Abstract
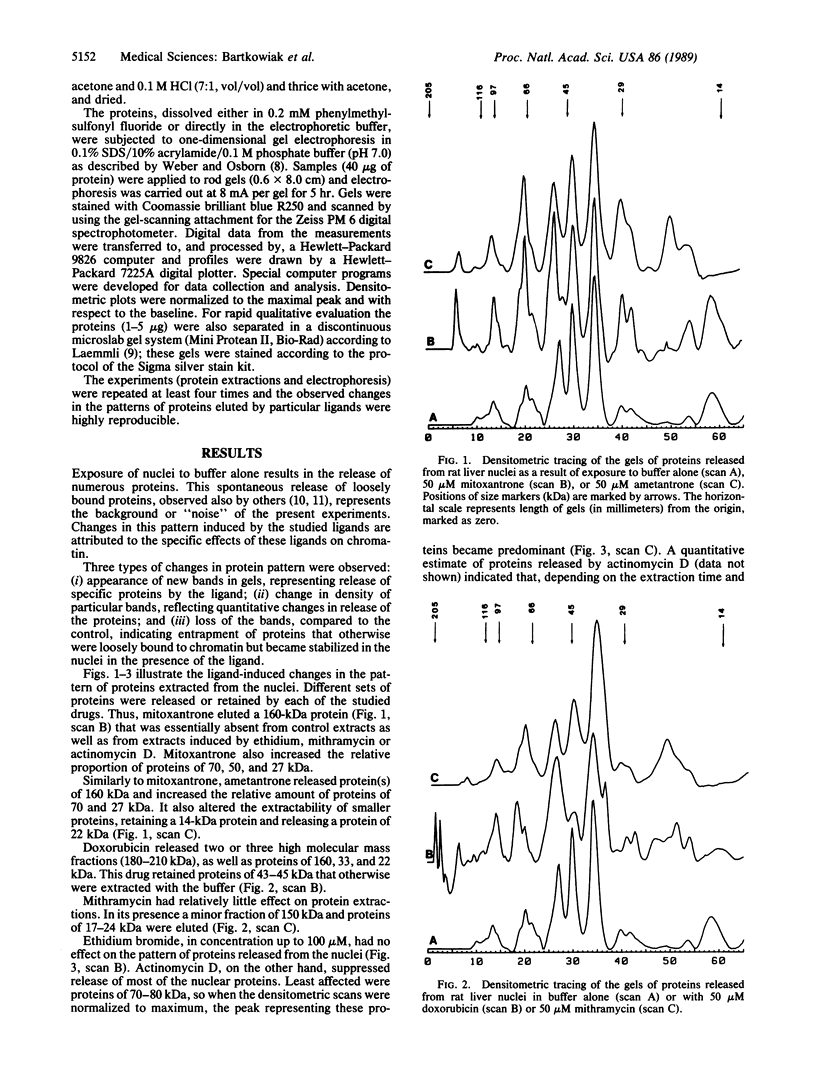
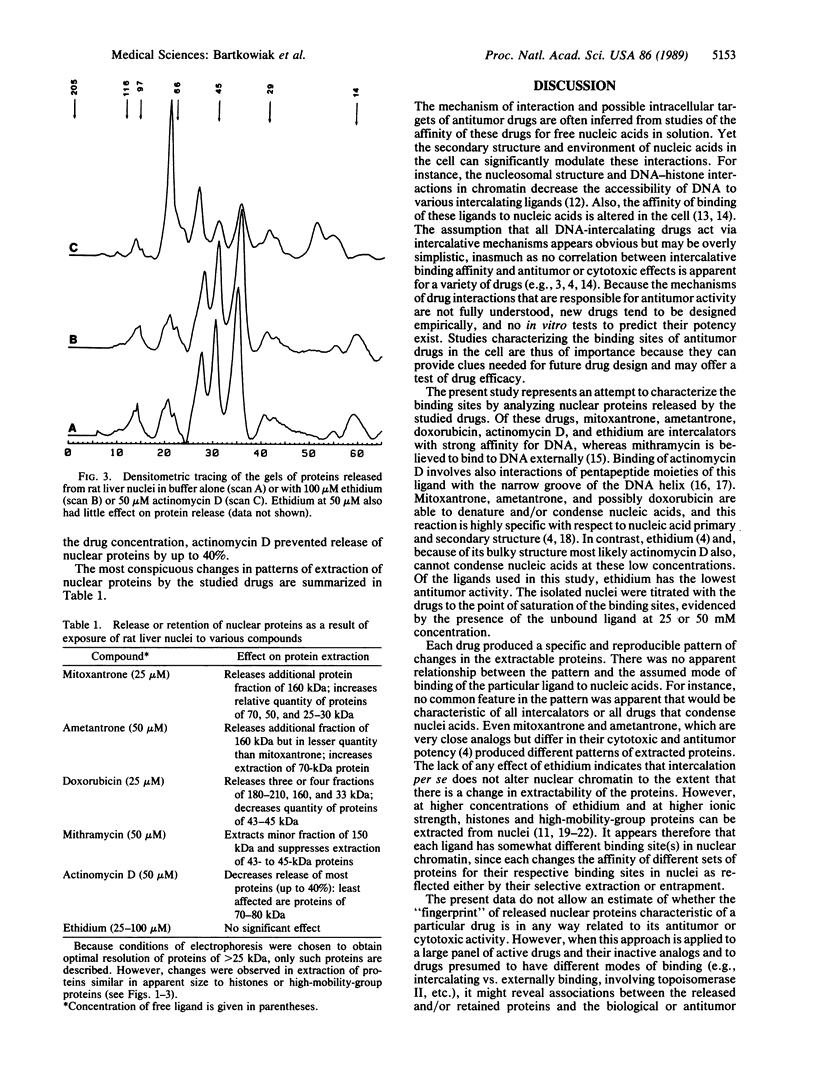
The nuclear chromatin binding sites of the antitumor drugs mitoxantrone, ametantrone, doxorubicin, mithramycin, and actinomycin D and the intercalating ligand ethidium were studied by polyacrylamide gel electrophoresis of the proteins released from rat liver nuclei in the presence and absence of these drugs in buffer of low ionic strength (10 mM NaCl). At 25-50 microM free ligand concentration, each drug produced a specific and reproducible pattern of extractable proteins of different molecular weight by (i) releasing new proteins, (ii) altering the quantity of particular extracted proteins, and/or (iii) selectively entrapping other proteins in the nuclei. Ethidium, up to 100 microM, did not affect release of proteins from the nuclei. These results indicate that each ligand either has different binding site(s) in chromatin or modulates chromatin structure in a specific way by changing the affinity of different sets of proteins for their respective binding sites, resulting in their selective extraction or entrapment. The lack of effect of ethidium indicates that intercalation of the ligand to DNA, per se, does not alter the release of nuclear proteins. If patterns of nuclear proteins selectively released or retained by antitumor drugs are found to correlate with biological activity, this type of analysis may be helpful in new drug design and screening.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Comings D. E., Okada T. A. Nuclear proteins. III. The fibrillar nature of the nuclear matrix. Exp Cell Res. 1976 Dec;103(2):341–360. doi: 10.1016/0014-4827(76)90271-8. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z., Traganos F., Kapuscinski J., Staiano-Coico L., Melamed M. R. Accessibility of DNA in situ to various fluorochromes: relationship to chromatin changes during erythroid differentiation of Friend leukemia cells. Cytometry. 1984 Jul;5(4):355–363. doi: 10.1002/cyto.990050411. [DOI] [PubMed] [Google Scholar]
