Abstract
Implicit learning, the non-conscious acquisition of sequential and spatial environmental regularities, underlies skills such as language, social intuition, or detecting a target in a complex scene. We examined relationships between a variation of the dopamine transporter (DAT1) gene (SLC6A3), which influences dopamine transporter expression in the striatum, and two forms of implicit learning that differ in the regularity to be learned and in striatal involvement. Participants, grouped as 9-repeat carriers or 10/10 homozygotes, completed the Triplets Learning Task (TLT) and the Spatial Contextual Cueing Task (SCCT). The TLT assesses sequence learning, recruiting the striatal system, particularly as training continues. In contrast, the SCCT assesses spatial context learning, recruiting medial temporal brain networks. For both tasks, participants demonstrated learning in faster and/or more accurate responses to repeating patterns or spatial arrays. As predicted, TLT learning was greater for the 9-repeat carriers than the 10/10 group (despite equal overall accuracy and response speed) whereas there were no significant group differences in SCCT. Thus, presence of the DAT1 9-repeat allele was beneficial only for implicit sequence learning, indicating the influence of DAT1 genotype on one form of implicit learning and supporting evidence that implicit learning of sequential dependencies and spatial layouts recruit different neural systems.
Keywords: Dopamine, Implicit Learning, Genetics
1. Introduction
Contemporary cognitive neuroscience increasingly incorporates genetics data to understand cognition [1]. However, our current understanding of genetic influences is limited because most research has focused on conscious, deliberate cognitive control [e.g., 2], while the genetics of non-conscious, implicit phenomena has been understudied. Implicit learning refers to the acquisition of information about environmental regularities without intending to learn or becoming aware of what has been learned [3]. Here, we investigated the effects of a polymorphism in the gene (SLC6A3) coding for the dopamine transporter (DAT1) on two different forms of implicit learning, namely sequence learning and spatial context learning.
Implicit sequence learning involves learning sequential dependencies among events, which is necessary for skills such as language [4] and social intuition [5]. To investigate sequence learning, we used the Triplets Learning Task [TLT; 6]. On each trial, participants encounter three sequentially presented stimuli, two red cues followed by a green target, each appearing in one of four spatial locations. Participants watch the red cues but respond only to the third, green target. Participants are unaware that certain series of events or “triplets” occur with greater frequency (high probability, HP) than others (low probability, LP). Nonetheless, with practice, they reveal sequence learning in the form of greater improvement in speed and/or accuracy for HP than LP triplets. Studies of Parkinson’s and Huntington’s disease patients with dopamine depletion and striatal pathology, respectively, have shown sequence learning deficits [e.g., 7, 8], indicating the involvement of the striatum and striatal dopamine system in this form of learning. Moreover, neuroimaging studies of implicit sequence learning in healthy young adults reveal striatal involvement that increases as practice continues [9–12].
Distinct from sequence learning, implicit spatial context learning refers to learning of regularities in spatial layouts, such as how context predicts where a target is likely to occur. For example, spatial context learning can help people anticipate where the stoplight is likely to be at an intersection. The Spatial Contextual Cueing Task [SCCT; 13] measures this type of learning. Participants search for a target amongst distractors in displays with spatial configurations that either repeat across trials or are novel. Repeated configurations provide contextual guidance for locating the target. Implicit spatial context learning is revealed by faster and/or more accurate responding to repeated vs. novel configurations without explicit awareness. Functional imaging of healthy subjects has shown medial temporal lobe (MTL) involvement during spatial context learning in the SCCT [14, 15]. In addition, impaired contextual learning has been observed in amnesics with MTL damage [16, 17].
Sensitive recognition tasks and post-experimental interviews with participants have reliably demonstrated that both the TLT and SCCT measure implicit learning [e.g., 6, 13, 16, 18]. Even after being told that regularities were present, participants cannot accurately describe their nature, nor can they discriminate between triplets that occur with HP vs. LP in the TLT and/or arrays that repeated vs. those that were novel in the SCCT.
Previous work has demonstrated double dissociations between implicit sequence learning and implicit spatial context learning by differentiating the brain regions involved. Specifically, reduced sequence learning but not spatial contextual learning was revealed in healthy older compared to younger adults [19], consistent with findings that healthy aging is associated with greater volume declines in the striatum than in the MTL [20]. This result was supported by Negash et al. [21] who found that spatial contextual learning, but not sequence learning, was reduced in older adults with mild cognitive impairment, a condition associated with MTL pathology, compared to age-matched controls. In addition, Negash showed that healthy older individuals carrying the ApoE-e4 allele, which is also associated with MTL atrophy [22], showed reduced spatial contextual learning, but not sequence learning, compared to non-e4 carriers. Thus, implicit sequence learning and implicit spatial context learning are dissociable and appear to be differentially influenced by individuals’ genetic profiles.
The present study examined relationships between these two forms of implicit learning and a polymorphism in DAT1, which influences DAT expression levels [23]. Although DAT1 is present throughout the brain [23, 24], it is expressed most abundantly in the striatum and plays a key role in striatal dopamine transmission, in that it is the primary means of clearing extracellular dopamine from the synaptic cleft [25], thereby regulating synaptic dopamine concentrations [26]. DAT1 displays a polymorphic 40-base pair variable number of tandem repeats in the 3′ untranslated region. The most common variations, the 9- and 10-repeat alleles, result in individual differences in DAT availability in the brain [27]. Most studies have found that the 10-repeat allele is associated with more DAT availability [28–32]. It has been suggested that individuals who carry two copies of this allele (i.e. 10/10 homozygotes) have higher DAT density and therefore less dopamine in the synapse than 9-repeat carriers [33]. However, other studies have reported greater DAT density in 9-repeat carriers [34–36], and no DAT density differences between genotypes [37, 38].
