Abstract
Three decades after the eradication of smallpox, the threat of bioterrorism and outbreaks of emerging diseases such as monkeypox have renewed interest in the development of safe and effective next-generation poxvirus vaccines and biodefense research. Current smallpox vaccines contain live virus and are contraindicated for a large percentage of the population. Safer, yet still effective inactivated and subunit vaccines are needed, and epitope identification is an essential step in the development of these subunit vaccines. In this study we focused on 4 vaccinia membrane proteins known to be targeted by humoral responses in vaccinees. In spite of the narrow focus of the study we identified 36 T cell epitopes, and provide additional support for the physical linkage between T and B epitopes. This information may prove useful in peptide and protein-based subunit vaccine development as well as in the study of CD4 responses to poxviruses.
Keywords: Vaccinia virus, Smallpox vaccine, Cellular immunity, T cell epitopes, CD4+ T cells
Introduction
Smallpox is a deadly disfiguring disease that was endemic worldwide and caused hundreds of millions of deaths in the last few centuries and countless deaths before that (Fenner et al., 1988). Edward Jenner's discovery of cowpox vaccination as a preventive measure (Jenner, 1798) led to the eradication of smallpox after a decade's long effort led by the WHO (Fenner, 1982). Thirty years after its eradication, variola virus is considered a biological weapon candidate and, given the possibility of bioterrorism or zoonotic poxvirus outbreaks, poxviruses remain a significant public health concern (Henderson, 1999, Henderson et al., 1999, Jahrling et al., 2005, Larkin, 2003, Mayr, 2003).
Control of this disease relied heavily on the availability of an extremely effective, live, viral vaccine based on the immunologically cross-protective vaccinia virus (Artenstein, 2008, Kennedy et al., 2009a, Kennedy et al., 2009b, Metzger & Mordmueller, 2007). Although its origin is unclear, vaccinia-based smallpox vaccines induce robust and protective humoral and cellular immune responses lasting for decades (Combadiere et al., 2004, Crotty et al., 2003, el-Ad et al., 1990, Ferrier-Rembert et al., 2008, Frey et al., 2003, Hammarlund et al., 2003, Kan et al., 2007). The vaccine also caused serious adverse events and routine vaccination was halted soon after the eradication (Fulginiti, 2003, Fulginiti et al., 2003, Goldstein et al., 1975, Lane et al., 1970, Morgan et al., 2008, Neff et al., 1967). Recent increased terrorist activity and outbreaks of monkeypox and other emerging infectious diseases have stimulated biodefense research and vaccine preparation in regard to smallpox. These efforts have led to renewed interest in the immunologic mechanisms behind smallpox vaccination, in particular: the identification of T and B cell epitopes, and the development of subunit vaccines (Golovkin et al., 2007, Heraud et al., 2006, Kennedy & Poland, 2007, Moutaftsi et al., 2010, Poland, 2005, Sakhatskyy et al., 2008).
Epitope identification for large, complex pathogens such as poxviruses is often a difficult and laborious process. To simplify the task, many groups turn to computer algorithms designed to predict peptide binding. Protein sequences can be scanned by computer algorithms to identify likely candidates, which are then synthesized and tested (Hammer et al., 1994, Larsen et al., 2007, Meister et al., 1995, Rammensee et al., 1995, Rammensee et al., 1999, Roomp et al., 2010, Tong et al., 2007). Even with the algorithms, comprehensive epitope identification requires that thousands of peptides be tested. Other groups use shotgun genomic approaches to express and test random protein sequences. Once hits are identified, gene sequencing is used to identify the protein targets from which individual peptides can then be tested. In spite of the complications involved in identifying immunogenic poxviruses epitopes, in the last several years a number of groups have identified hundreds of MHC I restricted peptides (Drexler et al., 2003, Jing et al., 2005, Johnson et al., 2005, Mathew et al., 2005, Moise et al., 2009, Moutaftsi et al., 2006, Oseroff et al., 2005, Pasquetto et al., 2005a, Pasquetto et al., 2005b, Sidney et al., 2008, Snyder et al., 2004, Terajima et al., 2003, Terajima et al., 2008, Tscharke et al., 2005, Tscharke et al., 2006, Walsh et al., 2009), and studies examining T helper epitopes are also being reported (Calvo-Calle et al., 2007, Jing et al., 2007, Jing et al., 2008, Mitra-Kaushik et al., 2007, Sette et al., 2008, Sirven et al., 2009, Strug et al., 2008, Tang et al., 2006, Wang et al., 2009). Our approach to the problem was to focus on potential interactions between antigen-specific B and T cells. Decades ago, B cells were shown to acquire antigen through surface bound immunoglobulin and present antigen to T cells (Lanzavecchia, 1985, Lanzavecchia & Bove, 1985). Similar antigen acquisition and presentation processes (involving uptake and internalization of MHC/peptide complexes) have been shown for T helper cells and may explain, in part, the close physical proximity of strong CTL and T helper epitopes (Adamopoulou et al., 2007, Davies et al., 2007, Huang et al., 1999, Kennedy et al., 2005, Umeshappa et al., 2009). We reasoned that poxvirus proteins targeted to B cells by antibody–antigen recognition would allow for the efficient presentation of any HLA class II epitopes within the same protein to vaccinia-specific T helper lymphocytes. With this hypothesis in mind we focused on four of the viral proteins known to be targeted by humoral immunity (A27L, A33R, B5R, and L1R) (Bell et al., 2004, Davies et al., 2007, Davies et al., 2008, Duke-Cohan et al., 2009, Galmiche et al., 1999, Golden et al., 2008, Lawrence et al., 2007, Wolffe et al., 1995).
Consistent with our hypothesis we identified multiple T cell epitopes within each of the four proteins tested. These results also provide additional evidence of what Sette et al. (2008) have labeled the “deterministic linkage of specificities” for poxvirus epitopes. This report adds to the growing list of HLA II-restricted poxvirus epitopes, and as with many of those other reports, we found no clear cut indications of immunodominance at the peptide level.
Results
Protein selection and library screening
Since robust humoral responses to vaccinia depend on T cell help, we hypothesized that the close physical proximity of T helper and B cell epitopes would provide the most efficient provision of the needed T cell help as has been seen in other model systems (Bernard et al., 2005, Bishop & Hostager, 2001, Lanzavecchia, 1985, Noelle & Snow, 1991). We initially compiled a list of viral proteins known to be targeted by humoral responses to vaccinia (Davies et al., 2007, Davies et al., 2008). From this list of over a dozen proteins we selected four (A33R, A27L, B5R, and L1R) for further study. Published literature for each of these four proteins indicates that they are targeted by T cell responses (Fogg et al., 2004, Hooper et al., 2000, Hooper et al., 2003, Tang et al., 2006). Overlapping peptide libraries spanning the amino acid sequence for each protein were synthesized and divided into pools using the Deconvolute This software (courtesy of Mario Roederer, NIH) (Roederer and Koup, 2003). Each peptide was placed into three separate pools to allow for more rapid deconvolution of positive responses. Twenty-nine individuals who had received the smallpox vaccine within the last 4 years were recruited.
Immune responses to vaccinia virus
Humoral immunity was measured using a reporter-based neutralizing antibody assay (R. Kennedy et al., 2009, Manischewitz et al., 2003). All of the subjects had detectable levels of vaccinia neutralizing antibody, with ID50 (the serum dilution which neutralizes 50% of viral activity) values ranging from 37.3 to 1631.6 (Fig. 1 ). Each of these values is significantly greater than those seen in vaccinia-naïve individuals (ID50 values routinely below 5.0) (Kennedy et al., 2009a). All but two of the 29 subjects enrolled in this study exhibited vaccinia-specific T cell responses (S.I. > 2) as measured by IFNγ ELISPOT assay. Spots per million cells in vaccinia stimulated wells were divided by the spots per million cells in background wells to provide the stimulation index (S.I.) which ranged from 1.22 to 444.0 (Fig. 1), reflecting a large spectrum of cellular immune responses.
Fig. 1.
