, DPPIV, and c-Met, whereas no HNF4α, which is essential for hepatocyte differentiation, but is not required in specification of the ventral foregut endoderm toward a hepatic fate (Li J et al., 2000). Moreover, hLBSC differ from MSC in their differentiation potential. While MSC could be induced into cartilage, our hLBSC could not (data not show). These data indicate that hLBSC may be an intermediate cell population in the differentiation from endoderm cells to hepatoblasts. Alternatively, based on the controversial hypothesis of a mesodermal origin of the liver parenchyma (Sell S, 2001), a mesendodermal origin of hLBSC cannot be excluded.
Here we report that in human fetal liver at early gestational stages before 8gw, αSMA and CK8 were co-expressed in most of the non-hematopoietic cells, suggesting a stage of fetal liver development with abundant EMT. By 8gw, only a few cells remain double positive for αSMA and CK8, and none were double positive at later gestational stages. Therefore the time course for human fetal liver EMT in our studies was not coincidental to the time course of hematopoiesis, as it was reported by Chagraoui J et al. (2003) to occur in murine fetal liver. Since Chargroui et al. did not show the data about EMT in earlier stages of mouse liver development (between 8.5dpc to 11.5dpc), we could not directly compare the roles of EMT between human and mouse liver development. However, it is reasonable to presume the presence of EMT cells during early mouse liver development, analogous to what we have found during early human liver development. EMT may be an essential and transient process in early human liver development, during the commitment of endoderm cells to differentiate into hepatoblasts.
Our data indicate that EMT occurs only in early development of the human liver. We were able to obtain a clonal culture of cells, which we termed hLBSC, with EMT-like properties including multilineage potential in vitro and the ability to repopulate in the injured SCID mouse liver model. In summary, the present study suggests that the hLBSC represent intermediate cells with features of EMT and stem cell along the differentiative pathway from endoderm to hepatoblasts, which may play an important role in the beginning of the spatial structure construction of the liver. hLBSC may have value for research on liver stem cell biology, in addition to potential value as cell therapy for liver diseases, drug screening and liver development.
Supplementary Material
Acknowledgements
The present study was financed by the National Basic Research Program of China (Grant Number: 2009CB941100, 2010CB945602), the National Natural Science Foundation of China (Grant Number: 30971462, 30800657, 30700400), and National Institutes of Health (NIH AI-056082, AI-078429).
Abbreviations
- ALB
albumin
- AFP
alpha-fetoprotein
- AP
alkaline phosphatase
- αSMA
alpha smooth muscle actin
- BME
β-mercaptoethanol
- CCL4
carbon tetrachloride
- CK
cytokeratin
- c-Kit
stem cell factor receptor
- c-Met
hepatocyte growth factor receptor
- DPPIV
dipeptidyl peptidase IV
- EGFP
enhanced green fluorescence protein
- FACS
fluorescence-activated cell sorting
- Fox
forkhead box factor
- GGT
gamma glutamyl transpeptidase
- H&E
hematoxylin and eosin
- HLA-DR
histocompatibility class II antigen
- HNF
hepatocyte nuclear factor
- hLBSC
human liver bud derived stem cells
- HSPC
hepatic stem/progenitor cells
- Oct4
octamer-binding protein 4
- PAS
Periodic acid–Schiff
- PBS
phosphate-buffered saline
- PPAR
Peroxisome Proliferators-Activated Receptor
- RT-PCR
reverse transcription polymerase chain reaction
- SB
sodium butyrate
- SCID
severe combined immunodeficiency
- SDF-1
Stromal Cell-Derived Factor-1
- TAT
tyrosine aminotransferase
- TGFβ
Transforming Growth Factor beta
- TO
tryptophan oxygenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biocarta Pathway Collections. [accessed 5 May 2010]; http://www.biocarta.com/genes/allPathways.asp.
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnoli C, Roberts IA, Kumar S, Choolani M, Bennett PR, Letsky E, et al. Identification of mesenchymal stem/progenitor cells in human first-trimester fetal blood, liver, and bone marrow. Blood. 2001;98:2396–2402. doi: 10.1182/blood.v98.8.2396. [DOI] [PubMed] [Google Scholar]
- Chagraoui J, Lepage-Noll A, Anjo A, Uzan G, Charbord P. Fetal liver stroma consists of cells in Epithelial-mesenchymal transition. Blood. 2003;101:2973–2982. doi: 10.1182/blood-2002-05-1341. [DOI] [PubMed] [Google Scholar]
- Choi SS, Diehl AM. Epithelial-to-mesenchymal transitions in the liver. Hepatology. 2009;50:2007–2013. doi: 10.1002/hep.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan YY, Riehle KJ, Lazaro C, Teoh N, Haque J, Campbell JS, et al. Isolation of multipotent progenitor cells from human fetal liver capable of differentiating into liver and mesenchymal lineages. Proc Natl Acad Sci U S A. 2006;103:9912–9917. doi: 10.1073/pnas.0603824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JA. Mesenchyme to epithelium transition during development of the mammalian kidney tubule. Acta Anat. 1996;156:187–201. doi: 10.1159/000147846. [DOI] [PubMed] [Google Scholar]