- Hsiang Y. H., Liu L. F. Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer Res. 1988 Apr 1;48(7):1722–1726. [PubMed] [Google Scholar]
- Johnson R. K., Zee-Cheng R. K., Lee W. W., Acton E. M., Henry D. W., Cheng C. C. Experimental antitumor activity of aminoanthraquinones. Cancer Treat Rep. 1979 Mar;63(3):425–439. [PubMed] [Google Scholar]
- Kapuscinski J., Darzynkiewicz Z. Condensation of nucleic acids by intercalating aromatic cations. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7368–7372. doi: 10.1073/pnas.81.23.7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuscinski J., Darzynkiewicz Z. Relationship between the pharmacological activity of antitumor drugs Ametantrone and mitoxantrone (Novatrone) and their ability to condense nucleic acids. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6302–6306. doi: 10.1073/pnas.83.17.6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuscinski J., Traganos F., Darzynkiewicz Z. Interactions of antitumor drug ditercalinium with nucleic acids in solutions and in situ. Cancer Lett. 1988 Nov;42(3):185–191. doi: 10.1016/0304-3835(88)90303-5. [DOI] [PubMed] [Google Scholar]
- LERMAN L. S. Structural considerations in the interaction of DNA and acridines. J Mol Biol. 1961 Feb;3:18–30. doi: 10.1016/s0022-2836(61)80004-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawrence J. J., Daune M. Ethidium bromide as a probe of conformational heterogeneity of DNA in chromatin. The role of histone H1. Biochemistry. 1976 Jul 27;15(15):3301–3307. doi: 10.1021/bi00660a021. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Studies of the binding of actinomycin and related compounds to DNA. J Mol Biol. 1968 Jul 28;35(2):251–290. doi: 10.1016/s0022-2836(68)80024-5. [DOI] [PubMed] [Google Scholar]
- Pani B., Plossi P., Russo E. HMG proteins released from the chromatin following incubation of mammalian nuclei with ethidium bromide. Exp Cell Res. 1989 Feb;180(2):557–562. doi: 10.1016/0014-4827(89)90083-9. [DOI] [PubMed] [Google Scholar]
