Summary
Proteins targeted for degradation by the Mycobacterium proteasome are post-translationally tagged with prokaryotic ubiquitin-like protein (Pup), an intrinsically disordered protein of 64 residues. In a process termed “pupylation”, Pup is synthesized with a terminal glutamine, which is deamidated to glutamate by Dop (deamidase of Pup) prior to attachment to substrate lysines by PafA (proteasome accessory factor A). Importantly, PafA was previously shown to be essential to cause lethal infections by Mycobacterium tuberculosis (Mtb) in mice. In this study we show that Dop, like PafA, is required for the full virulence of Mtb. Additionally, we show that Dop is involved not only in the deamidation of Pup, but is also needed to maintain wild type steady state levels of pupylated proteins in Mtb. Finally, using structural models and site-directed mutagenesis our data suggest Dop and PafA are members of the glutamine synthetase fold family of proteins.
Introduction
Proteasomes are self-compartmentalized proteases found in all eukaryotes, archaea and some bacteria [reviewed in (Cerda-Maira & Darwin, 2009)]. In eukaryotes, proteasomes are essential and are required for the turnover of numerous proteins, including damaged polypeptides and proteins involved in signal transduction. Proteins doomed to the proteasome are usually tagged with the small protein modifier ubiquitin (Ub), which forms isopeptide bonds to lysine (K, Lys) residues on substrate proteins. Ub is a highly structured protein of 76 amino acids and is the founding member of a growing group of small post-translational Ub-like modifiers (Ubls) in eukaryotes including SUMO (small Ub like modifier), ISG15 (interferon stimulated gene 15), and NEDD8 (neural precursor cell expressed, developmentally down regulated 8). All members share a common structural fold and a di-glycine motif at their carboxyl (C)-termini, where a series of enzymatic reactions ultimately conjugate Ub or other Ub-like modifiers (Ubl) to a lysine of a target protein [reviewed in (Hochstrasser, 2009)].
In contrast to the eukaryotic proteasome, bacterial proteasomes are not always essential for life (Burns et al., 2009, Knipfer & Shrader, 1997, Hong et al., 2005) but are required for the full virulence of one of the world’s most successful pathogens, Mtb (Darwin et al., 2003, Darwin et al., 2005, Gandotra et al., 2007, Lamichhane et al., 2006). Like eukaryotic proteasome substrates, proteins targeted to the Mtb proteasome are tagged with a small protein modifier named Pup (Pearce et al., 2008). Unlike eukaryotic Ub, Pup does not assume a Ub fold and does not conjugate to substrates via Gly but rather forms isopeptide bonds using a C-terminal glutamate (E, Glu) (Burns et al., 2009, Pearce et al., 2008, Striebel et al., 2009). Furthermore, homologues of canonical mammalian Ub conjugating enzymes do not appear to be involved in pupylation, thus the elucidation of the enzymatic mechanism of pupylation may provide novel targets for tuberculosis therapies.
Prior to ligation to substrate lysines by PafA Pup is translated with a C-terminal glutamine (Q, Gln) that must be deamidated by Dop (Imkamp et al., 2009, Striebel et al., 2009). Despite their distinct roles in pupylation, Dop and PafA are predicted to have structural similarity to glutamine synthetases (GS), in particular, the γ-glutamate-cysteine ligases (γ-GCS), which catalyze the formation of γ-glutamyl-cysteine from glutamate and cysteine (C, Cys) (Iyer et al., 2008). γ-glutamate-cysteine ligases use adenosine triphosphate (ATP) to phosphorylate the γ-carboxylate of glutamate, which primes it for nucleophilic attack by the amino group of cysteine. PafA catalyzes the ligation of deamidated Pup to proteasome substrates in a reaction that requires ATP hydrolysis (Pearce et al., 2008, Striebel et al., 2009). Thus, based on its predicted structural homology to GS/γ-GCS enzymes, PafA is proposed to phosphorylate deamidated Pup at the γ-carboxylate of its terminal glutamate (Striebel et al., 2009, Iyer et al., 2008). This intermediate would then be attacked by the amino group of the substrate lysine, forming an isopeptide bond. Consistent with this hypothesis, it was shown by nuclear magnetic resonance spectroscopy that PafA conjugates the γ-carboxylate of the terminal glutamate of Pup to a substrate lysine (Sutter et al., 2010). However, it is not understood how PafA facilitates Pup phosphorylation and ligation to substrates. Unlike PafA, Dop deamidates the C-terminal glutamine of Pup in a reaction that requires ATP binding, but not ATP hydrolysis (Striebel et al., 2009). Thus, in contrast to PafA, there does not appear to be an obvious link between GS/γ-GCS-like activity and Dop function.
In this work we show that disruption of dop sensitizes Mtb to reactive nitrogen intermediates (RNI) in vitro and severely attenuates Mtb growth in mice. We also show that Dop is not only required for Pup deamidation, but is also needed for maintaining steady-state levels of pupylated proteins in Mtb. We also introduced mutations at the C-terminus of Pup to test the importance of its terminal glutamine, as well as the penultimate di-glycine motif that is conserved in almost all conjugatable Ubls. Finally, we used site-directed mutagenesis to test the hypothesis that Dop and PafA have active sites with a GS/γ-GCS fold. Using structural prediction modeling and the mutagenesis data, we propose that Dop and PafA are members of the GS-fold superfamily with divergent activities.
Results
Dop is required for pupylation in Mtb
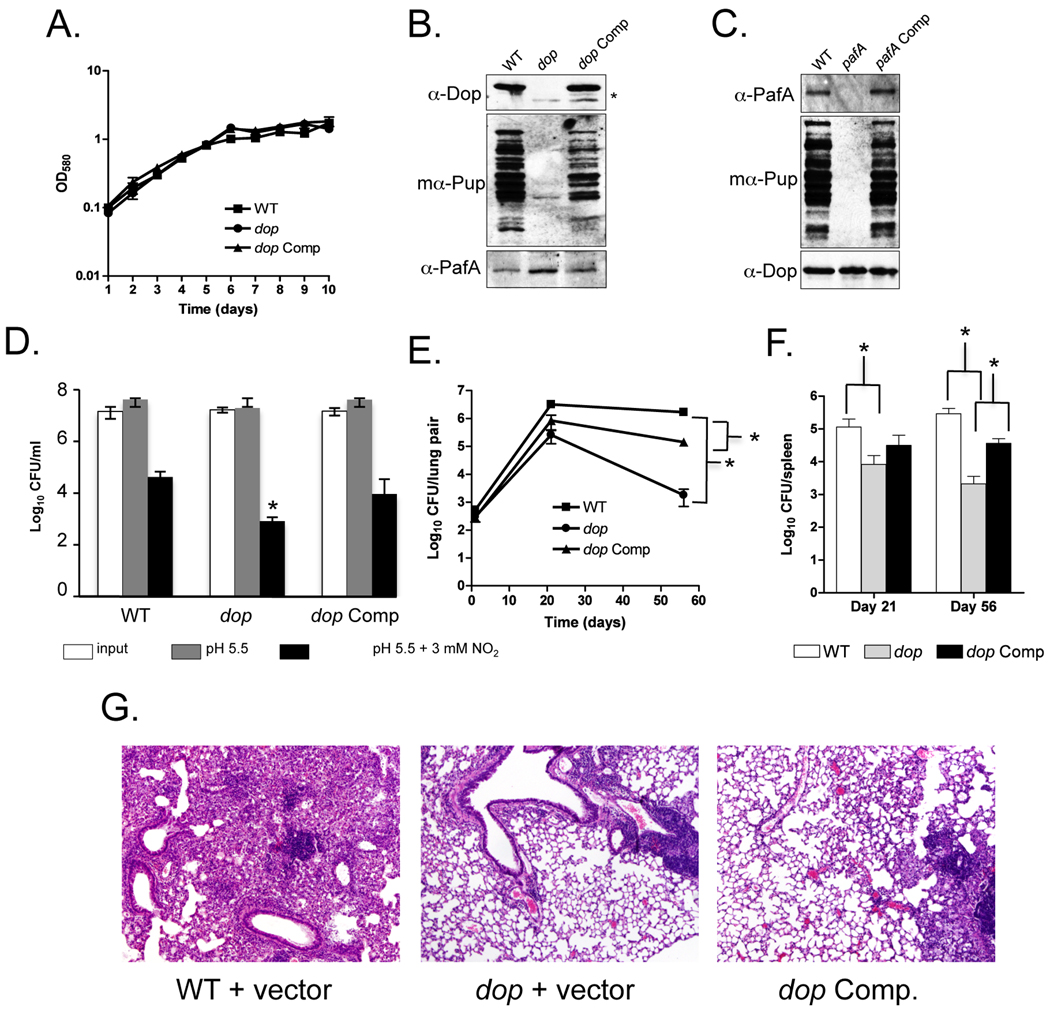
In vitro experiments showed that deamidation of the C-terminal glutamine of Pup is required prior to the attachment to two proteasome substrates, FabD and PanB (Striebel et al., 2009). In vivo, Dop was shown to be required for pupylation in Mycobacterium smegmatis (Msm), a non-pathogenic, fast growing mycobacterium species related to Mtb (Imkamp et al., 2009). In order to investigate the role of Dop in Mtb we characterized a dop∷ΦMycoMarT7 transposon disruption mutant and its isogenic parental wild type (WT) Mtb strain, CDC1551(Lamichhane et al., 2003). A growth-curve analysis showed that there were no growth-rate differences between the WT, dop and dop-complemented strains when cultured under standard conditions (see Experimental Procedures) (Fig. 1A). Furthermore no pupylation was apparent in the dop mutant (Fig. 1B). Because dop is encoded upstream of pup, we complemented the dop transposon mutation to determine if disruption of dop, and not polar effects on pup expression, resulted in the absence of pupylation. We were able to complement the pupylation defect to near WT levels using an integrative plasmid encoding dop expressed from its native promoter (Fig. 1B, Table S1).