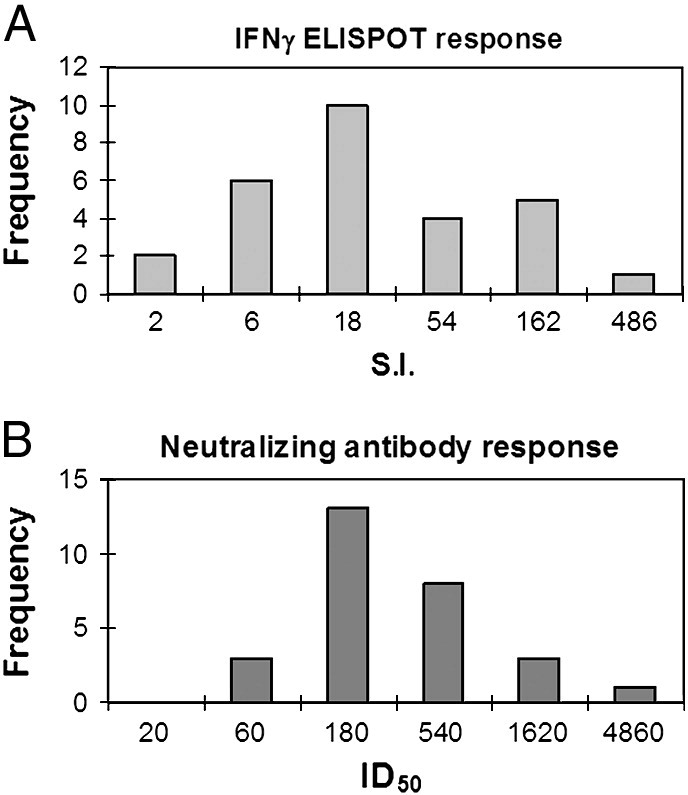
Cellular and humoral responses to vaccinia. A) The histogram on the left shows the range of IFNγ ELISPOT results as stimulation index (S.I.: average spot forming units in vaccinia stimulated wells divided by the average spot forming units in background wells). X-axis scale indicates upper bound of each bin, i.e. two subjects had an S.I. less than 2, and six subjects had S.I. values between 2.1 and 6. B) The histogram on the right shows the range of 50% inhibitory dose (ID50: reciprocal of the serum dilution which inhibits 50% of viral activity). X-axis scale indicates upper bound of each bin, i.e. no subjects had ID50 values less than 20, while 3 subjects had an ID50 between 21 and 60.
Immune responses to viral peptides and epitope identification
Immune responses to peptide pools were tested in IFNγ ELISPOT assays using cells from these individuals. A representative example of the immune response profile is shown in Fig. 2 . Positive pools were identified and the individual peptides potentially contributing to the response profile were identified using the Deconvolute This software. These potential epitopes were then screened individually or in smaller pools depending on the number of possible candidates (Fig. 3 ). Positive peptides were confirmed in follow-up experiments using cells depleted of CD8+ T lymphocytes (Fig. 4 ). One of the main benefits of using overlapping peptide libraries for epitope mapping is that it allows one to test all possible HLA class I and class II peptides without regard for allele-specific binding restrictions.
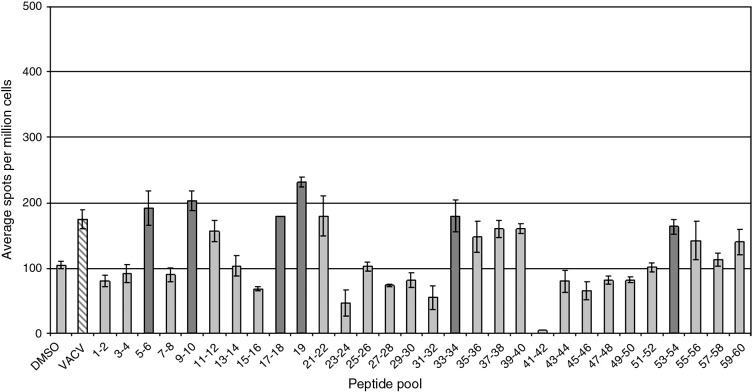
Fig. 2.
Immune responses to pooled peptides from a single individual are illustrated. Bars indicate the average number of IFNγ producing spots for wells (3–5 replicates) stimulated with the antigen indicated on the X-axis. Error bars represent the standard deviation. Dark gray bars indicate responses significantly above background (p < 0.05). Data illustrated in this figure to Fig.–4 come from the same individual.
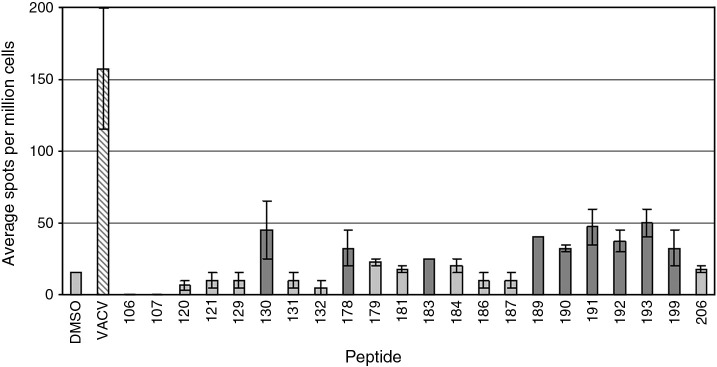
Fig. 3.
Immune responses to individual peptides from a single individual. The positive pools shown in Fig. 2 were used to select peptides for individual screening. Graph layout is as described in Fig. 2.
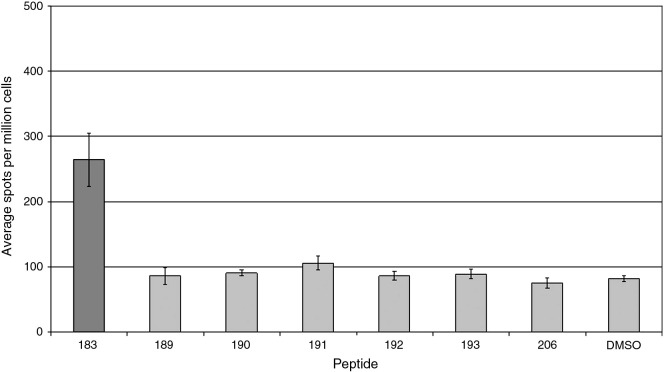
Fig. 4.
CD4+ T cell responses to selected peptides from a single individual. Prior to stimulation with the indicated peptides (X-axis) CD8+ T cells were removed by magnetic bead depletion. Resulting cell populations contained < 2% CD8+ T cells (data not shown). The graph layout is as described in Fig. 2.
Nearly all (28/29) subjects demonstrated significant T cell responses to peptide pools, indicative of cellular immune responses to these four proteins. Importantly, we were able to identify individual epitope-specific responses in nearly one-half of the responders (13/28). The remaining subjects had significant immune responses to pooled peptides but not to individual peptides, indicating that these initial responses were either false positives or that they represented the combined, synergistic effect of multiple epitope-specific T cell populations which, when analyzed individually, fell below the level of detection of our assays. The epitope mapping results are shown in Table 1, Table 2 . Table 1 shows peptides recognized by CD4+ T cells that are presumably presented by HLA class II molecules. Table 2 shows peptides recognized by PBMCs. These epitopes were not definitively linked to CD4+ T cell responses due to either a lack of sufficient cells to test the CD4+ T cell responses, or more commonly, the disappearance of responses to individual peptides when CD8+ T cells are removed from the ELISPOT assays. These results indicate that the peptides listed in Table 2 are likely presented by HLA class I molecules to CD8+ T cells. In support of this hypothesis, the sequence VLFRLENHA within peptide #21 (Table 2) has been identified as an HLA-A*0201 epitope (Otero et al., 2006), as has the sequence QTSVFSATV within peptide #22 (Sidney et al., 2008). Our results do not rule out the possible presence of HLA II epitopes eliciting minor responses below the level of sensitivity of our assays. For example, Sirven et al. (2009) have identified various DR binding epitopes within peptides #23 and #24. In some cases the first round peptide pool screening was immediately followed by individual peptide testing in CD8 T cell depleted PBMCs. Of the 36 peptides listed in Table 1, Table 2 only three were recognized by more than 1 individual. Peptide #6 was recognized by 3 subjects and peptides #15 and #20 were recognized by two individuals.
Table 1.