If, indeed, 10/10 homozygotes have more DAT expression and less dopamine, we would expect to see decreased learning in tasks facilitated by higher levels of striatal dopamine. Thus, we sought to determine the functional influence of DAT1 genotype on striatal-based implicit sequence learning vs. MTL-based implicit spatial context learning. Based on evidence that DAT1 expression is higher in the striatum than the MTL [23], we expected that DAT1 genotype would influence sequence learning but not spatial context learning. Specifically, we hypothesized that 9-repeat carriers would show greater sequence learning than 10/10 homozygotes in the later stages of sequence learning, given evidence that striatal involvement increases with training [e.g., 11]. In contrast, we expected that DAT1 genotype would be unrelated to spatial contextual learning, which relies on the integrity of the MTL.
2. Methods
2.1. Participants
Participants were 37 Georgetown University students who received either monetary compensation or course credit for their participation. Participants were grouped according to their DAT1 genotype. Participants in the 9-repeat/9-repeat homozygous group (n=7) and the 9-repeat/10-repeat heterozygous group (n=11) were combined, henceforth called 9-repeat carriers, and compared to 10/10 homozygotes (n=19). The genotype groups did not differ in age (9-repeat carriers: M = 20.2 ± 1.2; 10/10 homozygotes: M = 20.2 ± 1.2), years of education (9-repeat carriers: M = 14.1 ± 1.0; 10/10 homozygotes: M = 14.4 ± 1.0) or gender (9-repeat carriers: 15 females; 10/10 homozygotes: 12 females) (p’s > .17). Based on self-report, participants were without psychiatric disorder (e.g., ADHD) and did not use drugs known to influence cognitive functioning (e.g., dopaminergic medications). The Georgetown University Institutional Review Board approved all experimental procedures, and all participants gave informed consent.
2.2. Genotyping for DAT1
DNA was extracted from Oragene saliva kits (DNA Genotek Inc., Ottawa, Ontario, Canada). The 40 base pair VNTR polymorphism in the 3′ UTR of DAT1 was genotyped by PCR as previously described [39] using the following primers; Forward: 5′-TGTGGTGTAGGGAACGGCCTGAG-3′ Reverse: 5′-CTTCCTGGAGGTCACGGCTCAAGG-3′. PCR was performed using the AccuprimeTM Taq DNA polymerase system (Invitrogen) with the following PCR program: 94°C for 2 min, followed by 35 cycles of 94°C for 30 sec, 60°C for 30 sec, and 68°C for 1 min. The PCR products were then run out on a 2% agarose gel stained with ethidium bromide. A 100 bp DNA ladder was then used to identify the various repeat alleles by size: 7-repeat (360bp), 8-repeat (400bp), 9-repeat (440bp), 10-repeat (480bp), and 11-repeat (520bp).
2.3. Triplets Learning Task (TLT)
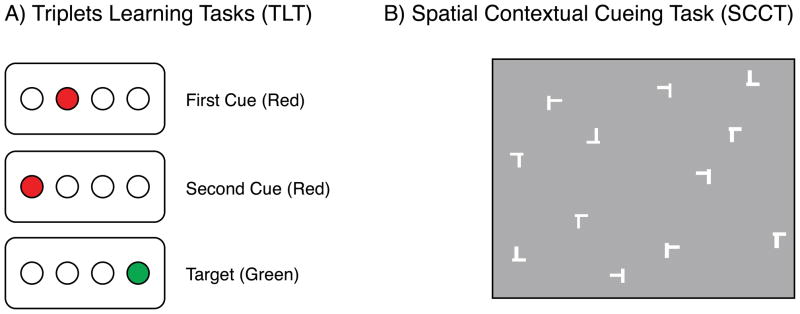
On a computer screen, participants viewed four open circles that filled in either red or green in discrete, sequentially ordered, three-event trials or “triplets” (see Figure 1a). For description purposes below, the four stimulus locations are referred to as 1, 2, 3, and 4, with 1 as the leftmost position and 4 as the rightmost position, though these numbers never appeared on the screen. Participants were asked to observe the first two consecutive red events and then to respond only to the third, green target event location, by pressing one of four corresponding buttons with their dominant hand. Red events were displayed for 120 ms each, and the green target remained in view until participants made a correct response to its location, with 650 ms separating the correct response and the first cue on the following trial [6].
Figure 1.
Sample displays from the Triplets Learning Task (A) and Spatial Contextual Cueing Task (B).
A randomly chosen set of 16 triplets occurred with high probability (HP) and the remaining possible 32 triplets occurred with low probability (LP). Repetitions (e.g. 111, 222) and trills (e.g. 141, 232) were not presented because studies have shown that they reflect pre-existing response tendencies, in addition to learning [40–42]. The frequency of HP to LP triplets was approximately nine-to-one. Each participant received the same set of HP and LP triplets, but their order of presentation was randomized within each block. Participants were not informed of any regularities; their only instruction was to respond as quickly and accurately as possible. Participants completed three testing sessions, each consisting of 5 blocks of 50 trials. During breaks after each block, participants viewed their mean reaction time and accuracy scores. These scores were presented in an attempt to maintain performance around 92% accuracy. Depending on their accuracy in the preceding block, participants were instructed in one of three ways via a message on the screen: “focus more on speed,” “focus more on accuracy” or “speed and accuracy are about right.”
2.4. Spatial Contextual Cueing Task (SCCT)
Participants viewed 12-item stimulus arrays that each contained a single target, a horizontal letter T (rotated 90°) and 11 distractors, which were rotated letter L’s (0°, 90°, 180°, or 270°) that were made to look more like the target by offsetting the point of intersection by 3 pixels [16; Expt. 2] (see Figure 1b). These visual arrays were presented in white against a gray background and were randomly generated by placing the 12 items into cells of an invisible grid (6 rows × 8 columns), with items repositioned by 63 pixels along each axis to avoid colinearity. Target location was balanced for distance from the screen’s center and screen half (left/right); no target appeared in the four center or corner cells.
On each trial, a white fixation dot appeared for 1 second, followed by a visual array that remained on the screen for up to 10 seconds until a response was made. Participants were asked to locate the target and to respond to its orientation as quickly and as accurately as possible by pressing the “z” (if the tail of the T was facing left) or “/” (if the tail of the T was facing right) on the keyboard, making no more than 1 or 2 errors per block. Once a response was detected, participants received auditory feedback of a high-pitch tone for a correct response or a low-pitch tone for an incorrect response. If no response was made, a low-pitch tone sounded after 10 seconds.