Fig. 1.
Mtb Dop is required for pupylation, resistance to RNI and virulence in mice. (A) Growth curve analysis of WT, dop and dop-complemented (“dop Comp”) Mtb CDC1551 strains. The strains were grown with no aeration and the optical density at absorbance 580 nm (OD580) of the cultures was measured daily. (B) An Mtb dop mutant is defective for pupylation. Total cell lysates from equivalent cell numbers of WT, dop, and dop-complemented ("dop Comp") CDC1551 strains were analyzed by immunoblotting. Asterisk (*) indicates endogenous truncated Dop resulting from the transposon insertion in dop. "mα-Pup" indicates the use of monoclonal antibodies to Pup. (C) Total cell lysates from equivalent cell numbers of WT, pafA, pafA-complemented ("pafA Comp") H37Rv strains. For (B) and (C), proteins were separated by 10% SDS-PAGE and analyzed with polyclonal antibodies to Dop or PafA, or monoclonal antibodies to Pup. (D) The Mtb dop mutant is hypersusceptible to RNI. Survival of WT, dop and dop-complemented Mtb CDC1551 strains was determined after six days of incubation in media pH 5.5 in the presence or absence of 3 mM NaNO2. Error bars indicate ± one standard deviation (SD). Asterisk (*) indicates the dop mutant was statistically significantly more sensitive to RNI than WT or the complemented strains (P < 0.05). Data are from two independent experiments each performed in triplicate, with an outlier excluded from each strain tested. (E, F) The dop Mtb mutant is severely attenuated for growth and survival in mice. Colony forming units (CFU) from lungs (E) and spleens (F) harvested on day 1 (n = 3), day 21 (n = 4) and day 56 (n = 4) from mice infected with WT, dop and dop-complemented Mtb CDC1551 strains. Error bars indicate ± one SD. Single asterisk (*) indicates a P value of < 0.05. The WT and dop-complemented strains were significantly different from each other in the lungs (as indicated) and spleens (not indicated due to space constraints), suggesting complementation was not complete. All statistics were performed using a nonparametric Student’s two-tailed t test. (G) Hematoxylin and eosin staining of formalin fixed lungs from mice infected with WT (MHD583), dop (MHD375) and dop-complemented (MHD376) Mtb CDC1551 strains. 4 5 magnification is shown.
The absence of dop could possibly result in reduced amounts of PafA or conversely, disruption of pafA could result in lower steady-state levels of Dop; in either case, pupylation would be abrogated. Therefore, we examined the steady state stability of PafA in the dop mutant, and of Dop in the pafA mutant. PafA and Dop were present at WT levels in the dop and pafA mutants, respectively, and pupylation was abolished in both strains (Figs. 1B and 1C). Taken together, these data show that dop is required for pupylation in Mtb, and that the absence of either Dop or PafA does not affect the steady-state stability of the other protein.
PafA, along with Mpa (Mycobacterium proteasomal ATPase), was identified in a screen for Mtb transposon mutants sensitive to RNI (Darwin et al., 2003). Similar to the pafA and mpa mutants, the Mtb dop mutant was hypersensitive to RNI produced by acidified sodium nitrite compared to WT Mtb (Fig. 1D). Complementation of the dop mutation restored resistance to RNI to near WT levels (Fig. 1D).
In addition to RNI resistance, proteasomal degradation and pupylation are essential for Mtb to cause lethal infections in mice (Darwin et al., 2003, Darwin et al., 2005, Gandotra et al., 2007, Lamichhane et al., 2006, Pearce et al., 2006). Using an aerosol model of infection, we showed that the dop mutant was also severely attenuated for survival and growth in mice (Fig. 1E, F). The degree of attenuation was similar to or greater than what was previously observed for the H37Rv mpa and pafA mutants (Darwin et al., 2003). Histopathology also revealed dramatic differences between WT and the dop mutant-infected mice, where WT Mtb-infected mice showed significant infiltration of lymphocytes and tissue destruction, which was not observed in the dop mutant infection (Fig. 1G). Although the growth defect of the dop mutant was partially restored in the dop-complemented strain, we did not observe restoration of the severe tissue destruction that was observed in the WT infection (Fig. 1G, right panels). This is most likely due to the lack of full complementation of the dop mutation. The relatively mild pathology may be due to lower bacterial burden or the deregulation of one or more proteins in the dop mutant required for causing severe pathology. Nonetheless, examination of lung sections revealed numerous foci of immune cells in the dop-complemented strain, but not in the dop mutant-infected lungs (not shown). This supports the observed increase in bacterial burden in the dop-complemented strain infection (Fig. 1E,F). Thus, these data strongly suggest that Dop is required for the full virulence of Mtb in an animal infection model.