Characteristics of newly identified MHC II-restricted T helper epitopes.
| Peptide #a | Sequenceb | Proteinc | VARV sequenced | Protein sequence homologye |
|||
|---|---|---|---|---|---|---|---|
| VACV | VARV | MPXV | CMLV | ||||
| 1 | KADEDDNEETLKQRLT | A27L | KADGDDNEETLKQRLT | 99.5 | 98.1 | 94.6 | 97.3 |
| 2 | VYSTCTVPTMNNAKLT | B5R | VYSTCTVPTMNNAKLT | 98.6 | 93.6 | 96.8 | 93.1 |
| 3 | CTVPTMNNAKLTSTET | B5R | CTVPTMNNAKLTSTET | 98.6 | 93.6 | 96.8 | 93.1 |
| 4 | LYNKPLYEVNSTMTLS | B5R | LYNKPLYEVNAIITLI | 98.6 | 93.6 | 96.8 | 93.1 |
| 5 | PNAVCETDKWKYENPC | B5R | PNAVCETDKWKYENPC | 98.6 | 93.6 | 96.8 | 93.1 |
| 6 | CYILHSDYQLFSDAKA | A33R | CYIFHSDYQLFSDAKA | 99.3 | 94 | 96.5 | 95 |
| 7 | AKLTSTETSFNNNQKV | B5R | AKLTSTETSFNDKQKV | 98.6 | 93.6 | 96.8 | 93.1 |
| 8 | CETDKWKYENPCKKMC | B5R | CETDKWKYENPCKKMC | 98.6 | 93.6 | 96.8 | 93.1 |
| 9 | TVYGDKIQGKNKRKRV | A33R | TVYGDKIQGKNKRKRV | 99.3 | 94 | 96.5 | 95 |
| 10 | KITNVTTKFEQIEKCC | A27L | KITNVTTKFEQIEKCC | 99.5 | 98.1 | 94.6 | 97.3 |
| 11 | AFLIVRLNQCMSANEA | A33R | AFLIVRLNQCMSANEA | 99.3 | 94 | 96.5 | 95 |
| 12 | SSTTQYDHKESCNGLY | A33R | SSTTQYKHQESCNGLY | 99.3 | 94 | 96.5 | 95 |
| 13 | SGSTFSIGGVIHLSCK | B5R | SGSTFSIGGVIHLSCK | 98.6 | 93.6 | 96.8 | 93.1 |
| 14 | CNLTVKNMCSADADAQ | L1R | CNLTVKNMCSADADAQ | 100 | 99.6 | 98.8 | 98.4 |
| 15 | NCAIKALMQLTTKATT | L1R | NCAIKALMQLTTKATT | 100 | 99.6 | 98.8 | 98.4 |
| 16 | KCDIEIGNFYIRQNHG | L1R | KCDIEIGNFYIRQNHG | 100 | 99.6 | 98.8 | 98.4 |
| 17 | QNVIIDECYGAPGSPT | L1R | QNVIIDECYGAPGSPT | 100 | 99.6 | 98.8 | 98.4 |
| 18 | GVIFLISVIVLVCSCD | B5R | GVIFLISVIVLVCSCN | 98.6 | 93.6 | 96.8 | 93.1 |
| 19 | AALFMYYAKRMLFTST | L1R | AALFMYYAKRMLFTST | 100 | 99.6 | 98.8 | 98.4 |
An arbitrarily assigned number, used to identify peptides discussed in the text.
Sequence of the identified epitope. The VACV-ACAM2000 sequence was used for peptide library synthesis.
Vaccinia protein name following VACV-Copenhagen designation.
Consensus variola sequence with differences indicated by bold, underlined font.
Average sequence homology between the protein listed in column c and the homologs from the indicated poxviruses (VACV = vaccinia, VARV = variola, MXPX = monkeypox, CMLV = camelpox).The VACV-ACAM2000 protein sequences were compared to sequence from each strain of the indicated poxvirus strains available from the Poxvirus Bioinformatics Resource Center (www.poxvirus.org). The average homology for each poxvirus was then calculated and presented in this table.
Table 2.
Characteristics of newly identified T cell epitopes.
| Peptide #a | Sequenceb | Proteinc | VARV sequenced | Protein sequence homologye |
|||
|---|---|---|---|---|---|---|---|
| VACV | VARV | MPXV | CMLV | ||||
| 20 | IRISMVISLLSMITMS | A33R | IRISMVISLLSMITMS | 99.3 | 94 | 96.5 | 95 |
| 21 | EVLFRLENHAETLRAA | A27L | DVLFRLENHAETLRAA | 99.5 | 98.1 | 94.6 | 97.3 |
| 22 | EQTSVFSATVYGDKIQ | A33R | EQTSVFSATVYGDKIQ | 99.3 | 94 | 96.5 | 95 |
| 23 | DKIQGKNKRKRVIGLC | A33R | DKIQGKNKRKRVIGIC | 99.3 | 94 | 96.5 | 95 |
| 24 | FSIGGVIHLSCKSGFI | B5R | FSIGGVIHLSCKSGFI | 98.6 | 93.6 | 96.8 | 93.1 |
| 25 | KLEQEANASAQTKCDI | L1R | KLEQEANASAQTKCDI | 100 | 99.6 | 98.8 | 98.4 |
| 26 | ITINCDVGYEVIGASY | B5R | ITINCDVGYEVIGASY | 98.6 | 93.6 | 96.8 | 93.1 |
| 27 | IIVALTIMGVIFLISV | B5R | IIVALTIMGVIFLISV | 98.6 | 93.6 | 96.8 | 93.1 |
| 28 | KATTQIAPRQVAGTGV | L1R | KATTQIAPRQVAGTGV | 100 | 99.6 | 98.8 | 98.4 |
| 29 | MYYAKRMLFTSTNDKI | L1R | MYYAKRMLFTSTNDKI | 100 | 99.6 | 98.8 | 98.4 |
| 30 | DTFFRTSPMVIATTDM | L1R | DTFFRTSPMVIATTDI | 100 | 99.6 | 98.8 | 98.4 |
| 31 | QIAPRQVAGTGVQFYM | L1R | QIAPRQVAGTGVQFYM | 100 | 99.6 | 98.8 | 98.4 |
| 32 | KRMLFTSTNDKIKLIL | L1R | KRMLFTSTNDKIKLIL | 100 | 99.6 | 98.8 | 98.4 |
| 33 | ANKENVHWTTYMDTFF | L1R | ANKENVHWTTYMDTFF | 100 | 99.6 | 98.8 | 98.4 |
| 34 | VIILAALFMYYAKRML | L1R | VIILAALFMYYAKRML | 100 | 99.6 | 98.8 | 98.4 |
| 35 | NVHWTTYMDTFFRTSP | L1R | NVHWTTYMDTFFRTSP | 100 | 99.6 | 98.8 | 98.4 |
| 36 | TTYMDTFFRTSPMVIA | L1R | TTYMDTFFRTSPMVIA | 100 | 99.6 | 98.8 | 98.4 |
Peptides listed in this table elicited significant IFNγ ELISPOT responses with total PBMCs and may represent CD4 and/or CD8 epitopes. Please refer to Table 1 for a description of each column.
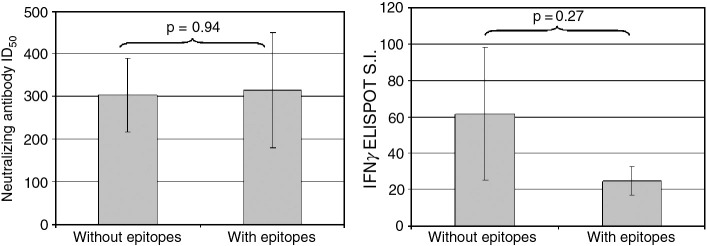
Epitope identification did not show any correlation with the magnitude of either humoral or cellular vaccinia-specific responses (Fig. 5 ), nor were there significant differences in immune response between individuals with identified peptide responses and those without (Fig. 6 ).
Fig. 5.