A spatial contextual regularity is embedded in this task, such that half the visual arrays repeated across blocks. In these repeated arrays, the distractor array predicted the location of the target, but not its orientation (i.e., the correct response). In other words, regularities in spatial arrays provided contextual guidance for the location of the target, but not whether the target was facing left or right. The remaining arrays for each block were randomly generated for each trial, creating a novel configuration that only appeared once throughout the experiment. Each participant received the same set of repeated and novel arrays, but their order was randomized within each block. Following 12 practice trials, participants completed 30 blocks of 12 trials each, with 6 repeated and 6 novel trials per block [43]. Participants were encouraged to take short breaks after each block.
2.5. Procedure
Participants first performed the TLT, followed by the SCCT within a single testing session. Including breaks, total testing time was approximately one hour. On a separate testing day, typically the next day but no more than a week later, participants completed two other unrelated implicit learning tasks that are not reported here. At this time, the experiment concluded with an interview as a probe of participants’ declarative knowledge in all the implicit learning tasks. Four increasingly specific questions were asked: (1) Did you notice anything about the tasks you have performed? (2) Did you notice any repeating patterns within the tasks? (3) Did you use any particular strategies? (4) There were, in fact, regularities in the tasks you just completed. Knowing this, could you describe any of these regularities? Both the TLT and SCCT have consistently yielded evidence of implicit learning on sensitive recognition tasks [6, 43]; therefore, we did not include such measures after each task in the current study in order to keep learning implicit in all tasks completed.
3. Results
3.1. Implicit sequence learning (TLT)
Median reaction times (RT) were determined for correct responses for each triplet type for each participant on each block. Overall accuracy was ~93%, so few trials were omitted. These data were then averaged across blocks to obtain a single mean RT for each participant for HP and LP triplets for each of the 3 testing sessions. A similar procedure was used to calculate the mean accuracy for each participant for each triplet type.
To assess potential group differences in implicit sequence learning, Genotype (9-repeat carriers, 10/10 homozygotes) × Triplet Type (HP, LP) × Session (1–3) mixed-design ANOVAs were conducted separately for accuracy and reaction time measures. Genotype varied between-subjects, and Triplet Type and Session varied within-subjects. For accuracy, sequence learning was revealed as a main effect of Triplet Type, F(1, 35) = 8.54, p = .006, reffect = .44, with more accurate responses to HP vs. LP triplets. There was a main effect of Session, F(2, 70) = 4.87, p = .01, reffect = .35, demonstrating that the feedback provided after every block guided participants to overall performance levels of 92% accuracy, as intended. No interactions reached significance (p’s > .26). The lack of a main effect or interaction with Genotype showed that feedback served to successfully match the groups on overall accuracy, which aids interpretation of reaction time data, in that there are no speed-accuracy trade offs.
For reaction time, skill learning can be seen in Figure 2, with overall RT (regardless of Triplet Type) decreasing across sessions, F(2,70) = 35.53 p < .0001, reffect = .71, reflecting overall skill and practice. Sequence learning was seen as a main effect of Triplet Type, F(1, 35) = 98.85, p < .0001, reffect = .86, with faster responding to HP relative to LP triplets. This sequence learning effect (or separation of HP and LP trials) increased with practice, as indicated by the significant Triplet Type × Session interaction, F(2,70) = 3.59, p = .03, reffect = .31. Finally, and most important, there was a Genotype × Triplet Type × Session interaction, F(2,70) = 5.19, p = .008, reffect = .36, indicating that there are group differences between genotypes which vary with practice. No other main effects or interactions reached significance (p’s > .26).
Figure 2.
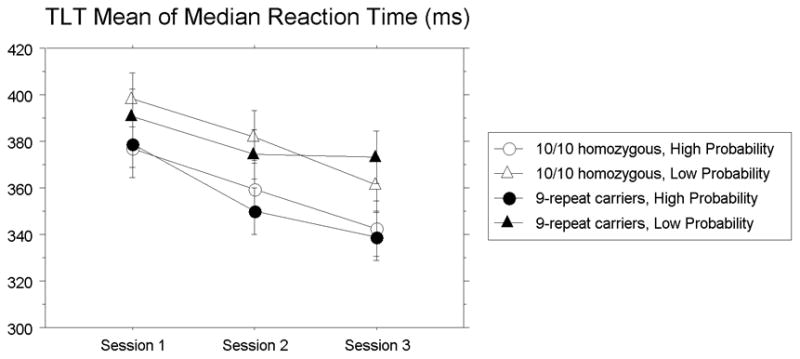
Triplets Learning Task (TLT). Mean of median reaction time (in milliseconds) over sessions for high- and low-probability triplets for 9-repeat carriers and 10/10 homozygotes. Error bars represent the standard error of the mean.
To explore this interaction more fully, we calculated sequence-specific learning scores, i.e., mean difference RT scores (LP – HP), which are shown in Figure 3a. Single sample t-tests indicated that these learning scores were significantly different from 0 for all three sessions (Session 1: t(37) = 4.93, p < .0001, reffect = .41; Session 2: t(37) = 7.99, p < .0001, reffect = .65; Session 3: t(37) = 7.55, p < .0001, reffect = .62), indicating sequence-specific learning. Most importantly, as predicted, when these scores were compared between the genotypes at each session, learning was significantly greater for the 9-repeat carriers than the 10/10 homozygotes in Session 3 only, t(35) = 2. 25, p = .03, reffect = .13.
Figure 3.