Dop is required to maintain WT steady state levels of the pupylome in Mtb
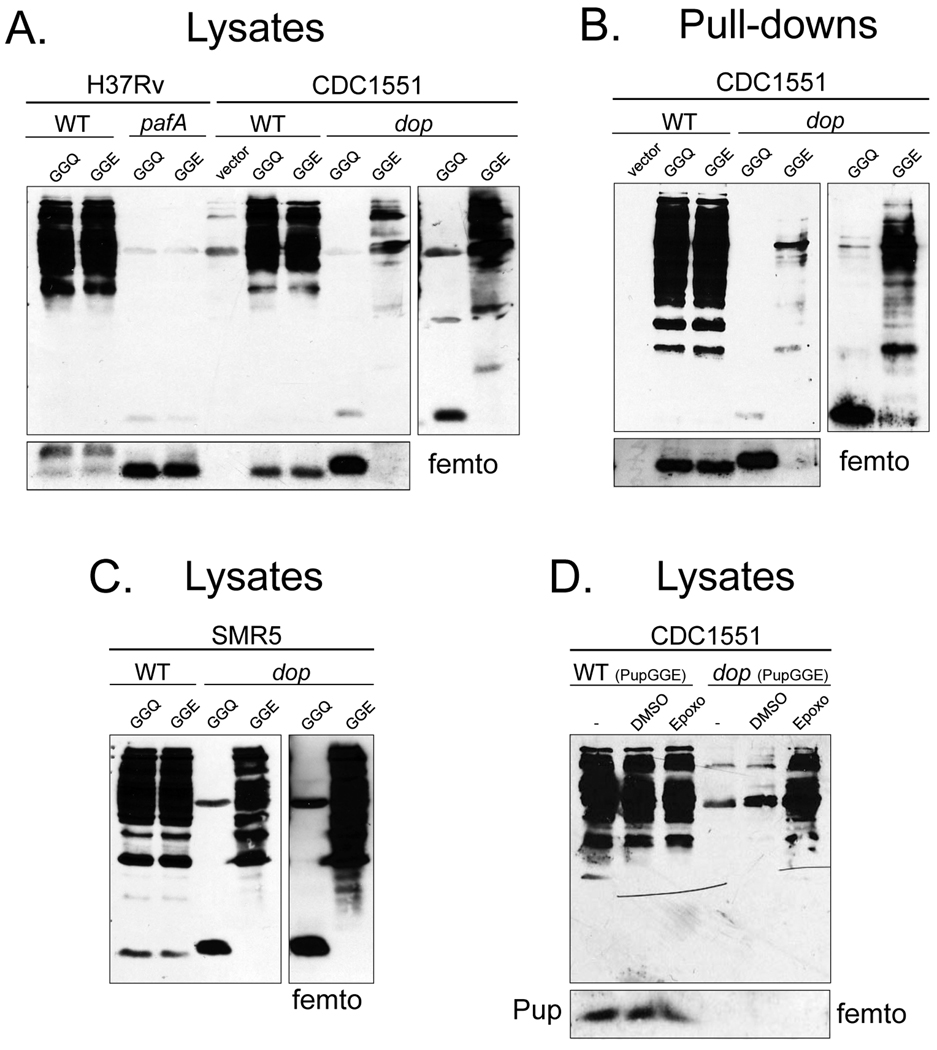
It was previously concluded that the function of Dop was to deamidate Pup, although it had been noted that bacteria that encode Pup terminating in glutamate also encode Dop (Striebel et al., 2009). Consistent with this, expression of pupGGE in an Msm dop mutant bypassed the need for Pup deamidation, and fully restored pupylation in vivo (Imkamp et al., 2009). This suggested that the primary function of Dop is to deamidate Pup in Msm. To determine if this were also true in Mtb, we transformed WT and dop mutant Mtb strains with plasmids encoding epitope-tagged pupGGQ or pupGGE under the control of a mycobacterial hsp60 promoter. As a negative control, we also transformed the pafA mutant strain with the same plasmids. Total cell lysates were analyzed with polyclonal antibodies to Pup. To our surprise, pupylation was not restored in the Mtb dop mutant expressing either his6-pupGGQ or his6-pupGGE (Fig. 2A). We next purified His6-pupylated proteins (the pupylome) (Festa et al., 2010) from these strains and examined pupylation by immunoblotting. As expected, we observed a robust pupylome purified from WT Mtb expressing either his6-pupGGQ or his6-pupGGE, but only a scant pupylome was purified from the dop mutant expressing his6-pupGGE (Fig. 2B). Furthermore, pupylation in the dop mutant could only be observed when the immunoblot was developed with a highly sensitive chemiluminescence detection reagent (Fig. 2A and B, right panels). In addition, free His6-PupGGE, but not His6-PupGGQ, was observed at far lower amounts in the dop mutant in comparison to the WT and pafA strains (Fig. 2A and B, lower panels).
Fig. 2.
Expression of pupGGE is not sufficient to bypass the requirement for Dop in Mtb. (A) Mtb H37Rv WT and pafA or CDC1551 WT and dop strains were transformed with plasmids encoding N-terminal His6 tagged pupGGQ, pupGGE or empty vector. Lysates with equivalent cell numbers from these strains were analyzed by 12% SDS-PAGE and immunoblotting with polyclonal antibodies to Pup. At least two transformants from each electroporation were characterized with similar results. (B) His6-PupGGQ or His6-PupGGE pupylomes from the same strains in (A) were purified. Proteins were analyzed by 12% SDS-PAGE and immunoblotting. (C) The same plasmids encoding pupGGQ and pupGGE were used to transform the previously reported Msm dop mutant and the parental WT strain. Total cell lysates with equivalent cell numbers were separated by 12% SDS-PAGE and analyzed with polyclonal antibodies to Pup. ThermoScientific Super Signal West Femto reagent (“femto”) was used to detect free Pup (lower panels) or the pupylomes in the dop mutant (right panels) for (A)–(C). (D) Proteasomal inhibition appeared to rescue pupylated proteins from degradation in the Mtb dop mutant expressing pupGGE.
It was possible that the plasmids used for expression in Mtb made less recombinant Pup than the plasmids used in the study by Imkamp and co-workers. To address this, we obtained the previously reported Msm dop mutant and determined if our pupGGE expressing plasmid could restore pupylation in Msm. Unlike our observations in Mtb, pupGGE could fully bypass the need for a deamidase (Fig. 2C), consistent with the previous report (Imkamp et al., 2009).
In eukaryotes, ubiquitin is removed and recycled from proteasome substrates prior to degradation [reviewed in (Finley, 2009)]. We wondered if Pup also needed to be removed from substrates and recycled in a Dop-dependent manner to maintain high steady state levels of Pup and the pupylome; the absence of Dop would result in “one way” trips for all pupylated proteins to the proteasome. To test this hypothesis we expressed pupGGE in the WT and dop strains in the presence of the specific proteasome inhibitor epoxomicin.. Indeed, a robust pupylome was observed after epoxomicin treatment of dop mutant Mtb expressing pupGGE (Fig. 2D). Thus this result suggests that Dop may be directly or indirectly involved in the removal and recycling of Pup from proteasome substrates.
C-terminal glutamine or glutamate is essential for pupylation
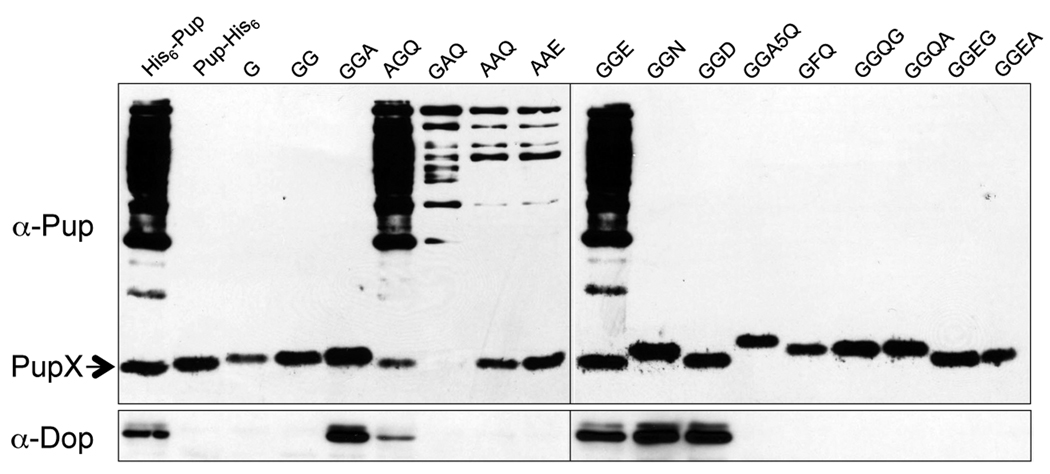
Almost all known Ubl modifiers attach to target proteins via a conserved di-glycine motif [reviewed in (Hochstrasser, 2009, Kerscher et al., 2006)]. Although Pup has this motif, it does not attach to proteins via a terminal glycine (Burns et al., 2009, Pearce et al., 2008). Additionally, all known Pup homologues either have a C-terminal glutamine or glutamate. We tested the importance of the Mycobacterium Pup di-glycine motif and the C-terminal glutamine or glutamate in pupylation. Msm was transformed with plasmids encoding an N-terminal His6-tagged Pup with various mutations at or near the C-terminus (Table S1). Recombinant His6-Pup was purified under non-denaturing conditions from Msm and the purified proteins were analyzed by immunoblotting using polyclonal antibodies to Pup.