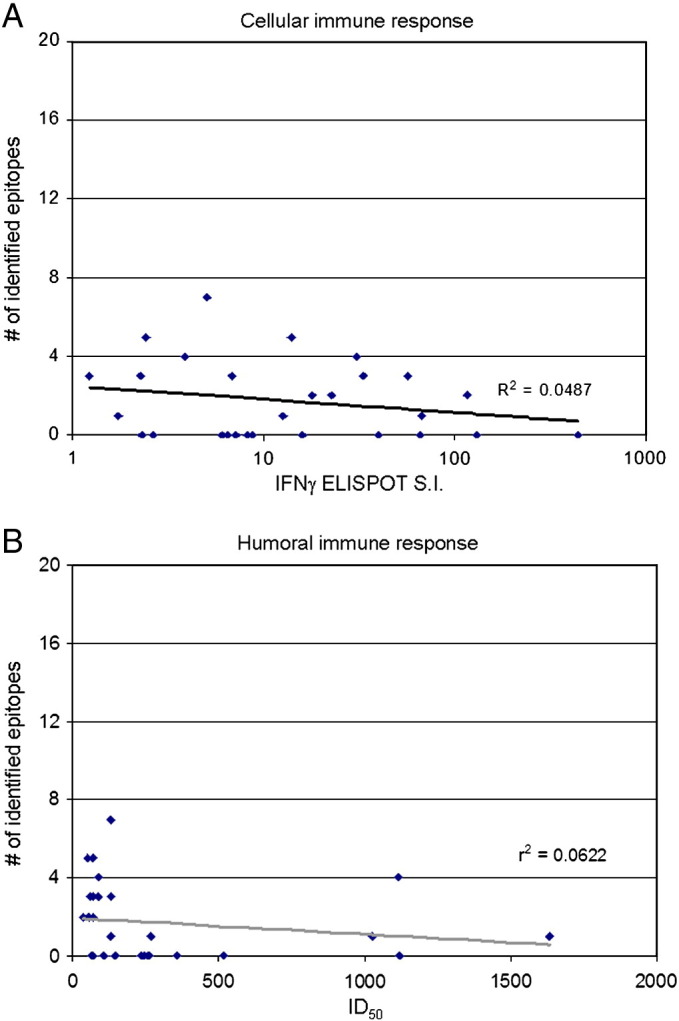
Correlation between immune response and number of epitopes identified. A) Scatter plot shows vaccinia-specific IFNγ ELISPOT S.I. on the X-axis and the number of identified epitopes on the Y-axis. The best fit line is depicted in gray with the r2 value indicated on the graph. Each diamond represents a single subject's response. B) Scatter plot shows the vaccinia-specific neutralizing antibody ID50 on the X-axis and the number of identified epitopes on the Y-axis. The best fit line is depicted in gray with the r2 value indicated on the graph. Each diamond represents a single subject's response.
Fig. 6.
Comparison of immune responses in subjects with and without identified epitopes. Average immune responses to vaccinia virus (left panel = humoral response represented by ID50 values, right panel = cellular immunity denoted by IFNγ ELISPOT S.I.) for subjects with identified T cell epitopes are compared to the anti-viral immune responses of subjects for which no epitopes were found. Student's t test comparison p-values are indicated on each graph.
Our results support our hypothesis that proteins targeted by B cell responses are likely to also contain T helper epitopes, as we have identified multiple peptides from each of the four target proteins.
Epitope characterization
As shown in Table 1, the identified vaccinia peptides show remarkable sequence homology (> 97%) with their variola counterparts. In fact, 27 of the 36 peptides were 100% conserved, of the 9 peptides with different protein sequences (Proteins #1, #4, #6, #7, #12, #18, #21, #23 and #30 from Table 1, Table 2): 6 peptides differed by only a single amino acid, 2 peptides had two amino acid differences and one peptide had 5 divergent amino acids. At the protein level, these 4 proteins show very high homology between vaccinia, variola, monkeypox and camelpox viruses. We synthesized several of the VARV homologs of the peptides in Table 1, Table 2 and detected an IFNγ response to the VARV version of peptide #18 but not to the peptides #1, #4, #6, #21, or #30 (Table 3 ).
Table 3.
Comparison of immune reactivity to VACV and VARV homologs.
| Peptide #a | Sequenceb | Virusc | S.I.d | p-valuee |
|---|---|---|---|---|
| 1 | KADEDDNEETLKQRLT | VACV | 1.5 | 0.01 |
| KADGDDNEETLKQRLT | VARV | 1 | 0.68 | |
| 4 | LYNKPLYEVNSTMTLS | VACV | 1.5 | 0.01 |
| LYNKPLYEVNAIITLI | VARV | 1 | 0.92 | |
| 6 | CYILHSDYQLFSDAKA | VACV | 3.5 | 0.006 |
| CYIFHSDYQLFSDAKA | VARV | 1.1 | 0.95 | |
| 18 | GVIFLISVIVLVCSCD | VACV | 2.1 | 0.012 |
| GVIFLISVIVLVCSCN | VARV | 2.3 | 0.02 | |
| 21 | EVLFRLENHAETLRAA | VACV | 2.5 | 0.04 |
| DVLFRLENHAETLRAA | VARV | 0.7 | 0.16 | |
| 30 | DTFFRTSPMVIATTDM | VACV | 2 | 0.008 |
| DTFFRTSPMVIATTDI | VARV | 1.1 | 0.89 |
Peptide sequence.
Virus containing the listed peptide sequence.
Stimulation index (mean spots per well containing peptide / mean spots per background wells).
p-Value comparing spots per well with peptide vs. spots per well with DMSO control. Positive responses are in bold font and represent peptides with a 1.5 fold increase in mean spots/well over background (S.I. ≥ 1.5) and a p-value of ≤ 0.05.
Similar to what other groups have found, we did not identify any clearly immunodominant peptides as most of the peptides were recognized by a single individual, in fact, of the 36 epitopes we identified only 3 were recognized by more than one individual. At the protein level we identified more epitopes from the B5R (11 epitopes) and L1R (15 epitopes) proteins than the A33R (7 epitopes) or A27L (3 epitopes) proteins.
Discussion
We initially hypothesized that T and B cell epitopes would reside within the same proteins or even within close physical proximity to one another as this would result in optimal activation of both CD4+ T cells and B cells and provision of the T cell help necessary for robust humoral responses to poxviruses. In support of this theory we identified 36 T cell epitopes within 4 viral proteins targeted by antibody responses.
A33R is a membrane glycoprotein found on the surface of EV particles, A27L is an MV membrane protein involved in cell attachment and fusion, B5R is an EV membrane protein required for the formation of EV particles, and L1R is a myristylated product of a vaccinia late gene and is essential for the formation of infectious MV (Chung et al., 1998, Isaacs et al., 1992, Roper et al., 1998, Wolffe et al., 1995). All of these proteins are the targets of neutralizing antibody responses, with B5R serving as the major target of EV neutralizing activity in serum samples from vaccine recipients (Bell et al., 2004). PBMCs from almost all of our subjects reacted to multiple peptide pools as demonstrated by IFNγ ELISPOT responses that were significantly higher than those seen in background wells. Unfortunately, this reactivity was lost in half of the subjects when we tested individual peptides. Sirven et al. (2009)) studied human CD4+ T cell responses to the B5R and A33R proteins among individuals with unknown smallpox vaccination history and found that multiple rounds of in vitro stimulation was required to detect T cell responses to HLA class II binding vaccinia peptides. The need for multiple peptide stimulation may have been due to the need to activate and expand a small number of naïve T cells or to activate cross-reactive memory cells, alternatively, repeat stimulation may have been required due to the low level of immune memory from vaccination 30+ years ago. In all of these cases, low level responses (due to either rare precursor frequency or low-affinity TCR/HLA interactions) are likely to require multiple rounds of stimulation for optimal response. While some of this loss in reactivity may be due to false positive responses, it is also possible that low frequencies of peptide-specific T cells resulted in suboptimal responses below the limit of detection in our assays. Another possibility is the fact that our epitope library consisted of 16mer peptides offset by 4 amino acids and likely did not capture all possible epitopes. As has been found in other studies examining CD4+ T cell responses to poxviruses, our results varied between individuals, likely a reflection of the large number of potential T cell epitopes within the ~ 250 ORFs of vaccinia virus, inter-individual HLA differences, and perhaps variations in time since vaccination. We had two individuals with undetectable responses to vaccinia virus and another 6 with extremely low vaccinia-specific responses (S.I. < 3), yet several of these subjects exhibited peptide-specific responses (Fig. 5). This discrepancy could be due to the fact that T cell responses to viral particles require infection, proteolytic cleavage of the antigen, peptide loading onto appropriate HLA molecules, and transport to the cell surface for presentation, an inherently less efficient process than exogenous loading of higher concentrations of soluble peptide onto surface HLA. These individuals may have very low levels of vaccinia-specific responses that are optimally detected using exogenous peptide rather than whole virus. Another possibility is that the epitope-specific responses are due to a population of cross-reactive T cells. Consistent with this hypothesis are the published reports documenting cross-reactivity between vaccinia and LCMV (Chen et al., 2001, Cornberg et al., 2007, Selin et al., 1998), or vaccinia and Pichinde virus (Cornberg et al., 2010). In humans similar cross-reactivity has been found between influenza and EBV (Clute et al., 2005) as well as between influenza and hepatitis C (Urbani et al., 2005). Considerable effort is being spent on developing subunit vaccines for select agents, including smallpox. This is especially important given the reactivity and morbidity of the current live viral vaccine. Our results provide additional support for the importance of these 4 membrane proteins in subunit based smallpox vaccines. Each protein contains multiple T and B cell epitopes and is widely targeted by both humoral and cellular immune responses in vaccine recipients. Although the proteins we tested are fairly well conserved among poxviruses, a small number of the peptides do have amino acid differences which could abrogate cross-reactivity. Of the six peptides that we were able to test, only one of the VARV homologs showed T cell reactivity. A similarly low level of cross-reactivity for CD8+ T cell epitopes has recently been reported by Sette et al. (2009) As subunit vaccines are being developed it will be important to minimize these differences through the use of whole protein-based vaccines, selective epitope choice and/or the inclusion of multiple epitopes in peptide-based vaccines in order to elicit population-wide, cross-protective immunity to the poxviruses constituting potential public health threats (variola, monkeypox and camelpox).
