Abstract
Publicly available genetic and expression data on lymphoblastoid cell lines (LCLs) make them a unique resource for understanding the genetic underpinnings of pharmacological outcomes and disease. LCLs have been used for pharmacogenomic discovery and validation of clinical findings associated with drug response. However, variation in cellular growth rate, baseline Epstein–Barr virus (EBV) copy number and ATP levels can all be confounders in such studies. Our objective is to better define confounding variables that affect pharmacological end points in LCLs. To this end, we evaluated the effect of these three variables on drug-induced cytotoxicity in LCLs. The drugs evaluated included daunorubicin, etoposide, carboplatin, cisplatin, cytarabine, pemetrexed, 5′-deoxyfluorouridine, vorinostat, methotrexate, 6-mercaptopurine, and 5-fluorouracil. Baseline ATP or EBV copy number were not significantly correlated with cellular growth rate or drug-induced cytotoxicity. In contrast, cellular growth rate and drug-induced cytotoxicity were significantly, directly related for all drugs except vorinostat. Importantly, cellular growth rate is under appreciable genetic influence (h2=0.30–0.39) with five suggestive linkage regions across the genome. Not surprisingly, a percentage of SNPs that significantly associate with drug-induced cytotoxicity also associate with cellular growth rate (P≤0.0001). Studies using LCLs for pharmacologic outcomes should therefore consider that a portion of the genetic variation explaining drug-induced cytotoxicity is mediated via heritable effects on growth rate.
Keywords: Cellular growth rate, lymphoblastoid cell lines, HapMap, chemotherapy, heritability, linkage
Introduction
The International HapMap Project (www.HapMap.org) was designed to provide a comprehensive collection of data to characterize sequence variation between populations and provide the research community with a unique resource to investigate genetics and cellular phenotypes that would be difficult to investigate in humans. The first phase of the project focused on 270 lymphoblastoid cell lines (LCLs), from three different ethnic populations: 90 Yoruba (YRI) from Ibadan, Nigeria, 90 Asian (ASN) consisting of 45 Japanese from Tokyo and 45 Han Chinese from Beijing, and 90 Caucasians from Utah, USA (CEU). These cell lines allow for the evaluation of cellular phenotypes because extensive genotyping and expression data are publicly available, making them a rich resource for genotype–phenotype studies. Furthermore, the Epstein–Barr virus (EBV) transformation established a set of cell lines that provides a renewable source of DNA and RNA.
There has been a considerable number of genetic, expression, and pharmacological studies using these LCLs as a model system. For example, genetic factors responsible for radiation sensitivity1,2 and variation in global gene expression3–9 have been reported. In addition, cell lines from the large Centre d’Etude du Polymorphisme Humain (CEPH) pedigrees were used to show that a significant genetic component contributes to susceptibility to the cytotoxic effects of chemotherapy.10–13 HapMap LCLs have been used to identify genetic determinants correlated with chemotherapeutic-induced cytotoxicity through their effect on gene expression in various populations.14–17 Furthermore, the associations of baseline gene expression and sensitivity to chemotherapeutic drugs were observed in the Human Variation LCL panel18 and single nucleotide polymorphisms (SNPs) associated with thiopurine methyltransferase activity19 were reported using the HapMap cell lines.
In our attempt to build cell-based models for pharmacogenomic discovery of SNPs associated with chemotherapeutic agents, we recognized the need to address cellular growth rate as a potential confounding variable. As many chemotherapeutic drugs target DNA, one would expect more rapidly dividing cells to be more sensitive to these agents. For example, Huang et al.16 identified 50 of the 208 significant SNPs associated with carboplatin-induced cytotoxicity were concomitantly associated with cellular growth rate. When cellular growth rate was incorporated in the model as a covariate, 179 SNPs remained significant predictors of carboplatin sensitivity. In another example, significant population differences in sensitivity to cytarabine of CEU and YRI cell lines were still present after incorporating cellular growth rate as a covariate in the study.17
Although LCLs have been shown to be a reasonable model for pharmacogenomic discovery and validation,20 there are limitations and unavoidable variables for any in vitro cell-based model. These confounders may affect the interpretation of pharmacogenetic findings. In a recent study, baseline EBV copy number, cellular growth rate and ATP levels present in LCLs were reported as potential confounding variables raising the question as to how best to account for these variables in pharmacogenomic studies.21 Our objective was to further characterize these potential variables and determine their relationship to pharmacological end points.