Expression of his6-pupGGQ or his6-pupGGE in WT Msm resulted in the production and purification of a robust pupylome (Fig. 3), consistent with data in Fig. 2 and a previous report (Imkamp et al., 2009). Additionally, deletion or replacement of glutamine with any amino acid other than glutamate eliminated pupylation, consistent with in vitro and in vivo Msm data (Striebel et al., 2009, Imkamp et al., 2009). Moreover, no pupylation was observed if a single amino acid was added after the terminal glutamine or glutamate. Interestingly, either or both glycines of the di-glycine motif could be replaced with alanine and still support a low level of pupylation (Fig. 3). Together, these results suggested that Pup deamidation or conjugation to substrates is not completely dependent upon the di-glycine motif, whereas glutamine or glutamate must be at the C-terminus in order to conjugate to proteins.
Fig. 3.
Mutation analysis of the C-terminus of Pup. WT Msm was transformed with plasmids expressing his6-pup with different C-terminal mutations. His6-Pup C-terminus mutant proteins were purified from soluble lysates and analyzed by 12% SDS-PAGE gel and immunoblotting with polyclonal antibodies to Pup. Only the terminal residues are indicated for the mutant proteins. “PupX” indicates unconjugated WT or C-terminal mutant Pup. Immunoblotting with antibodies to Dop reveals Dop requires the di-glycine motif for robust binding to Pup (lower panel).
We next determined if PafA or Dop could co-purify with the WT or mutant Pup derivatives. The same samples were examined using antibodies to PafA or Dop. When we examined these samples using antibodies to PafA, we observed several bands migrating around the molecular weight of PafA, making it difficult to discern which band corresponded to PafA (data not shown). Therefore, it was unclear if PafA could strongly interact with the C-terminus of Pup. In contrast, Dop co-purified robustly with Pup, but only if it had the di-glycine motif plus a terminal amino acid (Fig. 3). Replacement of one or both glycines with alanine dramatically reduced or eliminated Dop binding.
Because deamidated Pup (PupGGE) migrates more quickly than WT Pup (PupGGQ) through SDS-PAGE gels (Striebel et al., 2009) (Fig. 3), we could determine if Pup C-terminal mutants could be deamidated. Msm producing PupAAQ resulted in the production of a protein that co-migrated with PupAAE, suggesting PupAAQ can be deamidated, despite the loss of efficient Dop binding to PupAAQ (Fig. 3). Importantly, PupAAQ and PupAAE produced in Msm resulted in similar pupylomes. In contrast, PupGGD (D = aspartate) migrated more quickly than PupGGN (N = asparagine) in the protein gels, although both bound Dop (Fig. 3). This result indicated that despite effective binding of PupGGN to Dop, the side chain of asparagine was inaccessible to the deamidase activity of Dop. Taken together, these results suggest that although the di-glycine motif of Pup is required for tight binding to Dop, it is not essential for Pup deamidation or ligation. In contrast, the length of the terminal residue side chain is critical for Pup deamidation and substrate ligation.
PafA and Dop are predicted to have active sites with a GS fold
Several investigators reported the similarity of Dop and PafA to proteins of the GS-fold superfamily (Iyer et al., 2008, Striebel et al., 2009, Darwin, 2009). Using HHpred (Soding et al., 2005), sequence analyses indicated that the amino (N)-terminal half of PafA adopts a fold similar to that of E. coli YbdK, a γ-GCS (Lehmann et al., 2004). YbdK catalyzes the formation of γ-glutamyl-cysteine from glutamate and cysteine, but its physiological role is unknown. YbdK is believed to catalyze the ATP-dependent phosphorylation of the γ-carboxylate of glutamate, which generates an unstable phosphorylated intermediate that becomes a target for nucleophilic attack by the amino group of cysteine. Because PafA ligates Pup to proteasome substrates via the side chain carboxylate of the C-terminal glutamate (Sutter et al., 2010), it is presumed that the glutamate is phosphorylated prior to conjugation to a substrate lysine (Striebel et al., 2009, Iyer et al., 2008). Consistent with this hypothesis, PafA requires ATP hydrolysis for this reaction.
PafA is predicted to contain six β-strands (β1-6) with key residues conserved in members of the GS-fold family (Fig. 4A; Table 1) (Iyer et al., 2008). The conserved residues are primarily involved in the coordination of Mg2+, ATP, and glutamate. To determine if these residues were important for PafA activity, we mutated pafA encoded on an integrative plasmid previously utilized for complementation studies. The WT pafA plasmid restored substrate degradation (Festa et al., 2007) and pupylation in a pafA null mutant (Fig. 1C). The mutated genes were expressed in an Mtb pafA∷Φ MycoMarT7 null mutant (strain MHD2, Table S1) and pupylation was assessed.
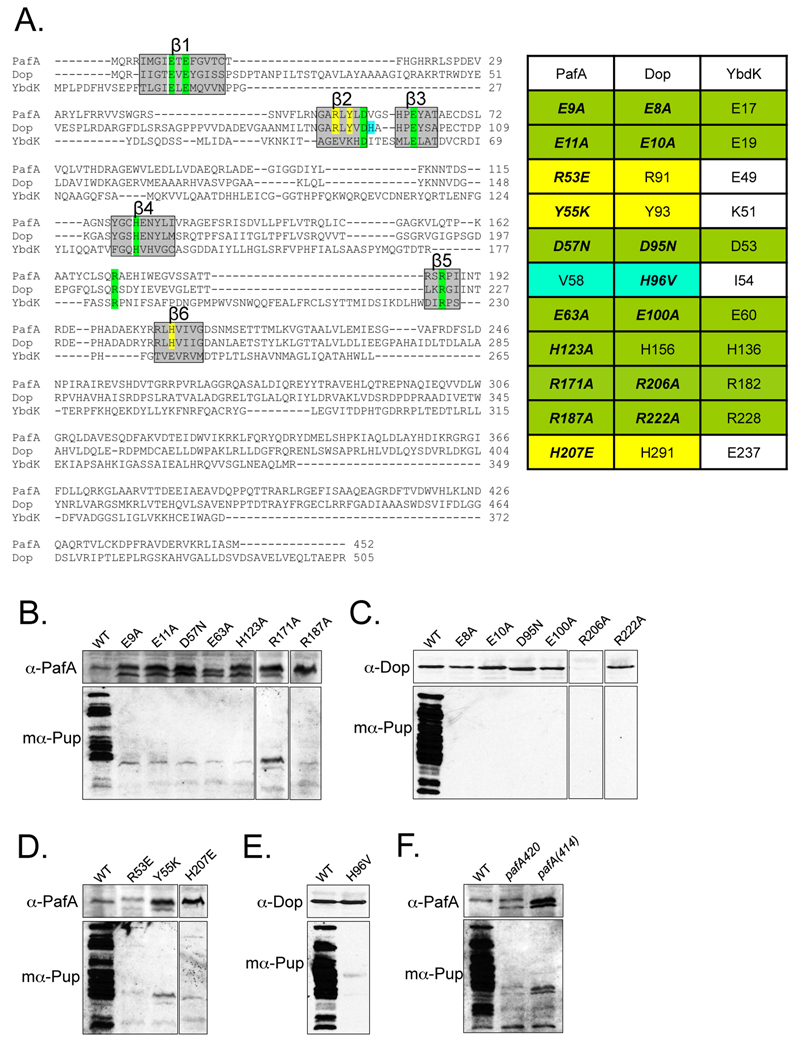
Fig. 4.