Our results also illustrate advantages of parallel efforts in epitope identification. For example, Jing et al. (2007) tested peptide responses from in vitro restimulated T cell cultures from recent vaccine recipients and identified a peptide from A33R (A33R160–173) and a peptide from L1R (L1R127–137) but did not identify any of our epitopes to either protein. Relying on their report or ours alone would present an incomplete picture of the T cell epitopes contained within these two proteins. Another example is the report by Calvo-Calle et al. (2007) which focused on DR1-binding peptide sequences across the viral genome. The authors did not find any epitopes from the A27L, A33R, or B5R proteins, however, they did identify two epitopes from L1R: L1R181–201 and L1R187–207 which overlap the sequences of peptides #19, #28, #31, #32, and #34 from Table 1, Table 2. In this instance the reports provide validation for the identified epitopes and complementary information, with Calvo-Calle providing evidence of DR1 restriction, while our report shows that 3 additional subjects recognize 5 epitopes within close physical proximity to the 2 epitopes recognized by the single subject from the Calvo-Calle report. Differences in approach (computer algorithm, overlapping peptides, and peptide elution from HLA molecules), subject selection (recent vaccinees vs. those immunized decades ago), vaccine studied (Dryvax, ACAM2000, MVA), immune assay selection (Flow cytometry, tetramer staining, ELISPOT, and ELISA) and even immune outcome (IL2 vs. TNFα vs. IFNγ vs. perforin and CD107a) may contribute to the distinctive results found by the different laboratory groups involved. Importantly, we feel that these parallel efforts allow redundancy in epitope identification, increase confidence in the results, provide complementary information regarding T cell responses, and frequently identify unique peptides thereby maximizing our chances of systematic and comprehensive epitope identification.
Conclusions
In this study we identified 36 viral epitopes in 4 selected vaccinia proteins, providing support for the hypothesis that T and B cell epitopes are found in close physical proximity, and that this close spatial linkage allows for the generation of robust immune responses. As seen in other reports, the CD4+ T cell response to these 4 proteins is diverse and varied, with only 3 of the 36 epitopes being recognized by more than one individual. While most of the vaccinia peptides were completely conserved in their variola counterparts, most of those epitopes with amino acid differences did not show cross-reactivity with variola sequences. Our results support using known immune interactions for a rational and directed epitope identification process and indicate that pathogen epitopes, once identified, must be carefully tested for reactivity and utility before incorporation into vaccine candidates.
Materials and methods
Subject recruitment
Healthy volunteers from Mayo Clinic who had participated in past smallpox vaccine trials or were part of the DHHS first responder's initiative were recruited for this study. All individuals provided informed consent and submitted to a single blood draw of 100 ml. Serum and PBMCs were isolated, aliquoted and frozen until use. Approval for the study was received from Mayo Clinic's Institutional Review Board.
Viruses and cell lines
Vaccinia viruses (both NYCBOH and WR strain) were grown in Hela S3 cells, titrated in Vero cells and stored at −70 °C until use. Virus was inactivated using Psoralen and UV light as described (Crotty et al., 2003) or heated to 52 °C for 1 h. DC2.4 and LB27.4 cells and allogeneic splenocytes were used as MHC II positive APCs where indicated.
Peptides and reagents
We designed a series of 16mer peptides, offset by 4 amino acids that spanned the length of each of the 4 selected viral proteins. Peptides for the overlapping library were purchased from Mimotopes (Clayton, Australia) and purified peptides were synthesized at Mayo Clinic or Mimotopes. All peptides were dissolved in DMSO at 20 mg/ml and stored at −20 °C until use. Peptide pools contained equal amounts of 9–11 peptides each.
ELISPOT assays
PBMCs were purified from whole blood using CPT tubes (Becton Dickinson Franklin Lake, NJ). After purification, PBMC were aliquoted and frozen in liquid nitrogen until use. Culture media consisted of RPMI (Invitrogen, Carlsbad, CA) supplemented with 10% FCS, penicillin/streptomycin, non-essential amino acids, sodium pyruvate and sodium bicarbonate and 50 μM beta-mercaptoethanol. Where indicated CD8+ T cells were removed using magnetic bead separation (Miltenyi Biotec Auburn, CA). IFNγ ELISPOT kits were obtained from BD Biosciences (San Diego, CA) and assays were conducted according to established protocols (Ryan et al., 2005a, Ryan et al., 2005b) adapted for monitoring vaccinia-specific responses (Ennis et al., 2002, Hammarlund et al., 2003). Briefly, 200,000 PBMCs were plated in each well. Peptide pools (consisting of 9–11 peptides each) were added at a final concentration of ~ 10 μg/ml, while individual peptides were added at concentration of 30 μg/ml. Vaccinia virus was inactivated with Psoralen and UV light (Crotty et al., 2003) and inactivated viral particles were added to individual wells at an MOI of 5.0 (based on pre-inactivation titration). Plates were incubated for 24 h, washed and developed as per manufacturer's instructions. All plates were then scanned and analyzed on an ImmunoSpot® S4 Pro Analyzer (Cellular Technology Ltd., Cleveland, OH, USA) using ImmunoSpot® version 4.0 software (Cellular Technology Ltd.).
Statistical analyses
For ELISPOT assays, the number of spots per million cells was calculated for each well. The average spot number per experimental group was compared to the negative control wells (medium alone or DMSO) using a two-tailed student's t test. Positive responses were defined as those with an average spot number 1.5 fold over background values and with p ≤ 0.05. The peptides reported here had positive results in at least two separate experiments. For the initial screening of peptide pools a more generous p-value of 0.10 was used. False positive results were minimized by the more rigorous statistical standards in the subsequent screening of individual peptides and the requirement for positive signals in multiple experiments.
Acknowledgments
This work was sponsored by the NIH/NIAID Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research (RCE) Program. The authors wish to acknowledge membership within and support from the Region V ‘Great Lakes’ RCE (NIH award 2-U54-AI-057153) as well as from NIH award AI40065.