Materials and methods
Cell lines
All LCLs were cultured in RPMI 1640 media containing 15% heat-inactivated fetal bovine serum (Hyclone, Logan, UT, USA) and 20mM L-glutamine. Cell lines were diluted three times per week at a concentration of 200 000–350 000 cells per ml and were maintained in a 37 °C, 5% CO2-humidified incubator. Medium and components were purchased from Cellgro (Herndon, VA, USA). All cell lines were purchased from Coriell Institute for Medical Research (http://ccr.coriell.org/).
Drugs
Pemetrexed disodium (CAS: 150399-23-8) was a gift from the Eli Lilly Corporation (Indianapolis, IN, USA) and prepared in PBS as a stock solution of 20mM. 5′-deoxyfluoruridine (5′-DFUR, an active form of capecitabine) was obtained from LKT Laboratories Inc. (St Paul, MN, USA) and prepared in equal amounts PBS (Invitrogen, Carlsbad, CA, USA) and DMSO (Sigma, St Louis, MO, USA) as a stock solution at a concentration of 32mM. Carboplatin,16 cisplatin,16 etoposide,16 daunorubicin16 and cytarabine (AraC)17 were prepared as previously described.
Cytotoxicity assay
The cytotoxic effect of carboplatin, cisplatin, etoposide, daunorubicin, AraC, 5′-DFUR and pemetrexed was determined using a short-term cellular growth inhibition assay. LCLs in the exponential growth phase with >85% viability, as determined using the Vi-Cell XR viability analyzer (Beckman Coulter, Fullerton, CA, USA), were plated in triplicate at a density of 1×105 cells per ml in 96-well round bottom plates (Corning Inc., Corning, NY, USA). Drug stock was prepared and immediately added to plates 24 h after plating. The different concentrations of drug were as follows: 5, 10, 20, 40, 80 μM carboplatin, 1, 2.5, 5, 10, 20 μM cisplatin, 0.02, 0.1, 0.5 and 2.5 μM etoposide, 0.0125, 0.025, 0.05, 0.1, 0.2 and 1 μM daunorubicin, 1, 5, 10, 40 and 80 μM cytarabine, 1, 2.5, 10, 20, 40, 80, and 160 μM 5′-DFUR and 0.02, 0.1, 0.5, 1, 5, 10 μM pemetrexed. Drug was left on cells for 72 h (except for cisplatin in which exposure was 48 h). AlamarBlue was added 24 h before absorbance reading at wavelengths of 570 and 600nm using the Synergy-HT multi-detection plate reader (BioTek, Winooski, VT, USA). Percent survival was quantified using manufacturers protocol (http://www.biotek.com/products/). Final percent survival was ascertained by averaging at least six replicates from two independent experiments.We then calculated a value to represent cellular sensitivity to the drug. For one set of drugs: cisplatin, carboplatin, etoposide and daunorubicin, we determined an IC50, the concentration of drug at which 50% cellular growth inhibition occurred, but for others: pemetrexed, cytarabine, and 5′-DFUR, area under the survival curve was used because the highest concentration of drug did not result in 50% cellular growth inhibition for some cell lines. The area under the survival curve was calculated for each cell line using the trapezoidal rule. All IC50 and area under the survival curve values were log2 transformed before statistical modeling, creating a dependent variable from an approximately normal distribution. For the final set of drugs, 5-fluorouracil, 6-mercaptopurine, methotrexate and vorinostat, the cytotoxicity data was collected from the recent publication of the cellular sensitivity.21
Cellular growth rate calculation
There were two estimates for growth rate: (1) a colorimetric assay using alamarBlue (Biosource International, Camarillo, CA, USA) in control (no drug addition) samples; and (2) a direct measurement by counting cells 48 h following seeding cells at 200 000 cells per ml. In the first estimate, measurements were taken from control wells in a series of drug-induced cytotoxicity experiments using a high throughput alamarBlue assay as described above. The growth rate estimate was determined using the alamarBlue equation. The alamarBlue equation used was: (117 216×Normalized absorbance at 570 nm)−(80 586×Normalized absorbance 600 nm). Absorbance readings at each wavelength were normalized to absorbance at the same wavelength for media without cells.