Dop and PafA share critical catalytic residues with GS-fold family members. (A) (Left) Alignment of PafA, Dop and E. coli YbdK shows conserved residues crucial for GS-fold protein function in β strands (boxed in grey). Highlighted residues were selected for mutagenesis. Sequences were compiled from the NCBI server and aligned in segments using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html). (Right) Residues conserved among PafA, Dop and GS-fold protein members are highlighted in green. Amino acids conserved among PafA and Dop homologues, but not in other GS-fold members are highlighted in yellow. A residue conserved in Dop homologues but not in PafA homologues is in cyan. (B)–(C) Residues conserved in GS/γ-GCS-fold proteins are important for PafA (B) and Dop (C) function. Site-directed mutation analysis of PafA and Dop. Null mutants of pafA or dop were transformed by electroporation with plasmids encoding pafA or dop, respectively, with single mutations. In some samples, a cross-reactive band is seen in the pafA mutant lysates below PafA. (D) Residues conserved in PafA and Dop homologues but not found in GS-fold proteins are critical for PafA function. (E) Replacement of a conserved histidine in Dop homologues for a valine conserved in PafA family members eliminates pupylation. (F) The C-terminus of PafA is required for pupylation. Endogenous PafA420 and recombinant PafA414 are indicated. For all experiments, total cell lysates of equivalent cell numbers were separated by 10% SDS-PAGE and analyzed by immunoblot analysis with antibodies to PafA, Dop or Pup as in Fig. 1.
β1 has a highly conserved GψExE motif (ψ indicates a hydrophobic residue; × indicates any amino acid) found in GS-fold proteins (Iyer et al., 2008), thus we introduced single point mutations at Glu9 and Glu11 in PafA. In addition, we replaced conserved residues in β2 (Asp57) and β3 (Glu63) (Iyer et al., 2008). β4 has a H/QxN/H motif that is likely to be equivalent to the H/DxN/H motif of characterized members of GS-fold proteins; this motif is required for metal ion coordination and for interaction with the phosphorylated glutamate. We thus mutated His123 to test its importance in PafA function. Finally, it was previously noted that PafA has conserved arginines that might be important for function: Arg187 in β5 and Arg171 present in a helical segment between the β4 and β5 strands were mutated (Iyer et al., 2008). As shown in Fig. 4B, mutagenesis of any residue in PafA that was predicted to coordinate Mg2+, ATP and glutamate abolished pupylation.
An alignment of PafA, Dop and YbdK shows that residues predicted to be critical for catalysis are conserved in all three proteins (Fig. 4A, in green). Therefore, several of the mutations made for PafA were also generated at equivalent positions in Dop. In β1 we mutated Glu8 and Glu10; in β2 Asp95; in β3 Glu100; and in β5 Arg222. None of the recombinant dop point mutants was able to restore pupylation in the Mtb dop strain (Fig. 4C). These results suggest that the coordination of metal ions and ATP are also crucial for Dop deamidase activity in vivo, despite a lack of a need for ATP hydrolysis (Striebel et al., 2009). We also mutated an arginine (Arg206) that is conserved in a helix at the N-terminal end of β5. Interestingly, Dop(R206A) could not be detected in the dop mutant, suggesting that this mutation resulted in the synthesis of an unstable protein (Fig. 4C). Thus similar to PafA, amino acids that are needed for catalytic activity in the GS fold are also critical for Dop activity in Mtb. Taken together these data strongly support the hypothesis that PafA and Dop are members of the GS-fold superfamily.
Amino acids unique to PafA and Dop are critical for function
In addition to targeting residues conserved among GS-fold members we replaced residues that are conserved only in PafA and Dop. We replaced two residues present in the sequence GARLY(L/V)D in β2, which is unique to and highly conserved in all PafA/Dop proteins (Fig. 4A, in yellow). Replacement of PafA Arg53 or Tyr55 with amino acids conserved in E. coli YbdK failed to facilitate pupylation in a pafA mutant (Fig. 4D). Thus these residues may be important for interacting with pupylation-specific intermediates.
Another strand with a distinct difference between PafA/Dop and GS-fold proteins is in β6. A Q/HxxxxD motif, which corresponds to the ExRxxD motif in the β6 strand of GS-fold members, was proposed to be critical for interacting with phosphorylated glutamate and metal ion in the active site (Iyer et al., 2008). Although it would seem that residues involved in glutamate and metal ion coordination would be conserved between PafA/Dop and GS-fold proteins, replacement of the first polar residue in this motif (His207 in PafA) to glutamate, which is the equivalent residue in GS-fold proteins, abolished PafA activity (Fig. 4D). Thus, it is not clear if His207 (or His291 in Dop) functions to coordinate a metal ion or phosphorylated glutamate. Like Arg53 and Tyr55, this residue may interact with pupylation-specific intermediates.
The last site-directed mutation was introduced at His96 in Dop, which is conserved in all Dop homologues but is a valine (V, Val) at the corresponding position in all PafA homologues. Replacement of Dop His96 with valine abolished Dop activity in vivo (Fig. 4E). Thus, His96 might play a specific role in Dop function.
The similarity of PafA and Dop to GS-fold proteins is only identified at the N-terminal halves of these proteins. The C-terminal domains of PafA and Dop are not similar to any protein of known function. Interestingly, of two independent pafA transposon insertion mutants, pafA282 and pafA420 (Table S1), the pafA420 mutant appeared to produce full length PafA (452 residues) but was defective for pupylation (Fig. 4F). Closer inspection revealed that the transposon insertion resulted in the production of a chimeric protein with the first 420 residues of PafA fused to 44 amino acids encoded by the transposon, resulting in a stable, nonfunctional protein of similar size to PafA (Fig. 4F). To further understand the role of the C-terminus of PafA, we designed a plasmid encoding a truncated PafA (PafA414) to try to complement the pafA282 null mutant. Consistent with the pafA420 allele, expression of pafA414 was unable to complement the pafA null mutation (Fig. 4F). Thus, our results suggest that, although the N-terminal half of PafA has the presumed catalytic domain, the C-terminus is also critical for pupylation in vivo.
Based on our mutagenesis data and structural modeling, we propose that the N-terminal domains of Dop and PafA adopt a GS fold that binds the C-terminus of Pup, Mg2+ and ATP (Fig. 5). In GS/γ-GCS proteins, glutamate is positioned such that the γ-carboxylate is phosphorylated by ATP. The phosphorylated intermediate is then primed for attack by ammonia (for GS) or an amino acid like cysteine (for γ-GCS). By analogy, it had been proposed that the PafA active site coordinates the C-terminal glutamate of Pup for phosphorylation by ATP followed by ligation to a substrate lysine (Iyer et al., 2008). Residues predicted to coordinate ATP, Mg2+ and glutamate in GS-fold proteins were indeed critical for PafA function. In contrast to PafA, we still do not understand how Dop, which is highly similar to PafA, deamidates Pup. The reaction may proceed through a Dop-Pup covalent intermediate, however, the model does not suggest any residues that could fulfill this function. Interestingly, Dop has a conserved histidine (His96) that is not found in PafA homologues, which might suggest that it, along with other residues, could be involved in the deamidation.