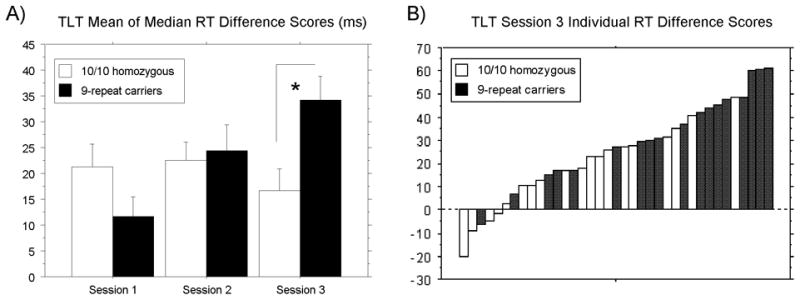
Triplets Learning Task (TLT). A) Mean of median reaction time difference scores (in milliseconds) between triplet types (i.e., low probability minus high probability) across sessions for each genotype. Error bars represent standard error of the mean. B) Mean of median reaction time difference scores (in milliseconds) for individual subjects for Session 3, with subjects ordered by the magnitude of their difference score.
The same effect can be seen in Figure 3b, which illustrates individual RT difference scores from Session 3. Using a median split on these difference scores, we classified any subject with a difference score greater than or equal to 27.4 as being a high learner and any subject with a difference score less than 27.4 as being a low learner (see Figure 3b). Of the 19 high learners, 13 were 9-repeat carriers and 6 were 10/10 homozygotes and of the 18 low learners, 5 were 9-repeat carriers and 13 were 10/10 homozygotes, χ2(2) = 6.11, p=.01. Thus, at the individual level, most 9-repeat carriers were high learners whereas most 10/10 homozygotes were low learners.
3.2. Implicit spatial learning (SCCT)
For each participant, median RT for correct trials and percentages of correct responses were calculated separately for each array type for each block and averaged into three 10-block sessions. Similar to TLT, mean of median RTs (see Figure 4) and mean accuracy were analyzed separately in Genotype (9-repeat carriers, 10/10 homozygotes) × Array (repeated, novel) × Session (1–3) mixed ANOVAs, with Genotype varied between-subjects, and Array and Session varied within-subjects. For accuracy, visual search skill learning was seen as a significant main effect of Session, F(2,70) = 5.72, p = .005, reffect = .38, showing that accuracy improved with practice. No other main effects or interactions reached significance (p’s > .12). Overall, accuracy was high (M = 94.9%, SD = .05).
Figure 4.
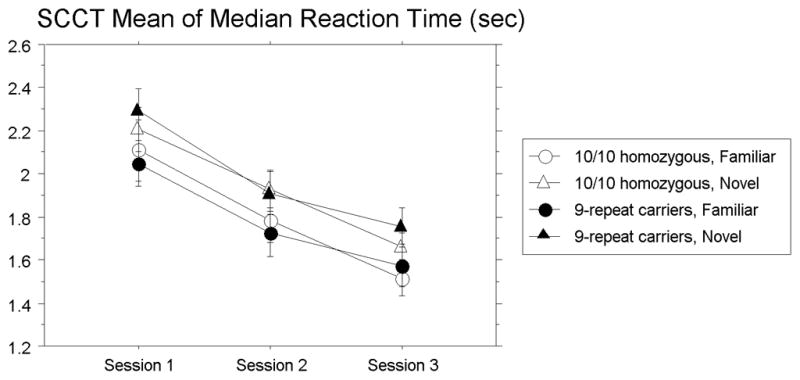
Spatial Contextual Cueing Task (SCCT). Mean of median reaction time (in seconds) over sessions for new and repeated configurations for both 9-repeat carriers and 10/10 homozygotes. Error bars represent the standard error of the mean.
For mean of median RT, shown in Figure 4, visual search skill learning was revealed by a significant main effect of Session, F(2, 70) = 94.01, p < .0001, reffect = .85, demonstrating that responses became faster with practice. Spatial context learning was revealed by a significant main effect of Array, F(1,35) = 18.00, p = .002, reffect = .58; participants responded faster to repeated vs. novel arrays. Finally, as predicted, no main effects or interactions with genotype approached significance, p’s > .35, indicating that contextual learning did not differ for 9-repeat carriers and 10/10 homozygotes.
Because there was no Array × Session interaction, we conducted an ANOVA across blocks (1–10) within the first session only, to ensure that the main effect of Array was reflecting learning. This revealed a significant Array × Block interaction, F(9, 324) = 2.04, p < .05, reffect = .23, with the Array effect significant on Block 10, t(36) = 2.32, p < .05, reffect = .13, but not on Block 1, t(36) = 1.66, p > .05, reffect = .07. No main effects or interactions with Genotype were significant (p’s > .12). Thus, learning occurred within the first session for both groups.
3.3. Implicitness
Comments from the post-experimental interviews were examined for insight into the regularities that may have been detected or the strategies people used. These revealed no evidence of declarative knowledge. No one could accurately describe the regularities from the TLT or the SCCT. In addition, no explicit strategies for these two learning tasks were reported.
4. Discussion
The present study investigated whether DAT1 genotype contributed to individual differences in implicit learning of sequential regularities and spatial contexts in healthy young adults. As predicted, DAT1 was related to sequence learning, with greater learning for the 9-repeat carriers than 10/10 homozygotes over time. In contrast, there were no significant group differences in spatial contextual learning. Because response speed and accuracy were nearly identical between the groups for both learning tasks, overall performance differences cannot account for the observed implicit sequence learning differences, suggesting specificity in how DAT1 contributes to neural and behavior functions. To our knowledge, this is the first study to reveal a relationship between DAT1 and implicit learning, indicating that some individual differences in implicit learning of sequential regularities are influenced by genotype.
Reduced sequence learning in 10/10 homozygotes likely reflects differences in striatal dopamine between DAT1 genotypes. Indeed, our results are consistent with the assumption that 10/10 homozygotes have greater levels of striatal DAT availability when compared to 9-repeat carriers [28–32] and that this higher gene expression leads to decreased levels of synaptic dopamine in that region [33]. Thus, our findings demonstrate the importance of dopamine for implicit sequence learning, even in healthy young adults. Reductions in striatal dopamine have previously been found to impair implicit sequence learning, as revealed by learning deficits in Parkinson’s disease patients [44]. Similarly, children with ADHD, a disorder well-characterized by dopamine dysfunction [45, 46], have shown sequence learning impairments in comparison to healthy controls [47]. Moreover, older adults show reduced implicit sequence learning relative to young adults [6, 48, 49] that may result from age-related decreases in striatal dopamine [50–52]. In fact, when using the same version of the TLT used here, older adults performed similarly to the present study’s young 10/10 homozygotes in that the learning deficit was revealed only in the third training session [53].