In the second method, ASN HapMap LCLs were diluted to 200 000 cells per ml at time zero and counted every 48 h using the ViCell Beckman-Coulter counter (Beckman-Coulter, Fullerton, CA, USA) once a week for a minimum of 3 different weeks. Each week’s counts were averaged together and divided by the cell count at time zero to give a proliferation factor, representing the average fold-growth of cells in 48 h. The two methods of measurements correlate with a P-value <0.0001 (r2=0.48; Supplementary Figure S1).
Baseline EBV measurement
Quantitative real-time PCR was performed to determine relative EBV copy number at baseline using TaqMan assay. Total DNA was isolated using QIAamp DNA Mini kit (QIAGEN, Valencia, CA, USA) from 120 HapMap LCLs (60 unrelated individuals from HapMap Phase II CEU and YRI) and 20 ng of DNA was used for each reaction. NRF1 was used as the endogenous control gene (NM_005011) and EBV was measured as described previously.21 A relative standard curve method was used to obtain the relative EBV copy number in the LCL samples. Each experiment was conducted a minimum of two times and samples were run in triplicate for each experiment.
Correlation of EBV copy number, ATP levels, growth rate and drug cytotoxicity
Linear regression was performed using the lm function in the R software package (http://www.r-project.org). A linear model was constructed by the least sum of squares. Pearson’s correlation coefficient was calculated for each examined relationship in the unrelated HapMap Phase II LCLs and each population was evaluated independently as well as together using population as a covariate.
Heritability analysis
Twenty-four families (328 individuals) or thirty-four families (444 individuals) were used to evaluate cellular growth rate for heritability and linkage analysis. Heritability analysis of growth rate was performed using Sequential Oligogenic Linkage Analysis Routines (SOLAR, http://solar.sfbrgenetics.org) computer software to estimate narrow sense heritability. 22 SOLAR uses likelihood ratio tests to evaluate heritability by comparing a purely polygenic model with a sporadic model in the case of testing heritability. Covariates such as age, sex and the age–sex interaction were tested as previously described.13
Linkage analysis
MERLIN23 was used to perform non-parametric quantitative trait locus linkage analysis, which is robust to non-normally distributed data. The genotypic data were downloaded from the CEPH database (http://www.cephb.fr/cephdb/) and the Marshfield map database (http://research.marshfieldclinic.org/genetics/GeneticResearch/compMaps.asp) and error-checked for Mendelian incompatibility, mis-specified relationships and unlikely recombinations using a platform for linkage analysis as previously described.10,13 The SNP data was downloaded from the SNP Consortium (http://snp.cshl.org). From the combined pool of SNP and microsatellite markers genotyped in the above databases, approximately 7209 non-redundant markers were selected based on the availability of genotypes in at least 50% of family members. Physical positions of selected microsatellite and SNP markers were found using Build 36 of the UCSC Genome Browser (http://www.genome.ucsc.edu). Genetic maps were constructed based on microsatellite and SNP positions in the Marshfield map. These highly heterozygous markers, yielding a dense genetic map, were used for the analysis.
Genome-wide association studies
SNP genotypes were downloaded from the International HapMap database, release 23a (www.HapMap.org) for the CEU and YRI populations. SNPs with evidence of Mendelian transmission errors and alleles with a minor allele frequency of <5% were removed, leaving a total of more than 2 million SNPs for the association analysis in each population. The quantitative transmission disequilibrium test (QTDT) was performed to identify SNPs associated with drug cytotoxicity as well as those associating with growth rate using QTDT software (www.sph.umich.edu/csg/abecasis/QTDT).24 Using gender as a covariate, the QTDT was performed separately in each population (CEU and YRI HapMap Phase II LCLs) and for each trait: drug cytotoxicity and growth rate. Significant P-values of less than or equal to 0.0001 were used to compare SNPs across the two traits within the same population.