Fig. 5.
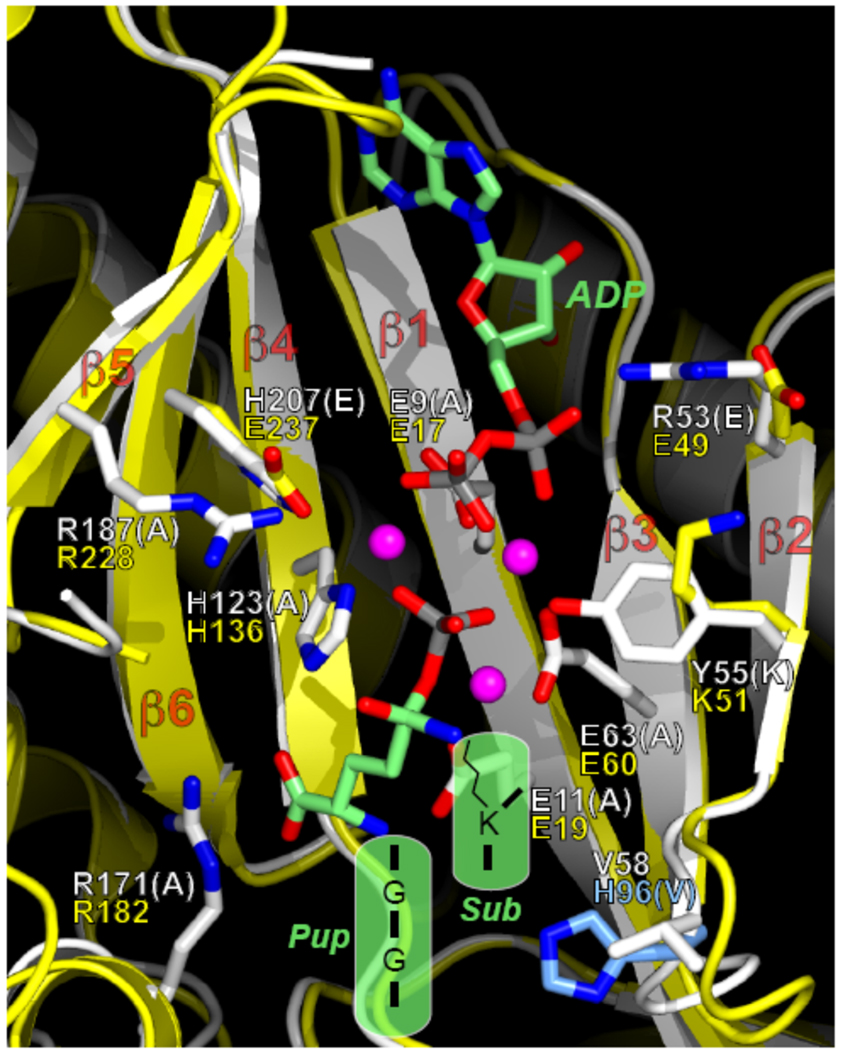
Proposed model of the PafA/Dop active site. Superimposition of the structural model of PafA with the template structure of E. coli YbdK (PDB code 1R8G) (Lehmann et al., 2004). Large insertions or deletions in PafA relative to YbdK were truncated due to the model’s poor reliability in these regions. The secondary-structure elements and side-chain carbon atoms of PafA are coloured white, and those for YbdK are coloured yellow. Select side chains for PafA and YbdK are shown in stick representation. When the residues differ between PafA and YbdK, both side chains are shown; otherwise, only the side chain in the PafA model is shown. The white residue labels (one-letter code) are for PafA, and the yellow ones are for YbdK. All of the PafA residues shown are conserved in Dop, except for Val58, which is a His (His96, coloured cyan). The residue type in parentheses indicates the residue to which WT PafA (or Dop) was mutated for functional tests. The transition-state analogue, ADP, and Mg2+ ions from the structure of E. coli γ-glutamyl-cysteine synthetase (Hibi et al., 2004) (PDB code 1VA6) were superimposed on the structure and are shown in stick or sphere (Mg2+ ions) representation. Carbon atoms of ADP and the analogue are coloured green, phosphorus atoms are coloured gray, and the Mg2+ ions are coloured magenta. For all molecules, oxygen atoms are coloured red and nitrogen atoms are coloured blue. Depicted in the semi-transparent green shapes are the donor (Pup) and the lysine-containing substrate (“Sub”) in the PafA-mediated ligation reaction. The C-terminal sequence of Pup is GGE upon deamidation by Dop (from GGQ). In the figure, the terminal glutamate is contained “within” the transition-state analogue (Arg171 of PafA is salt-bridged to the α-carboxylate group of the glutamate). For depiction purposes, some of the atoms in the transition-state analogue were switched to the appropriate atoms in the putative transition state, e.g., the tetrahedral sulfur atom was changed to a carbon atom, and the phosphate-bridging nitrogen atom was changed to an oxygen atom.
Discussion
In this study we show that Dop is a critical component of the Pup-proteasome system in Mtb. Importantly, Dop, like PafA, Mpa and the 20S proteasome, is required for resistance to RNI and virulence in mice. In addition, using site-directed mutagenesis, in vivo analysis and structural modeling, our data also strongly support the hypothesis that PafA and Dop belong to the GS-fold superfamily (Iyer et al., 2008).
We also tested the importance of the di-glycine motif ubiquitously conserved in Ubls and Pup. We found that replacement of either glycine with alanine did not abolish pupylation, but significantly reduced it (Fig. 3). Importantly, we demonstrated that the terminal amino acid must be glutamate or glutamine in order to conjugate to proteins in vivo. That neither PupGGN nor PupGGD could pupylate proteins (Fig. 3) and that PupGGN was not deamidated despite tight binding to Dop, strongly suggested that the length of the terminal side chain is critical for deamidation and ligation. We propose that the di-glycine motif in Pup provides either flexibility or minimal steric hindrance for the C-terminal glutamine/glutamate to interact with the active site of Dop and/or PafA. This is supported by the observation that the di-glycine motif is important for binding to Dop. Interestingly, although Dop binding was reduced in the absence of the di-glycine motif, it did not appear that deamidation was affected under the conditions tested, and low-level pupylation with PupAAQ and PupAAE looked equivalent to each other (Fig. 3). It is possible that Dop binding may be required for the transfer of Pup to PafA, thus poor Pup transfer could result in inefficient pupylation. Alternatively, the di-glycine motif may be required for PafA binding, which is presumably required for Pup attachment to substrates.
Our mutagenesis analysis supports earlier proposals that Dop and PafA appear to have active sites at their N-terminal halves that resemble those in the GS fold, in particular, the E. coli γ-GCS of unknown function, YbdK. Our data support a previous hypothesis that PafA phosphorylates the γ-carboxylate of the terminal Glu in deamidated Pup to prime it for attack by the amino group of a proteasome substrate lysine (Iyer et al., 2008). We also demonstrated that residues that are conserved in all PafA and Dop homologues, but not in other members of the GS-fold superfamily, were essential for PafA activity (Fig. 4D), suggesting that these residues are specific to PafA (and Dop) function. These residues could participate in substrate interactions that vary from YbdK, given that different ligation substrates are involved in the reaction (i.e. glutamate and cysteine for YbdK, and PupGGE and a substrate lysine for PafA).