- Paoletti J. Relaxation of chromatin structure induced by ethidium binding. Involvement of the intercalation process. Eur J Biochem. 1979 Oct 15;100(2):531–539. doi: 10.1111/j.1432-1033.1979.tb04199.x. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Zwelling L. A., Kao-Shan C. S., Whang-Peng J., Bradley M. O. Correlations between intercalator-induced DNA strand breaks and sister chromatid exchanges, mutations, and cytotoxicity in Chinese hamster cells. Cancer Res. 1985 Jul;45(7):3143–3149. [PubMed] [Google Scholar]
- Prentice D. A., Gurley L. R. Nuclease digestibility of chromatin is affected by nuclei isolation procedures. Biochim Biophys Acta. 1983 Jun 24;740(2):134–144. doi: 10.1016/0167-4781(83)90070-2. [DOI] [PubMed] [Google Scholar]
- Santi P., Papa S., del Coco R., Falcieri E., Zini N., Marinelli F., Maraldi N. M. Modifications of the chromatin arrangement induced by ethidium bromide in isolated nuclei, analyzed by electron microscopy and flow cytometry. Biol Cell. 1987;59(1):43–54. doi: 10.1111/j.1768-322x.1987.tb00514.x. [DOI] [PubMed] [Google Scholar]
- Schröter H., Maier G., Ponstingl H., Nordheim A. DNA intercalators induce specific release of HMG 14, HMG 17 and other DNA-binding proteins from chicken erythrocyte chromatin. EMBO J. 1985 Dec 30;4(13B):3867–3872. doi: 10.1002/j.1460-2075.1985.tb04159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M. Actinomycin and DNA transcription. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strätling W. H., Seidel I. Relaxation of chromatin structure by ethidium bromide binding: determined by viscometry and histone dissociation studies. Biochemistry. 1976 Nov 2;15(22):4803–4809. doi: 10.1021/bi00667a009. [DOI] [PubMed] [Google Scholar]
- Waring M. J. Complex formation between ethidium bromide and nucleic acids. J Mol Biol. 1965 Aug;13(1):269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]