- Hung SC, Cheng H, Pan CY, Tsai MJ, Kao LS, Ma HL. In vitro differentiation of size-sieved stem cells into electrically active neural cells. Stem Cells. 2002;20:522–529. doi: 10.1634/stemcells.20-6-522. [DOI] [PubMed] [Google Scholar]
- Inada M, Follenzi A, Cheng K, Surana M, Joseph B, Benten D, et al. Phenotype reversion in fetal human liver epithelial cells identifies the role of an intermediate meso-endodermal stage before hepatic maturation. J Cell Sci. 2008;121:1002–1013. doi: 10.1242/jcs.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119:1417–1419. doi: 10.1172/JCI39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiassov AP, Van Eyken P, van Pelt JF, Depla E, Fevery J, Desmet VJ, et al. Desmin expressing nonhematopoietic liver cells during rat liver development: an immunohistochemical and morphometric study. Differentiation. 1995;59:253–258. doi: 10.1046/j.1432-0436.1995.5940253.x. [DOI] [PubMed] [Google Scholar]
- Lemaigre FP. Mechanisms of liver development: concepts for understanding liver disorders and design of novel therapies. Gastroenterology. 2009;137:62–79. doi: 10.1053/j.gastro.2009.03.035. [DOI] [PubMed] [Google Scholar]
- Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- Li WL, Su J, Yao YC, Tao XR, Yan YB, Yu HY, et al. Isolation and characterization of mouse hepatic epithelial progenitor cell line from RS/PH mice. Stem cells. 2006;24:322–332. doi: 10.1634/stemcells.2005-0108. [DOI] [PubMed] [Google Scholar]
- Malhi H, Irani AN, Gagandeep S, Gupta S. Isolation of human progenitor liver epithelial cells with extensive replication capacity and differentiation into mature hepatocytes. J Cell Sci. 2002;115:2679–2688. doi: 10.1242/jcs.115.13.2679. [DOI] [PubMed] [Google Scholar]
- Mlecnik B, Scheideler M, Hackl H, Hartler J, Sanchez-Cabo F, Trajanoski Z. PathwayExplorer: web service for visualizing high-throughput expression data on biological pathways. Nucleic Acids Res. 2005;33:W633–W637. doi: 10.1093/nar/gki391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan R, Llobera M, Vilaró S. Epithelial-mesenchymal transition in cultured neonatal hepatocytes. Hepatology. 1995;21:820–831. [PubMed] [Google Scholar]
- Pagan R, Sánchez A, Martin I, Llobera M, Fabregat I, Vilaró S. Effects of growth and differentiation factors on the epithelial-mesenchymal transition in cultured neonatal rat hepatocytes. J Hepatol. 1999;31:895–904. doi: 10.1016/s0168-8278(99)80292-x. [DOI] [PubMed] [Google Scholar]
- Reyes M, Lund T, Lenvik T, Aguiar D, Koodie L, Verfaillie CM. Purification and ex vivo expansion of postnatal human marrow mesodermal progenitor cells. Blood. 2001;98:2615–2625. doi: 10.1182/blood.v98.9.2615. [DOI] [PubMed] [Google Scholar]
- Rogler LE. Selective bipotential differentiation of mouse embryonic hepatoblasts in vitro. Am J Pathol. 1997;150:591–602. [PMC free article] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BLM, Zaret KS. Distinctmesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Develop. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Sell S. Heterogeneity and plasticity of hepatocyte lineage cells. Hepatology. 2001;33:738–750. doi: 10.1053/jhep.2001.21900. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Nakauchi H. Identification and propagation of liver stem cells. Semin Cell Dev Biol. 2002;13:455–461. doi: 10.1016/s1084952102001349. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Zheng YW, Fukao K, Nakauchi H, Taniguchi H. Clonal expansion of hepatic stem/progenitor cells following flow cytometric cell sorting. Cell Transplant. 2001;10:393–396. [PubMed] [Google Scholar]
- Suzuki A, Zheng YW, Kaneko S, Onodera M, Fukao K, Nakauchi H, et al. Clonal identification and characterization of self-renewing pluripotent stem cells in the developing liver. J Cell Biol. 2002;156:173–184. doi: 10.1083/jcb.200108066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao XR, Li WL, Su J, Jin CX, Wang XM, Li JX, et al. Clonal mesenchymal stem cells derived from human bone marrow can differentiate into hepatocyte-like cells in injured livers of SCID mice. J Cell Biochem. 2009;108:693–704. doi: 10.1002/jcb.22306. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Timens W, Kamps WA. Hemopoiesis in human fetal and embryonic liver. Microsc Res Tech. 1997;39:387–397. doi: 10.1002/(SICI)1097-0029(19971201)39:5<387::AID-JEMT1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Valdes F, Alvarez AM, Locascio A, Vega S, Herrera B, Fernández M, et al. The epithelial mesenchymal transition confers resistance to the apoptotic effects of transforming growth factor Beta in fetal rat hepatocytes. Mol Cancer Res. 2002;1:68–78. [PubMed] [Google Scholar]
- Viebahn C. Epithelial-mesenchymal transformation during formation of the mesoderm in the mammalian embryo. Acta Anat. 1995;154:79–97. doi: 10.1159/000147753. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Quinn G, Asari A, Yamanokuchi H, Teratani T, Terada M, et al. Differentiation of embryonic stem cells into hepatocytes: Biological functions and therapeutic application. Hepatology. 2003;37:983–993. doi: 10.1053/jhep.2003.50202. [DOI] [PubMed] [Google Scholar]
- Zaret KS. Hepatocyte differentiation: from the endoderm and beyond. Curr Opin Genet Develop. 2001;11:568–574. doi: 10.1016/s0959-437x(00)00234-3. [DOI] [PubMed] [Google Scholar]
- Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.