References
- Adamopoulou E., Diekmann J., Tolosa E., Kuntz G., Einsele H., Rammensee H.G., Topp M.S. Human CD4+ T cells displaying viral epitopes elicit a functional virus-specific memory CD8+ T cell response. J. Immunol. 2007;178(9):5465–5472. doi: 10.4049/jimmunol.178.9.5465. [DOI] [PubMed] [Google Scholar]
- Artenstein A.W. New generation smallpox vaccines: a review of preclinical and clinical data. Rev. Med. Virol. 2008;18(4):217–231. doi: 10.1002/rmv.571. [DOI] [PubMed] [Google Scholar]
- Bell E., Shamim M., Whitbeck J.C., Sfyroera G., Lambris J.D., Isaacs S.N. Antibodies against the extracellular enveloped virus B5R protein are mainly responsible for the EEV neutralizing capacity of vaccinia immune globulin. Virology. 2004;325(2):425–431. doi: 10.1016/j.virol.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Bernard A., Coitot S., Bremont A., Bernard G. T and B cell cooperation: a dance of life and death. Transplantation. 2005;79(3 Suppl):S8–S11. doi: 10.1097/01.tp.0000153290.75695.31. [DOI] [PubMed] [Google Scholar]
- Bishop G.A., Hostager B.S. B lymphocyte activation by contact-mediated interactions with T lymphocytes. Curr. Opin. Immunol. 2001;13(3):278–285. doi: 10.1016/s0952-7915(00)00216-8. [DOI] [PubMed] [Google Scholar]
- Calvo-Calle J.M., Strug I., Nastke M.D., Baker S.P., Stern L.J. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 2007;3(10):1511–1529. doi: 10.1371/journal.ppat.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.D., Fraire A.E., Joris I., Brehm M.A., Welsh R.M., Selin L.K. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat. Immunol. 2001;2(11):1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- Chung C.S., Hsiao J.C., Chang Y.S., Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 1998;72(2):1577–1585. doi: 10.1128/jvi.72.2.1577-1585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute S.C., Watkin L.B., Cornberg M., Naumov Y.N., Sullivan J.L., Luzuriaga K., Welsh R.M., Selin L.K. Cross-reactive influenza virus-specific CD8+ T cells contribute to lymphoproliferation in Epstein–Barr virus-associated infectious mononucleosis. J. Clin. Invest. 2005;115(12):3602–3612. doi: 10.1172/JCI25078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combadiere B., Boissonnas A., Carcelain G., Lefranc E., Samri A., Bricaire F., Debre P., Autran B. Distinct time effects of vaccination on long-term proliferative and IFN-gamma-producing T cell memory to smallpox in humans. J. Exp. Med. 2004;199(11):1585–1593. doi: 10.1084/jem.20032083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornberg M., Sheridan B.S., Saccoccio F.M., Brehm M.A., Selin L.K. Protection against vaccinia virus challenge by CD8 memory T cells resolved by molecular mimicry. J. Virol. 2007;81(2):934–944. doi: 10.1128/JVI.01280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornberg M., Clute S.C., Watkin L.B., Saccoccio F.M., Kim S.K., Naumov Y.N., Brehm M.A., Aslan N., Welsh R.M., Selin L.K. CD8 T cell cross-reactivity networks mediate heterologous immunity in human EBV and murine vaccinia virus infections. J. Immunol. 2010;184(6):2825–2838. doi: 10.4049/jimmunol.0902168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S., Felgner P., Davies H., Glidewell J., Villarreal L., Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J. Immunol. 2003;171(10):4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- Davies D.H., Molina D.M., Wrammert J., Miller J., Hirst S., Mu Y., Pablo J., Unal B., Nakajima-Sasaki R., Liang X., Crotty S., Karem K.L., Damon I.K., Ahmed R., Villarreal L., Felgner P.L. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7(10):1678–1686. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- Davies D.H., Wyatt L.S., Newman F.K., Earl P.L., Chun S., Hernandez J.E., Molina D.M., Hirst S., Moss B., Frey S.E., Felgner P.L. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J. Virol. 2008;82(2):652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler I., Staib C., Kastenmuller W., Stevanovic S., Schmidt B., Lemonnier F.A., Rammensee H.G., Busch D.H., Bernhard H., Erfle V., Sutter G. Identification of vaccinia virus epitope-specific HLA-A*0201-restricted T cells and comparative analysis of smallpox vaccines. Proc. Natl. Acad. Sci. USA. 2003;100(1):217–222. doi: 10.1073/pnas.262668999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke-Cohan J.S., Wollenick K., Witten E.A., Seaman M.S., Baden L.R., Dolin R., Reinherz E.L. The heterogeneity of human antibody responses to vaccinia virus revealed through use of focused protein arrays. Vaccine. 2009;27(8):1154–1165. doi: 10.1016/j.vaccine.2008.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Ad B., Roth Y., Winder A., Tochner Z., Lublin-Tennenbaum T., Katz E., Schwartz T. The persistence of neutralizing antibodies after revaccination against smallpox. J. Infect. Dis. 1990;161(3):446–448. doi: 10.1093/infdis/161.3.446. [DOI] [PubMed] [Google Scholar]
- Ennis F.A., Cruz J., Demkowicz W.E., Jr., Rothman A.L., McClain D.J. Primary induction of human CD8+ cytotoxic T lymphocytes and interferon-gamma-producing T cells after smallpox vaccination. J. Infect. Dis. 2002;185(11):1657–1659. doi: 10.1086/340517. [DOI] [PubMed] [Google Scholar]
- Fenner F. A successful eradication campaign. Global eradication of smallpox. Rev. Infect. Dis. 1982;4(5):916–930. doi: 10.1093/clinids/4.5.916. [DOI] [PubMed] [Google Scholar]
- Fenner F., Henderson D.A., Arita I., Jezek Z., Ladnyi I.D. Smallpox and Its Eradication. Vol. 6. W.H.O.; Geneva: 1988. (History of International Public Health). [Google Scholar]
- Ferrier-Rembert A., Drillien R., Tournier J.N., Garin D., Crance J.M. Short- and long-term immunogenicity and protection induced by non-replicating smallpox vaccine candidates in mice and comparison with the traditional 1st generation vaccine. Vaccine. 2008;26(14):1794–1804. doi: 10.1016/j.vaccine.2007.12.059. [DOI] [PubMed] [Google Scholar]
- Fogg C., Lustig S., Whitbeck J.C., Eisenberg R.J., Cohen G.H., Moss B. Protective immunity to vaccinia virus induced by vaccination with multiple recombinant outer membrane proteins of intracellular and extracellular virions. J. Virol. 2004;78(19):10230–10237. doi: 10.1128/JVI.78.19.10230-10237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S.E., Newman F.K., Yan L., Lottenbach K.R., Belshe R.B. Response to smallpox vaccine in persons immunized in the distant past. JAMA. 2003;289(24):3295–3299. doi: 10.1001/jama.289.24.3295. [DOI] [PubMed] [Google Scholar]
- Fulginiti V.A. Risks of smallpox vaccination. JAMA. 2003;290(11):1452. doi: 10.1001/jama.290.11.1452-a. (author reply 1452) [DOI] [PubMed] [Google Scholar]
- Fulginiti V.A., Papier A., Lane J.M., Neff J.M., Henderson D.A. Smallpox vaccination: a review, part II. Adverse events. Clin. Infect. Dis. 2003;37(2):251–271. doi: 10.1086/375825. [DOI] [PubMed] [Google Scholar]
- Galmiche M.C., Goenaga J., Wittek R., Rindisbacher L. Neutralizing and protective antibodies directed against vaccinia virus envelope antigens. Virology. 1999;254(1):71–80. doi: 10.1006/viro.1998.9516. [DOI] [PubMed] [Google Scholar]
- Golden J.W., Josleyn M.D., Hooper J.W. Targeting the vaccinia virus L1 protein to the cell surface enhances production of neutralizing antibodies. Vaccine. 2008;26(27–28):3507–3515. doi: 10.1016/j.vaccine.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Goldstein J.A., Neff J.M., Lane J.M., Koplan J.P. Smallpox vaccination reactions, prophylaxis, and therapy of complications. Pediatrics. 1975;55(3):342–347. [PubMed] [Google Scholar]
- Golovkin M., Spitsin S., Andrianov V., Smirnov Y., Xiao Y., Pogrebnyak N., Markley K., Brodzik R., Gleba Y., Isaacs S.N., Koprowski H. Smallpox subunit vaccine produced in planta confers protection in mice. Proc. Natl. Acad. Sci. USA. 2007;104(16):6864–6869. doi: 10.1073/pnas.0701451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E., Lewis M.W., Hansen S.G., Strelow L.I., Nelson J.A., Sexton G.J., Hanifin J.M., Slifka M.K. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 2003;9(9):1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- Hammer J., Bono E., Gallazzi F., Belunis C., Nagy Z., Sinigaglia F. Precise prediction of major histocompatibility complex class II–peptide interaction based on peptide side chain scanning. J. Exp. Med. 1994;180(6):2353–2358. doi: 10.1084/jem.180.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D.A. The looming threat of bioterrorism. Science. 1999;283(5406):1279–1282. doi: 10.1126/science.283.5406.1279. [DOI] [PubMed] [Google Scholar]