If 9-repeat carriers are indeed learning more than 10/10 homozygotes, then they would be predicted to respond faster on HP triplets and slower on LP triplets than 10/10 homozygotes. This is because as people learn the predictive relationships within the triplets, they increasingly anticipate the correct target on trials with HP triplets. In contrast, on trials with LP triplets, people come to anticipate a different target than actually occurs. Thus, to avoid making too many errors (participants were guided to 92% accuracy), participants must first inhibit the expected, incorrect response. The results shown in Figure 2 are consistent with this, but with the group difference carried primarily by the LP trials. This pattern of results may be related to ceiling performance on the HP trials, in that participants were responding around 340 ms by Session 3 (M: 339.46, SE: 7.8), which is faster than what has been reported previously for HP triplets in the TLT [6, 18]. Thus, responses to the LP triplets may provide a more sensitive measure of triplet contingency learning in the present study. This interpretation is also consistent with reports that 9-repeat carriers have increased incidence of response inhibition when compared to 10/10 homozygotes [54, 55].
Though some studies of healthy young adults have revealed significant behavioral differences as a function of DAT1 genotype [56, 57], most work has revealed minimal or nonexistent behavioral differences between genotypes [58, 59, see also 60, for a review]. Here, we demonstrated a significant difference in sequence learning between DAT1 genotype that was not driven by just one or two subjects. This indicates that the TLT may be among the more sensitive measures to reveal behavioral effects of DAT1 genotype, perhaps because the task recruits fewer regions outside the DAT-rich striatum than previously employed tasks.
To our knowledge, only one previous study has examined the relationship between genotype at a polymorphism relevant to the dopamine system and implicit learning. Keri et al. [61] used a Weather Prediction Task that has been characterized by implicit striatal-based processes during early learning and explicit MTL-based processes later on [62]. Results revealed that a polymorphism of the dopamine D3 receptor (DRD3) gene, which is found in higher densities in the ventral striatum relative to the MTL [63], was associated with early, striatal-based learning, but not later, MTL-based learning. These findings, together with the present study, suggest that relationships between implicit learning and dopaminergic genes can provide additional insight into the neurochemical and neuroanatomical mechanisms of implicit learning. For example, our results provide evidence that contextual cueing and sequence learning involve different neural substrates [18, 19, 21, 47]. Because DAT1 has greater expression in the striatum relative to the MTL [23], our findings add to a body of work showing that implicit sequence learning requires striatal brain networks as training progresses, whereas implicit spatial context learning does not, reflecting instead the integrity of the MTL [e.g., 16].
Future research will be needed to overcome some limitations in the current study. First, the sample here is relatively small. Because the rate of false positive results is higher with smaller samples and less power [1], replication with a larger sample is in order.
Second, we included no measures of explicit knowledge of the presented spatial or temporal patterns, other than a brief post-experimental interview. Even so, extensive evidence, using similar participant populations and the same versions of the TLT and the SCCT as those used here, indicates that even when sensitive recognition tests are given, people are unable to discriminate between predictable and unpredictable items, thus showing no explicit knowledge [6, 43]. Further, people do not report adopting conscious, deliberate strategies for stimulus selection [13, 18, 19]. This stands in contrast to other learning tasks, like Weather Prediction [62], in which participants often develop explicit knowledge with practice and use hypothesis-testing strategies that make it difficult to dissociate implicit from explicit learning [64].
Third, we did not counterbalance task order; participants always completed the TLT before the SCCT. As a result, participants may have been more fatigued for the second task. However, this is unlikely to explain our pattern of results. Both implicit learning paradigms were relatively short (~30 minutes each), and thus, training was not significantly longer than typical testing sessions. Moreover, both groups demonstrated strong learning effects in both sequence and spatial context learning that are comparable to previous studies of similar populations [see 6, 43]. Further, it seems unlikely that genotype would interact with fatigue to confound the results presented here.
The present study extends evidence for genetic influences on cognition beyond the domain of explicit processes to implicit forms of learning. Future studies should explore the contributions of multiple dopaminergic genes for possible gene-gene interactions on implicit learning. For example, if there is a dopamine dosage effect, alleles that decrease dopamine availability might combine to further impair implicit sequence learning. Alternatively, there may be specificity in how different dopamine genes influence different forms of implicit learning, depending on the regions of the brain where genes are preferentially expressed and protein expression patterns. Sequence learning may be more sensitive to DAT1 genotype, whereas other forms of learning (e.g., reward-based learning) may be more sensitive to polymorphisms in other genes relevant to dopaminergic function (e.g., DARPP-32, DRD2) [65].
5. Conclusions
In sum, our findings revealed an association between a polymorphism of DAT1 and striatal-based implicit sequence learning, such that the presence of at least one 9-repeat allele was beneficial for detecting sequential regularities over time. In contrast, no relationship was observed between DAT1 genotype and implicit spatial context learning, a form of learning associated with the MTL. How these implicit learning differences influence real-world behaviors should be a topic of future research. The continued study of genetic influence on cognitive functioning among healthy adults may provide more insight into the neurochemical and anatomical correlates of memory, including various forms of implicit learning and processing.