Results
Effect of cellular growth rate on drug-induced cytotoxicity
We compared the rate of cellular growth to the drug-induced cytotoxicity for the following chemotherapeutic agents: carboplatin, cisplatin, pemetrexed, etoposide, daunorubicin, cytarabine (AraC) and 5′-deoxyfluoruridine (5′-DFUR, an active form of capecitabine). Significant correlations were observed for all drugs evaluated within each of the populations studied (Table 1). The indicated references describe the cytotoxicity for each of the drugs in publications focusing on drug response. Using previously reported results of both cellular growth rate and drug-induced cytotoxicity for 5-fluorouracil (5-FU), 6-mercaptopurine, methotrexate and vorinostat,21 we evaluated whether the sensitivity to these drugs was also related to the cellular growth rate. This previously published study described the two phenotypes, but did not analyze their relationship with one another. We found a significant correlation for 5-FU, 6-mercaptopurine and methotrexate in at least one tested population (Table 1). The scatter plots describing the correlation across all agents are described in Supplementary Figures S2–S4. Although the degree of correlation varies, the relationship was largely significant across different pharmacological agents and populations implying that cell growth inhibition by these drugs is at least partially dependent on the rate at which the cells divide. This is consistent with previous reports in tumor cell lines that show more rapidly growing cells are more responsive to chemotherapy.25
Table 1.
Relationship between cellular cytotoxicity and growth rate in three HapMap populations
| Drug | Population | r2 | P-value |
|---|---|---|---|
| Etoposide16 | CEU | 0.167 | 1.96 × 10−3 |
| YRI | 0.385 | 1.59 × 10−7 | |
| Daunorubicin16 | CEU | 0.0565 | 7.80 × 10−2 |
| YRI | 0.482 | 1.10 × 10−9 | |
| Cytarabine17 | CEU | 0.229 | 1.90 × 10−4 |
| YRI | 0.142 | 3.50 × 10−3 | |
| ASN | 0.360 | 4.09 × 10−10 | |
| Pemetrexed | CEU | 0.418 | 9.77 × 10−8 |
| YRI | 0.410 | 4.85 × 10−8 | |
| 5-Deoxyfluorouridine | CEU | 0.385 | 4.27 × 10−7 |
| YRI | 0.121 | 1.10 × 10−2 | |
| ASN | 0.499 | 8.82 × 10−14 | |
| Carboplatin16 | CEU | 0.0755 | 4.50 × 10−2 |
| YRI | 0.421 | 2.70 × 10−8 | |
| ASN | 0.246 | 6.70 × 10−7 | |
| Cisplatin16 | CEU | 0.249 | 1.10 × 10−4 |
| YRI | 0.458 | 4.04 × 10−9 | |
| ASN | 0.344 | 1.25 × 10−9 | |
| 5-Fluorouracil21 | CEU | 0.00117 | 8.45 × 10−1 |
| YRI | 0.0416 | 1.32 × 10−1 | |
| ASN | 0.0652 | 1.64 × 10−2 | |
| 6-Mercaptopurine21 | CEU | 0.0394 | 2.53 × 10−1 |
| YRI | 0.0606 | 6.75 × 10−2 | |
| ASN | 0.0482 | 3.98 × 10−2 | |
| Methotrexate21 | CEU | 0.0169 | 4.63 × 10−1 |
| YRI | 0.0452 | 1.16 × 10−1 | |
| ASN | 0.128 | 8.90 × 10−4 | |
| Vorinostat21 | CEU | 0.0174 | 4.51 × 10−1 |
| YRI | 0.0128 | 4.06 × 10−1 | |
| ASN | 0.0233 | 1.56 × 10−1 | |
A linear regression was performed between growth rate and cellular sensitivity to multiple drugs. Footnote refers to the publication where the cytotoxicity response was described previously.
Variation in cellular growth rate between passages
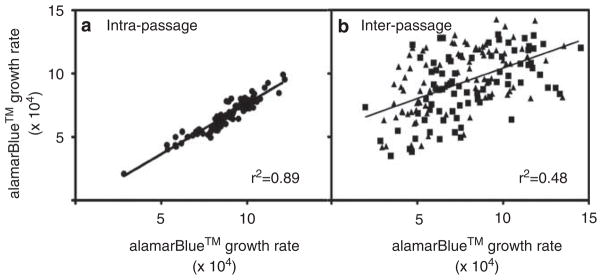
Cellular growth rate for the same cell line was calculated over multiple time periods. Two of these growth rate estimates were generated from two separate experiments completed during the same time period on 90 HapMap LCL lines derived from the International HapMap Asian populations. For cell lines derived from the same freeze–thaw, the growth rate was relatively stable (Figure 1a, r2=0.89); however, date of passage (that is, passage number) affects the growth rate estimates within a given cell line (Figure 1b). Each cell line had its cellular growth rate compared from two different time periods and we found significant differences in both the CEU and YRI population using the Wilcoxon’s rank sum test (YRI W=2171, P=4.87×10−7 CEU W=2501 P=9.07×10−3). It is clear that cellular growth rate varies over time for at least some cell lines, which may relate to the cellular conditions, freeze/thaw cycle and type/batch of culture media.