Despite the similarities to proteins of known structure, it remains to be determined how PafA mediates phosphorylation of Pup, or how PafA coordinates the nucleophilic attack of the phosphorylated γ-carboxylate by the ε-amino group of a substrate lysine. Interestingly, Vibrio cholerae multifunctional-autoprocessing repeats-in-toxin (MARTX) was indentified to cross-link actin monomers in a mechanism predicted to be similar if not identical to PafA-mediated pupylation. MARTX is a large protein (over 2000 amino acids) with a domain predicted to have a GS-fold that crosslinks Lys50 with Glu270 on two actin monomers (Geissler et al., 2009). As for PafA, it is not known how MARTX catalyzes this reaction.
The enzymology of Dop deamidase activity is far from clear. Site-directed mutagenesis revealed that Dop shares the same GS-fold active-site residues as PafA, however, unlike PafA and GS/γ-GCS proteins, Dop does not require ATP hydrolysis for function (Striebel et al., 2009). It is interesting that two proteins with very different enzymatic activities (deamidation v. ligation) have such similar active sites. However, our results revealed a potentially critical difference between the Dop and PafA families: a histidine residue adjacent to β2 is unique to Dop homologues. Replacement of this histidine with valine, which is conserved in all PafA homologues, completely abrogated Dop activity (Fig. 4E). Perhaps this histidine, along with other residues, participates in the deamidation of glutamine.
Although it appears that the active sites of PafA and Dop are at the N-terminal domains, we cannot rule out the possibility that important catalytic residues are present at the C-terminal domains. The C-terminal halves of PafA and Dop lack homology to proteins of known function, and could not be modeled based on an existing structure. Removal of residues from the C-terminus of PafA (Fig. 4F) or Dop (Fig. 1B) eliminated pupylation or destabilized the protein, thus it is possible that amino acids in this uncharacterized domain are important for catalysis. It seems more likely that the role of the C-terminal domains is to interact with substrates or additional co-factors required for pupylation or proteasomal degradation. In particular, because Mtb appears to have a single Pup ligase to mediate all pupylation, perhaps other proteins interact with the C-terminus of PafA to mediate the specificity of how substrates are chosen for modification.
Dop is present in all bacteria that have pup (Darwin, 2009), including species that are predicted to encode PupGGE rather than PupGGQ. If the only role of Dop were to deamidate Pup, one would predict that PupGGE could bypass the need for Dop in Mtb. However, when PupGGE was produced in an Mtb dop mutant, little pupylation was observed, and free PupGGE was undetectable (Fig. 2). Treatment with a proteasome protease inhibitor of the dop mutant Mtb producing PupGGE restored a robust pupylome but still no free PupGGE could be detected; this suggested that although pupylated proteins could be rescued from proteasomal degradation PupGGE was not released. Thus we propose that Dop is involved in the direct or indirect “de-pupylation” of substrates to recycle Pup and maintain WT steady-state levels of Pup and pupylated proteins in Mtb.
In contrast to what was observed in Mtb, expression of pupGGE in an Msm dop mutant restored pupylation to WT levels (Fig. 2C) (Imkamp et al., 2009). Although we do not understand why this difference occurs, one possibility is that PupGGE levels in Msm are sufficiently high enough to maintain a WT pupylome whereas this is not the case for Mtb. It may be necessary to produce less PupGGE in Msm or examine different growth conditions to determine if Dop affects PupGGE levels.
Taken together, we demonstrated that PafA and Dop are candidate members of the GS-fold superfamily, and that C-terminal residues of Pup are critical for interacting with Dop and covalently attaching to proteins. Our results also suggest Dop has functions beyond Pup deamidation. Importantly, disruption of dop or pafA in Mtb results in severe attenuation in an animal model of infection (Fig. 1) (Darwin et al., 2003). Because Dop and PafA do not significantly resemble any mammalian enzymes to date, both may prove to be ideal targets for future anti-tuberculosis therapies.
Experimental Procedures
Bacterial strains, plasmids and culture conditions
Bacterial strains and plasmids used in this study are listed in Table S1. E. coli strains were grown in Luria-Bertani (LB) broth (Difco) or LB agar at 37°C. E. coli strains were chemically transformed as previously described (Sambrook et al., 1989). Mycobacteria were transformed as described elsewhere (Hatfull & W.R. Jacobs, 2000). Msm strains were grown in Middlebrook 7H9 broth (Difco) supplemented with 0.2% glycerol and 0.05% Tween-80. Cultures were grown at 37°C with aeration on an orbital shaker or roller drum. Mtb cultures were grown in Middlebrook 7H9 broth supplemented with 0.5% bovine serum albumin (BSA), 0.2% dextrose and 0.085% sodium chloride (ADN). Mtb cultures were grown without aeration in 25 or 75 cm2 vented flasks (Corning) at 37°C. For solid media, Middlebrook 7H11 supplemented with oleic acid, dextrose and catalase (OADC; BBL) was used. Antibiotics were used at the following concentrations: 150 µg/ml (E. coli) or 50 µg/ml (Msm/Mtb) hygromycin; 100 µg/ml (E. coli) or 25 µg/ml (Mtb) kanamycin.
For growth curve analysis, WT, dop and dop-complemented Mtb CDC1551 strains were grown without aeration and the OD580 of the cultures was measured daily for 10 days. Nitrite sensitivity assays were performed exactly as previously described (Darwin et al., 2003).
As previously reported, WT Mtb Dop was incorrectly annotated to have 49 extra residues at the amino (N)-terminus (Imkamp et al., 2009). We confirmed that Dop migrates at approximately 55 kD (data not shown) and a new annotation for Mtb dop (Rv2112c/MT2172) has been submitted (GenBank accession number HM004510).
dop and pafA414, were amplified from pET24b(+)-dop-His6 and pET24b(+)-pafA-His6 using primer pair XbaI-pafDf1 and HindIII-pafD1 and primer pair pMV306F and 414PafA-ClaIr, respectively. The resulting fragments were ligated into pMV306 to generate pMV-dop and pMV-pafA414-His6, respectively. The pafA and dop mutations were constructed by sewing overlap extension PCR (Horton et al., 1989). The resulting products were cloned into pMV306 as a KpnI-ClaI (pafA) or HindIII-XbaI (dop) fragments. For Pup C-terminal mutation analysis we introduced mutations into pMN-NL-His6-pup, which was constructed by PCR amplifying His6-pup from pET24b(+)-His6-pup using His-prcC-Nde_F2 and HIII_prcC-p300_R. The fragment obtained was ligated into pUV15+ using the NdeI and HindIII sites, and then subcloned into pMN402 to reduce expression. The C-terminal mutants KGG, KG, KGAQ, and KGGA were cloned in a similar manner, but using a reverse primer specific for each mutation with an EcoRV site rather than HindIII site. The remaining C-terminal mutations were constructed by PCR amplifying the genes and directly cloning them into pMN402 using the same restriction sites. pMN-NL-pup-His6 was generated by cloning pup-his6 into the NdeI and HindIII sites of pMN402. All plasmids were sequenced (GENEWIZ, Inc.) before transformation of Mtb or Msm.
Mouse infections
Mouse infections were performed essentially as described previously (Darwin et al., 2003). Briefly, 6–8 week old C57BL6/J female mice (Jackson Laboratories) were infected using a Glas-Col Inhalation Exposure System (Terre Haute, IN). Mtb was nebulized to administer ~200–400 CFU/mouse. Mice were humanely sacrificed using a protocol approved by the Institutional Animal Care and Use Committee. Bacteria were recovered from homogenized lungs and spleens and plated onto 7H11-OADC plates. Formalin fixed lungs were sectioned and stained at the NYU Research Histopathology Core.