Acknowledgments
This research has been supported by NIA/NIH grants R37AG15450, R01AG036863, and F31AG034691, Canadian Institutes for Health Research (CIHR), NIMH R01 MH065395-01, NIH IDDRC P30HD4067, and NIH NCMRR/NINDS 5R24 HD050846. The authors want to thank Adam Green for helpful comments. Preliminary findings from this project were presented at the Cognitive Neuroscience Society conference in Montreal, Canada in April 2010.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Green AE, Munafò MR, DeYoung CG, Fossella JA, Fan J, Gray JR. Using genetic data in cognitive neuroscience: from growing pains to genuine insights. Nature Reviews Neuroscience. 2008;9:710–20. doi: 10.1038/nrn2461. [DOI] [PubMed] [Google Scholar]
- 2.Fossella J, Sommer T, Fan J, Wu Y, Swanson JM, Pfaff DW, et al. Assessing the molecular genetics of attention networks. BMC Neurosci. 2002;3:14. doi: 10.1186/1471-2202-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frensch PA. One concept, multiple meanings: On how to define the concept of implicit learning. In: Stadler MA, Frensch PA, editors. Handbook of implicit learning. Thousand Oaks, CA: Sage Publications, Inc; 1998. pp. 47–104. [Google Scholar]
- 4.Kuhl PK. Early language acquisition: cracking the speech code. Nat Rev Neurosci. 2004;5:831–43. doi: 10.1038/nrn1533. [DOI] [PubMed] [Google Scholar]
- 5.Lieberman MD. Intuition: a social cognitive neuroscience approach. Psychol Bull. 2000;126:109–37. doi: 10.1037/0033-2909.126.1.109. [DOI] [PubMed] [Google Scholar]
- 6.Howard JH, Jr, Howard DV, Dennis NA, Kelly AJ. Implicit learning of predictive relationships in three-element visual sequences by young and old adults. Journal of Experimental Psychology: Learning, Memory and Cognition. 2008;34:1139–57. doi: 10.1037/a0012797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson GM, Jackson SR, Harrison J, Henderson L, Kennard C. Serial reaction time learning and Parkinson’s disease: evidence for a procedural learning deficit. Neuropsychologia. 1995;33:577–93. doi: 10.1016/0028-3932(95)00010-z. [DOI] [PubMed] [Google Scholar]
- 8.Willingham DB, Koroshetz WJ. Evidence for dissociable motor skills in Huntington’s disease patients. Psychobiology. 1993;21:173–82. [Google Scholar]
- 9.Rose M, Haider H, Weiller C, Buchel C. The role of medial temporal lobe structures in implicit learning: an event-related FMRI study. Neuron. 2002;36:1221–31. doi: 10.1016/s0896-6273(02)01105-4. [DOI] [PubMed] [Google Scholar]
- 10.Schendan HE, Searl MM, Melrose RJ, Stern CE. An FMRI study of the role of the medial temporal lobe in implicit and explicit sequence learning. Neuron. 2003;37:1013–25. doi: 10.1016/s0896-6273(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 11.Rauch SL, Whalen PJ, Savage CR, Curran T, Kendrick A, Brown HD, et al. Striatal recruitment during an implicit sequence learning task as measured by functional magnetic resonance imaging. Hum Brain Mapp. 1997;5:124–32. [PubMed] [Google Scholar]
- 12.Reiss JP, Campbell DW, Leslie WD, Paulus MP, Stroman PW, Polimeni JO, et al. The role of the striatum in implicit learning: a functional magnetic resonance imaging study. Neuroreport. 2005;16:1291–5. doi: 10.1097/01.wnr.0000175615.93312.1a. [DOI] [PubMed] [Google Scholar]
- 13.Chun MM, Jiang Y. Contextual cueing: implicit learning and memory of visual context guides spatial attention. Cogn Psychol. 1998;36:28–71. doi: 10.1006/cogp.1998.0681. [DOI] [PubMed] [Google Scholar]
- 14.Greene AJ, Gross WL, Elsinger CL, Rao SM. Hippocampal differentiation without recognition: an fMRI analysis of the contextual cueing task. Learn Mem. 2007;14:548–53. doi: 10.1101/lm.609807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preston AR, Gabrieli JD. Dissociation between explicit memory and configural memory in the human medial temporal lobe. Cereb Cortex. 2008;18:2192–207. doi: 10.1093/cercor/bhm245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun MM, Phelps EA. Memory deficits for implicit contextual information in amnesic subjects with hippocampal damage. Nat Neurosci. 1999;2:844–7. doi: 10.1038/12222. [DOI] [PubMed] [Google Scholar]
- 17.Manns JR, Squire LR. Perceptual learning, awareness, and the hippocampus. Hippocampus. 2001;11:776–82. doi: 10.1002/hipo.1093. [DOI] [PubMed] [Google Scholar]
- 18.Bennett IJ, Romano JC, Howard JHJ, Howard DV. Two forms of implicit learning in young adults with dyslexia. Annals of the New York Academy of Sciences. 2008:184–98. doi: 10.1196/annals.1416.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard JH, Jr, Howard DV, Dennis NA, Yankovich H, Vaidya CJ. Implicit spatial contextual learning in healthy aging. Neuropsychology. 2004b;18:124–34. doi: 10.1037/0894-4105.18.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raz N, Lindenberger U, Rodrigue KM, Kennedy KM, Head D, Williamson A, et al. Regional brain changes in aging healthy adults: general trends, individual differences and modifiers. Cereb Cortex. 2005;15:1676–89. doi: 10.1093/cercor/bhi044. [DOI] [PubMed] [Google Scholar]
- 21.Negash S, Petersen LE, Geda YE, Knopman DS, Boeve BF, Smith GE, et al. Effects of ApoE genotype and mild cognitive impairment on implicit learning. Neurobiol Aging. 2007;28:885–93. doi: 10.1016/j.neurobiolaging.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Lehtovirta M, Laakso MP, Frisoni GB, Soininen H. How does the apolipoprotein E genotype modulate the brain in aging and in Alzheimer’s disease? A review of neuroimaging studies. Neurobiology of Aging. 2000;21:293–300. doi: 10.1016/s0197-4580(00)00120-2. [DOI] [PubMed] [Google Scholar]