Figure 1.
Growth rate between and within passages. Growth rate estimates from the same lines were evaluated for correlation across different passage times. (a) Plots of the estimates of growth rate at 72 h for the same cell lines within the same passage and treatment time using the Asian HapMap cells. The r2 correlation is 0.89 with a slope of 0.80. (b) Plots of the estimates of growth rate at 72 h for the same cell lines across different passages, with years serving as a proxy for different passages using the CEU and YRI HapMap cells. The triangles represent the YRI lines whereas the squares represent the CEU lines. There was no difference across the two populations. The r2 correlation is 0.274 with a slope of 0.48.
Effect of baseline EBV and ATP on cellular growth rate
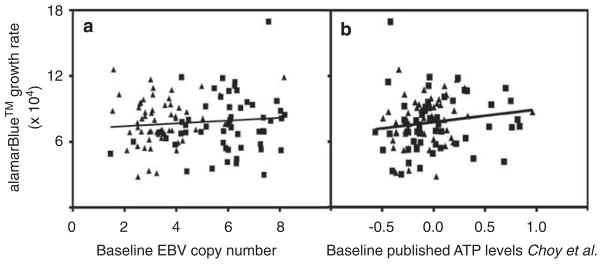
We measured baseline EBV copy number on 120 unrelated LCLs from the CEU and YRI HapMap populations. As illustrated in Figure 2, we found no relationship between cellular EBV copy number and cellular growth rate (r2=0.0097). Consistent with our results, we also found no relationship between publicly available baseline EBV copy number from 251 LCLs21 and cellular growth rate (Supplementary Figure S5). Baseline EBV copy number was also found not to correlate with drug-induced cytotoxicity (data not shown).
Figure 2.
Correlation between growth rate and baseline EBV copy number. (a) Shows growth rate estimates calculated from alamarBlue growth inhibition assay correlated against baseline EBV copy number in 107 unrelated HapMap lymphoblastoid cell lines. The correlation (r2=0.00969) is not significant (P=0.30). (b) Uses the same growth rate estimates to show no significant relationship with baseline ATP levels as measured in Choy et al.20
We next evaluated the effect of cellular ATP levels on cellular phenotypes, using recently published baseline ATP cellular levels by Choy et al.21 We found no significant correlation in any of the HapMap populations between these parameters (Table 2, Figure 2b) nor did we find a relationship with drug-induced cytotoxicity.
Table 2.
Relationship between baseline ATP levels and cellular growth rate using previously published ATP levels in Choy et al.21
| Population | r2 | P-value |
|---|---|---|
| CEU | 0.0053 | 0.674 |
| YRI | 2.05 × 10−6 | 0.992 |
| ASN | 0.031 | 0.101 |
Heritability of cellular growth rate
Because cellular growth rate was the single confounding variable that was related to drug-induced cytotoxicity, the question arose as to whether this was a heritable trait. To increase power for the estimate of heritability, we calculated cellular growth rate from our previously published cytotoxicity data13,14,26 on cell lines derived from large CEPH pedigrees. Cellular growth rates were obtained through three independent experiments and found to have significant heritability with h2 ranging from 0.30 to 0.39 with P-values ranging from 2.0×10−7 to 1.7×10−11 (Table 3), implying a significant genetic contribution to the growth rate phenotype.
Table 3.