- Henderson D.A., Inglesby T.V., Bartlett J.G., Ascher M.S., Eitzen E., Jahrling P.B., Hauer J., Layton M., McDade J., Osterholm M.T., O'Toole T., Parker G., Perl T., Russell P.K., Tonat K. Smallpox as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 1999;281(22):2127–2137. doi: 10.1001/jama.281.22.2127. [DOI] [PubMed] [Google Scholar]
- Heraud J.M., Edghill-Smith Y., Ayala V., Kalisz I., Parrino J., Kalyanaraman V.S., Manischewitz J., King L.R., Hryniewicz A., Trindade C.J., Hassett M., Tsai W.P., Venzon D., Nalca A., Vaccari M., Silvera P., Bray M., Graham B.S., Golding H., Hooper J.W., Franchini G. Subunit recombinant vaccine protects against monkeypox. J. Immunol. 2006;177(4):2552–2564. doi: 10.4049/jimmunol.177.4.2552. [DOI] [PubMed] [Google Scholar]
- Hooper J.W., Custer D.M., Schmaljohn C.S., Schmaljohn A.L. DNA vaccination with vaccinia virus L1R and A33R genes protects mice against a lethal poxvirus challenge. Virology. 2000;266(2):329–339. doi: 10.1006/viro.1999.0096. [DOI] [PubMed] [Google Scholar]
- Hooper J.W., Custer D.M., Thompson E. Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology. 2003;306(1):181–195. doi: 10.1016/S0042-6822(02)00038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.F., Yang Y., Sepulveda H., Shi W., Hwang I., Peterson P.A., Jackson M.R., Sprent J., Cai Z. TCR-mediated internalization of peptide–MHC complexes acquired by T cells. Science. 1999;286(5441):952–954. doi: 10.1126/science.286.5441.952. [DOI] [PubMed] [Google Scholar]
- Isaacs S.N., Wolffe E.J., Payne L.G., Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 1992;66(12):7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahrling P.B., Fritz E.A., Hensley L.E. Countermeasures to the bioterrorist threat of smallpox. Curr. Mol. Med. 2005;5(8):817–826. doi: 10.2174/156652405774962326. [DOI] [PubMed] [Google Scholar]
- Jenner E. Law; London: 1798. An inquiry into the causes and effects of the variolae vaccinae, a disease discovered in some of the western counties of England, particularly Gloucestershire, and known by the name of the cow pox. [Google Scholar]
- Jing L., Chong T.M., McClurkan C.L., Huang J., Story B.T., Koelle D.M. Diversity in the acute CD8 T cell response to vaccinia virus in humans. J. Immunol. 2005;175(11):7550–7559. doi: 10.4049/jimmunol.175.11.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing L., Chong T.M., Byrd B., McClurkan C.L., Huang J., Story B.T., Dunkley K.M., Aldaz-Carroll L., Eisenberg R.J., Cohen G.H., Kwok W.W., Sette A., Koelle D.M. Dominance and diversity in the primary human CD4 T cell response to replication-competent vaccinia virus. J. Immunol. 2007;178(10):6374–6386. doi: 10.4049/jimmunol.178.10.6374. [DOI] [PubMed] [Google Scholar]
- Jing L., Davies D.H., Chong T.M., Chun S., McClurkan C.L., Huang J., Story B.T., Molina D.M., Hirst S., Felgner P.L., Koelle D.M. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J. Virol. 2008;82(14):7120–7134. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.L., Ovsyannikova I.G., Madden B.J., Poland G.A., Muddiman D.C. Accurate mass precursor ion data and tandem mass spectrometry identify a class I human leukocyte antigen A*0201-presented peptide originating from vaccinia virus. J. Am. Soc. Mass Spectrom. 2005;16(11):1812–1817. doi: 10.1016/j.jasms.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Kan V.L., Manischewitz J., King L.R., Golding H. Durable neutralizing antibodies after remote smallpox vaccination among adults with and without HIV infection. AIDS. 2007;21(4):521–524. doi: 10.1097/QAD.0b013e32802f7d7c. [DOI] [PubMed] [Google Scholar]
- Kennedy R., Poland G.A. T-Cell epitope discovery for variola and vaccinia viruses. Rev. Med. Virol. 2007;17(2):93–113. doi: 10.1002/rmv.527. [DOI] [PubMed] [Google Scholar]
- Kennedy R., Undale A.H., Kieper W.C., Block M.S., Pease L.R., Celis E. Direct cross-priming by th lymphocytes generates memory cytotoxic T cell responses. J. Immunol. 2005;174(7):3967–3977. doi: 10.4049/jimmunol.174.7.3967. [DOI] [PubMed] [Google Scholar]
- Kennedy R., Pankratz V.S., Swanson E., Watson D., Golding H., Poland G.A. Statistical approach to estimate vaccinia-specific neutralizing antibody titers using a high throughput assay. Clin. Vaccine Immunol. 2009;16(8):1105–1112. doi: 10.1128/CVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy R.B., Ovsyannikova I., Poland G.A. Smallpox vaccines for biodefense. Vaccine. 2009;27(Suppl 4):D73–D79. doi: 10.1016/j.vaccine.2009.07.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy R.B., Ovsyannikova I.G., Jacobson R.M., Poland G.A. The immunology of smallpox vaccines. Curr. Opin. Immunol. 2009;21(3):314–320. doi: 10.1016/j.coi.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J.M., Ruben F.L., Abrutyn E., Millar J.D. Deaths attributable to smallpox vaccination, 1959 to 1966, and 1968. JAMA. 1970;212(3):441–444. [PubMed] [Google Scholar]
- Lanzavecchia A. Antigen-specific interaction between T and B cells. Nature. 1985;314(6011):537–539. doi: 10.1038/314537a0. [DOI] [PubMed] [Google Scholar]
- Lanzavecchia A., Bove S. Specific B lymphocytes efficiently pick up, process and present antigen to T cells. Behring Inst. Mitt. 1985;(77):82–87. [PubMed] [Google Scholar]
- Larkin M. Monkeypox spreads as US public-health system plays catch-up. Lancet Infect. Dis. 2003;3(8):461. doi: 10.1016/S1473-3099(03)00713-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence S.J., Lottenbach K.R., Newman F.K., Buller R.M., Bellone C.J., Chen J.J., Cohen G.H., Eisenberg R.J., Belshe R.B., Stanley S.L., Jr., Frey S.E. Antibody responses to vaccinia membrane proteins after smallpox vaccination. J. Infect. Dis. 2007;196(2):220–229. doi: 10.1086/518793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manischewitz J., King L.R., Bleckwenn N.A., Shiloach J., Taffs R., Merchlinsky M., Eller N., Mikolajczyk M.G., Clanton D.J., Monath T., Weltzin R.A., Scott D.E., Golding H. Development of a novel vaccinia-neutralization assay based on reporter-gene expression. J. Infect. Dis. 2003;188(3):440–448. doi: 10.1086/376557. [DOI] [PubMed] [Google Scholar]
- Mathew A., Terajima M., West K., Green S., Rothman A.L., Ennis F.A., Kennedy J.S. Identification of murine poxvirus-specific CD8+ CTL epitopes with distinct functional profiles. J. Immunol. 2005;174(4):2212–2219. doi: 10.4049/jimmunol.174.4.2212. [DOI] [PubMed] [Google Scholar]
- Mayr A. Smallpox vaccination and bioterrorism with pox viruses. Comp. Immunol. Microbiol. Infect. Dis. 2003;26(5–6):423–430. doi: 10.1016/S0147-9571(03)00025-0. [DOI] [PubMed] [Google Scholar]
- Meister G.E., Roberts C.G., Berzofsky J.A., De Groot A.S. Two novel T cell epitope prediction algorithms based on MHC-binding motifs; comparison of predicted and published epitopes from Mycobacterium tuberculosis and HIV protein sequences. Vaccine. 1995;13(6):581–591. doi: 10.1016/0264-410x(94)00014-e. [DOI] [PubMed] [Google Scholar]
- Metzger W., Mordmueller B.G. Vaccines for preventing smallpox. Cochrane Database Syst. Rev. 2007;(3) doi: 10.1002/14651858.CD004913.pub2. (CD004913) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra-Kaushik S., Cruz J., Stern L.J., Ennis F.A., Terajima M. Human cytotoxic CD4+ T cells recognize HLA-DR1-restricted epitopes on vaccinia virus proteins A24R and D1R conserved among poxviruses. J. Immunol. 2007;179(2):1303–1312. doi: 10.4049/jimmunol.179.2.1303. [DOI] [PubMed] [Google Scholar]