- 23.Lewis DA, Melchitzky DS, Sesack SR, Whitehead RE, Auh S, Sampson A. Dopamine transporter immunoreactivity in monkey cerebral cortex: regional, laminar, and ultrastructural localization. J Comp Neurol. 2001;432:119–36. doi: 10.1002/cne.1092. [DOI] [PubMed] [Google Scholar]
- 24.Dahlin A, Xia L, Kong W, Hevner R, Wang J. Expression and immunolocalization of the plasma membrane monoamine transporter in the brain. Neuroscience. 2007;146:1193–211. doi: 10.1016/j.neuroscience.2007.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madras BK, Miller GM, Fischman AJ. The dopamine transporter and attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1397–409. doi: 10.1016/j.biopsych.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Deutch AY, Roth RH. Neurotransmitters. In: Zigmond MJ, Bloom FE, Landis SC, Roberts JL, Squire LR, editors. Functional Neuroscience. San Diego: Academic Press; 1999. pp. 193–234. [Google Scholar]
- 27.Vandenbergh DJ, Persico AM, Hawkins AL, Griffin CA, Li X, Jabs EW, et al. Human dopamine transporter gene (DAT1) maps to chromosome 5p15.3 and displays a VNTR. Genomics. 1992;14:1104–6. doi: 10.1016/s0888-7543(05)80138-7. [DOI] [PubMed] [Google Scholar]
- 28.VanNess SH, Owens MJ, Kilts CD. The variable number of tandem repeats element in DAT1 regulates in vitro dopamine transporter density. BMC Genet. 2005;6:55. doi: 10.1186/1471-2156-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brookes KJ, Neale BM, Sugden K, Khan N, Asherson P, D’Souza UM. Relationship between VNTR polymorphisms of the human dopamine transporter gene and expression in postmortem midbrain tissue. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:1070–8. doi: 10.1002/ajmg.b.30572. [DOI] [PubMed] [Google Scholar]
- 30.Fuke S, Suo S, Takahashi N, Koike H, Sasagawa N, Ishiura S. The VNTR polymorphism of the human dopamine transporter (DAT1) gene affects gene expression. The Pharmacogenomics Journal. 2001;1:152–6. doi: 10.1038/sj.tpj.6500026. [DOI] [PubMed] [Google Scholar]
- 31.Mill J, Asherson P, Browes C, D’Souza U, Craig I. Expression of the dopamine transporter gene is regulated by the 3′ UTR VNTR: Evidence from brain and lymphocytes using quantitative RT-PCR. Am J Med Genet. 2002;114:975–9. doi: 10.1002/ajmg.b.10948. [DOI] [PubMed] [Google Scholar]
- 32.Heinz A, Goldman D, Jones DW, Palmour R, Hommer D, Gorey JG, et al. Genotype influences in vivo dopamine transporter availability in human striatum. Neuropsychopharmacology. 2000;22:133–9. doi: 10.1016/S0893-133X(99)00099-8. [DOI] [PubMed] [Google Scholar]
- 33.Swanson JM, Flodman P, Kennedy J, Spence MA, Moyiz R, Schuck S, et al. Dopamine genes and ADHD. Neurosci Biobehav Rev. 2000;24:21–5. doi: 10.1016/s0149-7634(99)00062-7. [DOI] [PubMed] [Google Scholar]
- 34.Jacobsen LK, Staley JK, Zoghbi SS, Seibyl JP, Kosten TR, Innis RB, et al. Prediction of dopamine transporter binding availability by genotype: a preliminary report. Am J Psychiatry. 2000;157:1700–3. doi: 10.1176/appi.ajp.157.10.1700. [DOI] [PubMed] [Google Scholar]
- 35.van de Giessen EM, de Win MM, Tanck MW, van den Brink W, Baas F, Booij J. Striatal dopamine transporter availability associated with polymorphisms in the dopamine transporter gene SLC6A3. J Nucl Med. 2009;50:45–52. doi: 10.2967/jnumed.108.053652. [DOI] [PubMed] [Google Scholar]
- 36.van Dyck CH, Malison RT, Jacobsen LK, Seibyl JP, Staley JK, Laruelle M, et al. Increased dopamine transporter availability associated with the 9-repeat allele of the SLC6A3 gene. J Nucl Med. 2005;46:745–51. [PubMed] [Google Scholar]
- 37.Krause J, Dresel SH, Krause KH, La Fougere C, Zill P, Ackenheil M. Striatal dopamine transporter availability and DAT-1 gene in adults with ADHD: no higher DAT availability in patients with homozygosity for the 10-repeat allele. World J Biol Psychiatry. 2006;7:152–7. doi: 10.1080/15622970500518444. [DOI] [PubMed] [Google Scholar]
- 38.Martinez D, Gelernter J, Abi-Dargham A, van Dyck CH, Kegeles L, Innis RB, et al. The variable number of tandem repeats polymorphism of the dopamine transporter gene is not associated with significant change in dopamine transporter phenotype in humans. Neuropsychopharmacology. 2001;24:553–60. doi: 10.1016/S0893-133X(00)00216-5. [DOI] [PubMed] [Google Scholar]
- 39.Daly G, Hawi Z, Fitzgerald M, Gill M. Mapping susceptibility loci in attention deficit hyperactivity disorder: preferential transmission of parental alleles at DAT1, DBH and DRD5 to affected children. Molecular Psychiatry. 1999;4:192–6. doi: 10.1038/sj.mp.4000510. [DOI] [PubMed] [Google Scholar]
- 40.Cleeremans A, McClelland JL. Learning the structure of event sequences. J Exp Psychol Gen. 1991;120:235–53. doi: 10.1037//0096-3445.120.3.235. [DOI] [PubMed] [Google Scholar]
- 41.Remillard G, Clark JM. Implicit learning of first-, second-, and third-order transition probabilities. J Exp Psychol Learn Mem Cogn. 2001;27:483–98. doi: 10.1037/0278-7393.27.2.483. [DOI] [PubMed] [Google Scholar]
- 42.Boyer M, Destrebecqz A, Cleeremans A. Processing abstract sequence structure: learning without knowing, or knowing without learning? Psychol Res. 2005;69:383–98. doi: 10.1007/s00426-004-0207-4. [DOI] [PubMed] [Google Scholar]
- 43.Bennett IJ, Barnes KA, Howard JHJ, Howard DV. An abbreviated implicit spatial context learning task that yields greater learning. Behavior Research Methods. 2009;41:391–5. doi: 10.3758/BRM.41.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegert RJ, Taylor KD, Weatherall M, Abernethy DA. Is implicit sequence learning impaired in Parkinson’s disease? A meta-analysis. Neuropsychology. 2006;20:490–5. doi: 10.1037/0894-4105.20.4.490. [DOI] [PubMed] [Google Scholar]