Heritability of growth rate calculated from three experiments using large CEPH pedigrees
| Experiment | Number of families | Number of individuals | h2 | P-value |
|---|---|---|---|---|
| 1 | 34 | 444 | 0.39 | 1.7 × 10−11 |
| 2 | 24 | 328 | 0.31 | 1.0 × 10−7 |
| 3 | 24 | 328 | 0.30 | 2.0 × 10−7 |
Growth rate and genome-wide strategies for pharmacogenetic phenotypes
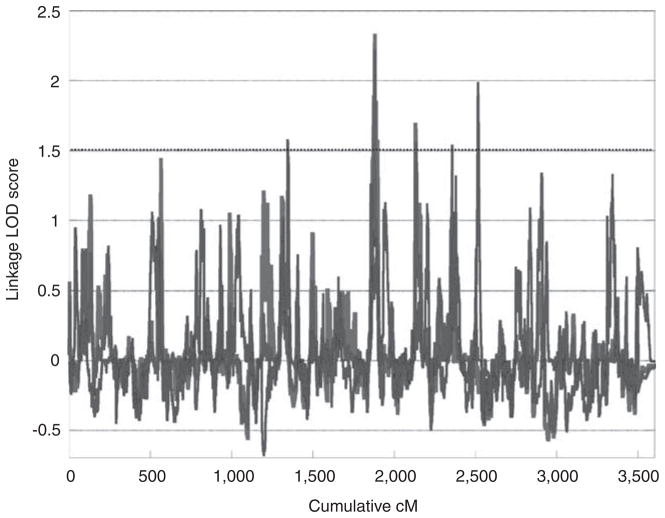
We performed both genome-wide linkage and association analysis using cellular growth rate as a phenotype. Two of the three genome-wide linkage scans peaked at the same location at 120cM on chromosome 9 (1881cM cumulative) (Figure 3). Although no linkage peak in any of the analyses reached the LOD threshold of 3 for genome-wide significance, five unique peaks achieved suggestive significance with a LOD score greater than 1.5.
Figure 3.
Genome-wide linkage scan for growth rate in lymphoblastoid cell lines. Growth rate was calculated from alamarBlue 72 h growth inhibition experiments using cell lines from the CEPH large pedigrees. Experiment 1 used 444 individuals from 34 families and experiments 2 and 3 used 328 individuals in 24 families. Five peaks reached a LOD score of 1.5, genome-wide suggestive significance.
Next, genome-wide association analysis with growth rate was performed with the same QTDT parameters as was used for cytotoxic response to 5′-DFUR in both the CEU and YRI cell lines. At the significance threshold of P≤0.0001 that has been used for previous analyses,14,15 273 and 554 SNPs significantly associated with growth rate in the CEU and YRI populations, respectively (Table 4) compared with 324 and 360 SNPs associating with cytotoxicity. The overlap of SNPs at this significance threshold between drug cytotoxicity and growth rate varies between the populations with 9% overlap in CEU and 0.003% overlap in YRI.
Table 4.
Genome-wide association of growth rate and 5′-DFUR cytotoxicity in two populations
| CEU | YRI | |
|---|---|---|
| SNPs associated at 0.0001 threshold | SNPs associated at 0.0001 threshold | |
| Growth rate estimate | 273 | 554 |
| 5′-DFUR cytotoxicity | 324 | 360 |
| Overlap | 29 | 1 |
| Percent overlap | 8.95% | 0.0028% |
Discussion
Over the past several years, there has been a surge in the number of studies employing LCLs for pharmacogenomic discovery and validation.20 While LCLs are a promising model for genotype–phenotype studies, there are some artifacts that may reduce power and have the potential to create spurious association due to confounding.21 For these reasons, we considered the effect of baseline EBV copy number, baseline ATP levels and cellular growth rate on cellular sensitivity to drugs. There was no relationship between EBV copy number or cellular ATP levels with either cellular growth rate or drug-induced cytotoxicity, indicating that neither of these variables are important confounders. In contrast, cellular growth rate and cytotoxicity were highly correlated, suggesting that cellular response to chemotherapeutic drugs is significantly greater for more rapidly growing cells. In addition, cellular growth rate was found to be a heritable trait with linkage analysis revealing five peaks above a “suggestive” LOD of 1.5.
The correlation between cellular growth rate and sensitivity to chemotherapeutic drugs was expected given that the mechanism of action of many of these agents is through their interaction with DNA. Measurements of cell growth inhibition following treatment with cytotoxic drugs in a short-term assay, by its nature, would have a greater effect in cells with a higher turnover rate of DNA (that is, no cell division in the time frame of the assay would result in no drug effect). In this study, we chose agents that were previously evaluated for pharmacogenomic outcomes in LCLs in our laboratory including daunorubicin,13 etoposide,14 cisplatin,15 carboplatin,16 cytarabine,17 pemetrexed and 5′-DFUR as well as those evaluated in LCLs by others: 5-FU,11,21 6-mercaptopurine, 21 methotrexate21 and vorinostat.21 The mechanism of action of cytarabine, a nucleoside analogue, is incorporation into DNA and inhibition of DNA synthesis;27 carboplatin and cisplatin form inter- and intra-strand DNA crosslinks,28 5-FU and 5′-DFUR inhibit thymidine synthesis, and subsequently affect DNA synthesis,29 etoposide and daunorubicin inhibit topoisomerase II and produce DNA double-stranded breaks.30,31 Therefore, one must consider that the markers identified in cell based models using whole genome approaches may be a combination of drug-specific markers and those contributing to cell growth.