PafA/Dop model
The program Modeller (Sali & Blundell, 1993) as implemented at http://toolkit.tuebingen.mpg.de/ was used to generate a three-dimensional structural model of PafA, residues 1–308. The template structure was that of the E. coli protein YbdK (Lehmann et al., 2004) (PDB code 1R8G). A sequence alignment of PafA with YbdK, which served as the input to Modeller, was based largely on the alignment of Iyer et al. (Iyer et al., 2008).
Isolation of pupylated substrates using Pup C-terminal mutants
WT Msm strain mc2155 was transformed with the plasmids encoding the Pup C-terminus mutants. Cultures of 30 ml were grown to OD580 of ~2.0 and the cells were collected by centrifugation and resuspended in 1 ml of Ni-NTA lysis buffer (The QIAexpressionist, Qiagen). The resuspended cells were transferred to bead beating tubes each with 250 µl of zirconia silica beads (BioSpec Products). Cells were lysed by bead beating four times for 1 min each in a BioSpec Mini Bead Beater. Samples were clarified by microcentrifugation at top speed for 90 s and then soluble proteins were passed through a 0.2 μ filter. The soluble lysates were quantified using a NanoDrop spectrophotometer and 10 mg of total proteins were incubated with 35 µl of Ni-NTA agarose for 2 h at 4°C with agitation. The agarose was washed four times with 500 µl Ni-NTA wash buffer. Proteins were eluted with 60 µl of Ni-NTA elution buffer.
PupGGQ and PupGGE derivatives in Mtb and Msm
The pMN-his6-pup-KGGQ and pMN-his6-pup-KGGE plasmids were electroporated into Mtb and Msm. For Msm, total cell lysates from equivalent cell numbers (optical density A580 = 1.0–1.5) were harvested by centrifugation and washed once with phosphate-buffered saline (PBS) (Cellgro) with 0.05% Tween-80. The cells were collected again, resuspended in TE buffer (10 mM Tris-Cl, pH 7.5, 1 mM EDTA) and lysed by bead beating.
For Mtb, total cell lysates were essentially examined as described for Msm. In addition, the recombinant His6-pupylomes were purified from Mtb to better examine the extent of pupylation. 30 ml cultures were grown to an OD A580 of ~2.0 and the cells were collected by centrifugation and resuspended in 1 ml of Ni-NTA lysis buffer. The resuspended cells were lysed by bead beating four times for 1 min each. Samples were clarified by centrifugation at 13,000 rpm for 90 sec and passed through a 0.2 μ filter to sterilize. The lysates were quantified using a NanoDrop spectrophotometer and 5.2 mg of total proteins were incubated with 35 µl of Ni-NTA agarose for 2 h at 4°C with agitation The agarose was washed four times with 500 µl Ni-NTA wash buffer. Proteins were eluted with 60 µl of Ni-NTA elution buffer.
WT and dop Mtb strains expressing pupGGE were treated with the proteasome inhibitor epoxomicin similarly as previously described (Pearce et al., 2006). Briefly, strains were grown in 7H9-ADN to an OD580 of ~0.4. Epoxomicin (Boston Biochem) in DMSO was then added to a final concentration of 50 µM. Control cultures either had an equal volume of DMSO or nothing added. The cultures were incubated for two days at 37°C, and total cell lysates were examined as described above.
Immunoblot analysis
For all immunoblots, cell lystes or purified proteins were separated by either 10% or 12% SDS-PAGE, transferred to nitrocellulose, incubated with either rabbit polyclonal antibodies to Dop, PafA or Pup; or mouse monoclonal antibodies to Pup. All primary antibodies were produced by Covance (Denver, PA), and visualized using horseradish peroxidase (HRP) coupled anti-rabbit or anti-mouse secondary antibodies (Pierce). Detection of HRP was performed using either SuperSignal West Pico or West Femto Chemiluminescent Substrate (ThermoScientific).
Supplementary Material
Acknowledgements
We are grateful to Kristin Burns and Kaye Horstman for critical review of this manuscript. We thank Frank Imkamp, Eilika Weber-Ban and Peter Sander for the Msm WT and dop strains used in this work. This work was supported by NIH grants HL92774 awarded to K.H.D. and AI 30036, 37856, and 36973 awarded to W.R.B. K.H.D is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease.
References
- Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., 3rd Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Maira F, Darwin KH. The Mycobacterium tuberculosis proteasome: more than just a barrel-shaped protease. Microbes Infect. 2009;11:1150–1155. doi: 10.1016/j.micinf.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH. Prokaryotic Ubiquitin-Like Protein, Proteasomes, and Pathogenesis. Nat. Rev. Microbiol. 2009;7:485–491. doi: 10.1038/nrmicro2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Weich N, Gutierrez-Ramos J-C, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- Festa RA, McAllister F, Pearce MJ, Mintseris J, Burns KE, Gygi SP, Darwin KH. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis. PLoS One. 2010;5:e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa RA, Pearce MJ, Darwin KH. Characterization of the proteasome accessory factor (paf) operon in Mycobacterium tuberculosis. J Bacteriol. 2007;189:3044–3050. doi: 10.1128/JB.01597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra D, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Bonebrake A, Sheahan KL, Walker ME, Satchell KJ. Genetic determination of essential residues of the Vibrio cholerae actin cross-linking domain reveals functional similarity with glutamine synthetases. Mol Microbiol. 2009;73:858–868. doi: 10.1111/j.1365-2958.2009.06810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Jacobs JWR. Molecular Genetics of Mycobacteria. Washington, DC: ASM Press; 2000. [Google Scholar]
- Hibi T, Nii H, Nakatsu T, Kimura A, Kato H, Hiratake J, Oda J. Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis. Proc Natl Acad Sci U S A. 2004;101:15052–15057. doi: 10.1073/pnas.0403277101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Wang L, Lammertyn E, Geukens N, Van Mellaert L, Li Y, Anne J. Inactivation of the 20S proteasome in Streptomyces lividans and its influence on the production of heterologous proteins. Microbiology. 2005;151:3137–3145. doi: 10.1099/mic.0.28034-0. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Imkamp F, Rosenberger T, Striebel F, Keller PM, Amstutz B, Sander P, Weber-Ban E. Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol Microbiol. 2009;75:744–754. doi: 10.1111/j.1365-2958.2009.07013.x. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Knipfer N, Shrader TE. Inactivation of the 20S proteasome in Mycobacterium smegmatis. Mol Microbiol. 1997;25:375–383. doi: 10.1046/j.1365-2958.1997.4721837.x. [DOI] [PubMed] [Google Scholar]
- Lamichhane G, Raghunand TR, Morrison NE, Woolwine SC, Tyagi S, Kandavelou K, Bishai WR. Deletion of a Mycobacterium tuberculosis proteasomal ATPase homologue gene produces a slow-growing strain that persists in host tissues. J Infect Dis. 2006;194:1233–1240. doi: 10.1086/508288. [DOI] [PubMed] [Google Scholar]
- Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, Broman KW, Bishai WR. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2003;100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann C, Doseeva V, Pullalarevu S, Krajewski W, Howard A, Herzberg O. YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays. Proteins. 2004;56:376–383. doi: 10.1002/prot.20103. [DOI] [PubMed] [Google Scholar]
- Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH. Identification of substrates of the Mycobacterium tuberculosis proteasome. EMBO J. 2006;25:5423–5432. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Maniatis T, Fritsch E. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- Sutter M, Damberger FF, Imkamp F, Allain FH, Weber-Ban E. Prokaryotic Ubiquitin-like Protein (Pup) Is Coupled to Substrates via the Side Chain of Its C-Terminal Glutamate. J Am Chem Soc. 2010 doi: 10.1021/ja910546x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.