- 45.Krause KH, Dresel SH, Krause J, Kung HF, Tatsch K. Increased striatal dopamine transporter in adult patients with attention deficit hyperactivity disorder: effects of methylphenidate as measured by single photon emission computed tomography. Neurosci Lett. 2000;285:107–10. doi: 10.1016/s0304-3940(00)01040-5. [DOI] [PubMed] [Google Scholar]
- 46.Dougherty DD, Bonab AA, Spencer TJ, Rauch SL, Madras BK, Fischman AJ. Dopamine transporter density in patients with attention deficit hyperactivity disorder. Lancet. 1999;354:2132–3. doi: 10.1016/S0140-6736(99)04030-1. [DOI] [PubMed] [Google Scholar]
- 47.Barnes KA, Howard JH, Jr, Howard DV, Kenealy L, Vaidya CJ. Two forms of implicit learning in childhood ADHD. Developmental Neuropsychology. doi: 10.1080/87565641.2010.494750. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howard DV, Howard JH, Jr, Japikse K, DiYanni C, Thompson A, Somberg R. Implicit sequence learning: effects of level of structure, adult age, and extended practice. Psychol Aging. 2004a;19:79–92. doi: 10.1037/0882-7974.19.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howard JH, Jr, Howard DV. Age differences in implicit learning of higher order dependencies in serial patterns. Psychol Aging. 1997;12:634–56. doi: 10.1037//0882-7974.12.4.634. [DOI] [PubMed] [Google Scholar]
- 50.Volkow ND, Gur RC, Wang GJ, Fowler JS, Moberg PJ, Ding YS, et al. Association between decline in brain dopamine activity with age and cognitive and motor impairment in healthy individuals. Am J Psychiatry. 1998;155:344–9. doi: 10.1176/ajp.155.3.344. [DOI] [PubMed] [Google Scholar]
- 51.Rieckmann A, Backman L. Implicit learning in aging: extant patterns and new directions. Neuropsychol Rev. 2009;19:490–503. doi: 10.1007/s11065-009-9117-y. [DOI] [PubMed] [Google Scholar]
- 52.Backman L, Nyberg L, Lindenberger U, Li SC, Farde L. The correlative triad among aging, dopamine, and cognition: current status and future prospects. Neurosci Biobehav Rev. 2006;30:791–807. doi: 10.1016/j.neubiorev.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Simon JR, Howard JH, Jr, Howard DV. Age differences in implicit learning of probabilistic, unstructured sequences. J Gerontol B Psychol Sci Soc Sci. doi: 10.1093/geronb/gbq066. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JW, Kim BN, Cho SC. The dopamine transporter gene and the impulsivity phenotype in attention deficit hyperactivity disorder: a case-control association study in a Korean sample. J Psychiatr Res. 2006;40:730–7. doi: 10.1016/j.jpsychires.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Colzato LS, Pratt J, Hommel B. Dopaminergic Control of Attentional Flexibility: Inhibition of Return is Associated with the Dopamine Transporter Gene (DAT1) Front Hum Neurosci. 2010;4:53. doi: 10.3389/fnhum.2010.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brehmer Y, Westerberg H, Bellander M, Fürth D, Karlsson S, Bäckman L. Working memory plasticity modulated by dopamine transporter genotype. Neuroscience Letters. 2009;467:117–20. doi: 10.1016/j.neulet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 57.Garcia-Garcia M, Barcelo F, Clemente C, Escera C. The role of the dopamine transporter DAT1 genotype on the neural correlates of cognitive flexibility. Eur J Neurosci. 2010;31:754–60. doi: 10.1111/j.1460-9568.2010.07102.x. [DOI] [PubMed] [Google Scholar]
- 58.Dreher J, Kohn P, Kolachana B, Weinberger DR, Berman KF. Variation in dopamine genes influences responsivity of the human reward system. Proc Natl Acad Sci USA. 2009;106:617–22. doi: 10.1073/pnas.0805517106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bertolino A, Blasi G, Latorre V, Rubino V, Rampino A, Sinibaldi L, et al. Additive effects of genetic variation in dopamine regulating genes on working memory cortical activity in human brain. J Neurosci. 2006:3918–22. doi: 10.1523/JNEUROSCI.4975-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rommelse NNJ, Altink ME, Arias-Vàsquez A, Buschgens CJM, Fliers E, Faraone SV, et al. A review and analysis of the relationship between neuropsychological measures and DAT1 in ADHD. Am J Med Genet B: Neuropsychiatr Genet B. 2008:147. doi: 10.1002/ajmg.b.30848. [DOI] [PubMed] [Google Scholar]
- 61.Keri S, Juhasz A, Rimanoczy A, Szekeres G, Kelemen O, Cimmer C, et al. Habit learning and the genetics of the dopamine D3 receptor: evidence from patients with schizophrenia and healthy controls. Behav Neurosci. 2005;119:687–93. doi: 10.1037/0735-7044.119.3.687. [DOI] [PubMed] [Google Scholar]
- 62.Knowlton BJ, Squire LR, Gluck MA. Probabilistic classification learning in amnesia. Learn Mem. 1994;1:106–20. [PubMed] [Google Scholar]
- 63.Meador-Woodruff JH. Dopamine Receptor Transcript Localization in Human Brain. In: Watson SJ, editor. Psychopharmacology: The Fourth Generation Of Progress. Philadelphia: Lippincott-Raven Publishers; 1998. [Google Scholar]
- 64.Meeter M, Myers CE, Shohamy D, Hopkins RO, Gluck MA. Strategies in probabilistic categorization: results from a new way of analyzing performance. Learn Mem. 2006;13:230–9. doi: 10.1101/lm.43006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frank MJ, Moustafa AA, Haughey HM, Curran T, Hutchison KE. Genetic triple dissociation reveals multiple roles for dopamine in reinforcement learning. Proc Natl Acad Sci U S A. 2007;104:16311–6. doi: 10.1073/pnas.0706111104. [DOI] [PMC free article] [PubMed] [Google Scholar]