Several previous studies by our laboratory considered growth rate as a covariate and evaluated the role of growth rate in significant association signals and population differences in sensitivity to the drug.16,17 This current study shows that neither extrinsic factor, baseline ATP or EBV, influence growth rate; however, there is some variation in the growth rate observed between passages. Although we observe a very tight correlation of cell growth rate when evaluating cells in the same passage, there is some variation observed over time (taking cells out of freeze years later). This variation in growth rate over time is a limitation of using drug-induced cytotoxicity as a phenotype for pharmacogenomic discovery because drug sensitivity and growth rate are related. However, the correlation coefficients between growth rate and cytotoxicity are quite low suggesting growth rate has only a modest effect on cellular sensitivity in LCLs. Further work should be done to comprehensively evaluate the effects of passaging, freezing and thawing on LCL growth rate.
Although environment and experimental variation were obvious contributors of cellular growth rate, the extent to which genetics had a role was not clear. The use of large pedigrees allowed us to determine that 30–39% of the variation in cellular growth rate is because of genetic factors. Importantly, the confounding effects of different passage numbers likely underestimates, rather than overestimates the heritability of cellular growth rate because families within each experiment were phenotyped over a period of several years. The significant heritability of growth rate and use of CEPH pedigrees allowed us the opportunity to map the trait using genome-wide methods of linkage analysis and to evaluate for overlap with drug-associated linkage peaks.10–13,32 For example, the overlap between the peak identified for 5-FU and docetaxel-induced cytotoxicity on chromosome 911 is at the same location as a peak observed for growth rate alone identified in this study. Therefore, genes in this region may act through proliferation modification rather than mechanisms specific to 5-FU or docetaxel.
While growth rate is an independently heritable variable in pharmacological studies in LCLs, the model remains an essential in vitro model to investigate toxic drugs that would be unethical or difficult to study in humans. The most appropriate way to incorporate growth rate into pharmacogenomic studies is through measurement of the cellular growth rate at the same time that the phenotype of interest is being characterized followed by the use of growth rate as a covariate with the trait of interest. This may be a means of “isolating” the other genetic effects that are modulating sensitivity to a given drug. However, from a translational/clinical perspective, variants or genes important in chemotherapeutic-induced cytotoxicity may be relevant regardless of their mechanism. Although variants identified as significant for chemotherapeutic-induced sensitivity due to their effect on cellular growth rate may still have utility for affecting clinical outcomes, there are questions that remain. For example, we do not know how growth rates in vitro compare with the growth rates in vivo or whether the same variables affect growth rates similarly in vitro and in vivo. The growth limiting mechanisms may be substantially different and thus the variants that affect growth in vitro may not be equally important in an in vivo setting. In summary, studies using LCLs for pharmacological outcomes should consider that a portion of the genetic variation explaining drug-induced cytotoxicity is mediated via heritable effects on cellular growth rate. Researchers using LCLs for pharmacogenomic studies must determine the variants of greatest interest and address the confounding effects of growth rate in the most appropriate manner for their specific study.
Supplementary Material
Acknowledgments
This work was supported by the Pharmacogenetics of Anticancer Agents Research Group by the National Institute of Health/National Institute of General Medical Sciences Grant U01GM61393, data deposits are supported by UO1GM61374 (http://pharmgkb.org/) and NIH/NCI Breast SPORE P50 CA125183. We acknowledge Dr Sunita J Shukla, Dr Christine M Hartford, Ms Bridget E McIlwee, and Mr Wasim Bleibel for results from their cytotoxicity growth inhibition experiments and Mr Steven J Stark for excellent technical assistance.
Abbreviations
- 5′-DFUR
5′-deoxyfluorouridine
- 5-FU
5-fluorouracil
- AraC
cytarabine
- ASN
Asian
- ATP
adenosine tri-phosphate
- CEPH
Centre d’Etude du Polymorphisme Humain
- CEU
Caucasians
- DNA
deoxyribonucleic acid
- EBV
Epstein–Barr virus
- h2
heritability
- HapMap
International HapMap Project
- LOD
logarithm of odds
- LCL/s
lymphoblastoid cell line/s
- QTDT
Quantitative Transmission Disequilibrium Test
- RNA
ribonucleic acid
- SNP/s
single nucleotide polymorphism/s
- YRI
Yoruba
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
References
- 1.Gipps EM, Kidson C. Cellular radiosensitivity: expression of an MS susceptibility gene? Neurology. 1984;34:808–811. doi: 10.1212/wnl.34.6.808. [DOI] [PubMed] [Google Scholar]
- 2.Jen KY, Cheung VG. Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Res. 2003;13:2092–2100. doi: 10.1101/gr.1240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 4.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM. Gene-expression variation within and among human populations. Am J Hum Genet. 2007;80:502–509. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 11.Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc Natl Acad Sci USA. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla SJ, Duan S, Badner JA, Wu X, Dolan ME. Susceptibility loci involved in cisplatin-induced cytotoxicity and apoptosis. Pharmacogenet Genomics. 2008;18:253–262. doi: 10.1097/FPC.0b013e3282f5e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan S, Bleibel WK, Huang RS, Shukla SJ, Wu X, Dolan ME. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang RS, Duan S, Bleibel WK, Kistner EO, Zhang W, Clark TA, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci USA. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang RS, Duan S, Shukla SJ, Kistner EO, Clark TA, Chen TX, et al. Identification of genetic variants contributing to Cisplatin-induced cytotoxicity by use of a genome-wide approach. Am J Hum Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang RS, Kistner EO, Bleibel WK, Shukla SJ, Dolan ME. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol Cancer Ther. 2007;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartford CM, Duan S, Delaney SM, Mi S, Kistner EO, Lamba JK, et al. Populuation-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood. 2009;113:2145–2153. doi: 10.1182/blood-2008-05-154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 2008;68:7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones TS, Yang W, Evans WE, Relling MV. Using HapMap tools in pharmacogenomic discovery: the thiopurine methyltransferase polymorphism. Clin Pharmacol Ther. 2007;81:729–734. doi: 10.1038/sj.clpt.6100135. [DOI] [PubMed] [Google Scholar]
- 20.Welsh MM, Mangravite L, Medina MW, Tantisira K, Zhang W, Huang RS, et al. Pharmacogenomic discovery using cell-based models. Pharmacol Rev. 2009;61:413–429. doi: 10.1124/pr.109.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choy E, Yeleknsy R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 24.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tveit KM, Fodstad O, Pihl A. The usefulness of human tumor cell lines in the study of chemosensitivity. A study of malignant melanomas. Int J Cancer. 1981;28:403–408. doi: 10.1002/ijc.2910280402. [DOI] [PubMed] [Google Scholar]
- 26.Shukla SJ, Duan S, Wu X, Badner JA, Kasza K, Dolan ME. Whole-genome approach implicates CD44 in cellular resistance to carboplatin. Hum Genomics. 2009;3:128–142. doi: 10.1186/1479-7364-3-2-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia. 2001;15:875–890. doi: 10.1038/sj.leu.2402114. [DOI] [PubMed] [Google Scholar]
- 28.Rabik CA, Njoku MC, Dolan ME. Inactivation of O6-alkylguanine DNA alkyltransferase as a means to enhance chemotherapy. Cancer Treat Rev. 2006;32:261–276. doi: 10.1016/j.ctrv.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Walko CM, Lindley C. Capecitabine: a review. Clin Ther. 2005;27:23–44. doi: 10.1016/j.clinthera.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Meresse P, Dechaux E, Monneret C, Bertounesque E. Etoposide: discovery and medicinal chemistry. Curr Med Chem. 2004;11:2443–2466. doi: 10.2174/0929867043364531. [DOI] [PubMed] [Google Scholar]
- 31.Wang B, Perchellet EM, Wang Y, Tamura M, Hua DH, Perchellet JP. Antitumor triptycene bisquoinones: a novel synthetic class of dual inhibitors of DNA topoisomerase I and II activities. Anticancer Drugs. 2003;14:503–514. doi: 10.1097/00001813-200308000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Bleibel WK, Duan S, Huang RS, Kistner EO, Shukla SJ, Wu X, et al. Identification of genomic regions contributing to etoposide-induced cytototoxicity. Hum Genet. 2009;125:173–180. doi: 10.1007/s00439-008-0607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.