- Moise L., McMurry J.A., Buus S., Frey S., Martin W.D., De Groot A.S. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine. 2009 doi: 10.1016/j.vaccine.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J., Roper M.H., Sperling L., Schieber R.A., Heffelfinger J.D., Casey C.G., Miller J.W., Santibanez S., Herwaldt B., Hightower P., Moro P.L., Hibbs B.F., Levine N.H., Chapman L.E., Iskander J., Lane J.M., Wharton M., Mootrey G.T., Swerdlow D.L. Myocarditis, pericarditis, and dilated cardiomyopathy after smallpox vaccination among civilians in the United States, January–October 2003. Clin. Infect. Dis. 2008;46(Suppl 3):S242–S250. doi: 10.1086/524747. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M., Peters B., Pasquetto V., Tscharke D.C., Sidney J., Bui H.H., Grey H., Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24(7):817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M., Tscharke D.C., Vaughan K., Koelle D.M., Stern L., Calvo-Calle M., Ennis F., Terajima M., Sutter G., Crotty S., Drexler I., Franchini G., Yewdell J.W., Head J.W., Blum J., Peters B., Sette A. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future Microbiol. 2010;5(2):221–239. doi: 10.2217/fmb.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff J.M., Lane J.M., Pert J.H., Moore R., Millar J.D., Henderson D.A. Complications of smallpox vaccination. I. National survey in the United States, 1963. N. Engl. J. Med. 1967;276(3):125–132. doi: 10.1056/NEJM196701192760301. [DOI] [PubMed] [Google Scholar]
- Noelle R.J., Snow E.C. T helper cell-dependent B cell activation. FASEB J. 1991;5(13):2770–2776. doi: 10.1096/fasebj.5.13.1833257. [DOI] [PubMed] [Google Scholar]
- Oseroff C., Kos F., Bui H.H., Peters B., Pasquetto V., Glenn J., Palmore T., Sidney J., Tscharke D.C., Bennink J.R., Southwood S., Grey H.M., Yewdell J.W., Sette A. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc. Natl. Acad. Sci. USA. 2005;102(39):13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero M., Calarota S.A., Dai A., De Groot A.S., Boyer J.D., Weiner D.B. Efficacy of novel plasmid DNA encoding vaccinia antigens in improving current smallpox vaccination strategy. Vaccine. 2006;24(21):4461–4470. doi: 10.1016/j.vaccine.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Pasquetto V., Bui H.H., Giannino R., Banh C., Mirza F., Sidney J., Oseroff C., Tscharke D.C., Irvine K., Bennink J.R., Peters B., Southwood S., Cerundolo V., Grey H., Yewdell J.W., Sette A. HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J. Immunol. 2005;175(8):5504–5515. doi: 10.4049/jimmunol.175.8.5504. [DOI] [PubMed] [Google Scholar]
- Pasquetto V., Bui H.H., Giannino R., Mirza F., Sidney J., Oseroff C., Tscharke D.C., Irvine K., Bennink J.R., Peters B., Southwood S., Cerundolo V., Grey H., Yewdell J.W., Sette A. HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J. Immunol. 2005;175(8):5504–5515. doi: 10.4049/jimmunol.175.8.5504. [DOI] [PubMed] [Google Scholar]
- Poland G.A. Smallpox vaccines: from first to second to third generation. Lancet. 2005;365(9457):362–363. doi: 10.1016/S0140-6736(05)17840-4. [DOI] [PubMed] [Google Scholar]
- Rammensee H.G., Friede T., Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41(4):178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- Rammensee H., Bachmann J., Emmerich N.P., Bachor O.A., Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3–4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Roederer M., Koup R.A. Optimized determination of T cell epitope responses. J. Immunol. Methods. 2003;274(1–2):221–228. doi: 10.1016/s0022-1759(02)00423-4. [DOI] [PubMed] [Google Scholar]
- Roomp K., Antes I., Lengauer T. Predicting MHC class I epitopes in large datasets. BMC Bioinformatics. 2010;11:90. doi: 10.1186/1471-2105-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper R.L., Wolffe E.J., Weisberg A., Moss B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 1998;72(5):4192–4204. doi: 10.1128/jvi.72.5.4192-4204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J.E., Ovsyannikova I.G., Dhiman N., Pinsky N.A., Vierkant R.A., Jacobson R.M., Poland G.A. Inter-operator variation in ELISPOT analysis of measles virus-specific IFN-gamma-secreting T cells. Scand. J. Clin. Lab. Invest. 2005;65(8):681–689. doi: 10.1080/00365510500348252. [DOI] [PubMed] [Google Scholar]
- Ryan J.E., Ovsyannikova I.G., Poland G.A. Detection of measles virus-specific interferon-gamma-secreting T-cells by ELISPOT. Methods Mol. Biol. 2005;302:207–218. doi: 10.1385/1-59259-903-6:207. [DOI] [PubMed] [Google Scholar]
- Sakhatskyy P., Wang S., Zhang C., Chou T.H., Kishko M., Lu S. Immunogenicity and protection efficacy of subunit-based smallpox vaccines using variola major antigens. Virology. 2008;371(1):98–107. doi: 10.1016/j.virol.2007.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selin L.K., Varga S.M., Wong I.C., Welsh R.M. Protective heterologous antiviral immunity and enhanced immunopathogenesis mediated by memory T cell populations. J. Exp. Med. 1998;188(9):1705–1715. doi: 10.1084/jem.188.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Moutaftsi M., Moyron-Quiroz J., McCausland M.M., Davies D.H., Johnston R.J., Peters B., Rafii-El-Idrissi Benhnia M., Hoffmann J., Su H.P., Singh K., Garboczi D.N., Head S., Grey H., Felgner P.L., Crotty S. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28(6):847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Grey H., Oseroff C., Peters B., Moutaftsi M., Crotty S., Assarsson E., Greenbaum J., Kim Y., Kolla R., Tscharke D., Koelle D., Johnson R.P., Blum J., Head S., Sidney J. Definition of epitopes and antigens recognized by vaccinia specific immune responses: their conservation in variola virus sequences, and use as a model system to study complex pathogens. Vaccine. 2009;27(Suppl 6):G21–G26. doi: 10.1016/j.vaccine.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Assarsson E., Moore C., Ngo S., Pinilla C., Sette A., Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirven P., Castelli F.A., Probst A., Szely N., Maillere B. In vitro human CD4+ T cell response to the vaccinia protective antigens B5R and A33R. Mol. Immunol. 2009;46(7):1481–1487. doi: 10.1016/j.molimm.2008.12.016. [DOI] [PubMed] [Google Scholar]
- Snyder J.T., Belyakov I.M., Dzutsev A., Lemonnier F., Berzofsky J.A. Protection against lethal vaccinia virus challenge in HLA-A2 transgenic mice by immunization with a single CD8+ T-cell peptide epitope of vaccinia and variola viruses. J. Virol. 2004;78(13):7052–7060. doi: 10.1128/JVI.78.13.7052-7060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strug I., Calvo-Calle J.M., Green K.M., Cruz J., Ennis F.A., Evans J.E., Stern L.J. Vaccinia peptides eluted from HLA-DR1 isolated from virus-infected cells are recognized by CD4+ T cells from a vaccinated donor. J. Proteome Res. 2008;7(7):2703–2711. doi: 10.1021/pr700780x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Murtadha M., Schnell M., Eisenlohr L.C., Hooper J., Flomenberg P. Human T-cell responses to vaccinia virus envelope proteins. J. Virol. 2006;80(20):10010–10020. doi: 10.1128/JVI.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terajima M., Cruz J., Raines G., Kilpatrick E.D., Kennedy J.S., Rothman A.L., Ennis F.A. Quantitation of CD8+ T cell responses to newly identified HLA-A*0201-restricted T cell epitopes conserved among vaccinia and variola (smallpox) viruses. J. Exp. Med. 2003;197(7):927–932. doi: 10.1084/jem.20022222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terajima M., Orphin L., Leporati A.M., Pazoles P., Cruz J., Rothman A.L., Ennis F.A. Vaccinia virus-specific CD8(+) T-cell responses target a group of epitopes without a strong immunodominance hierarchy in humans. Hum. Immunol. 2008;69(12):815–825. doi: 10.1016/j.humimm.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J.C., Tan T.W., Ranganathan S. Methods and protocols for prediction of immunogenic epitopes. Brief. Bioinform. 2007;8(2):96–108. doi: 10.1093/bib/bbl038. [DOI] [PubMed] [Google Scholar]
- Tscharke D.C., Karupiah G., Zhou J., Palmore T., Irvine K.R., Haeryfar S.M., Williams S., Sidney J., Sette A., Bennink J.R., Yewdell J.W. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 2005;201(1):95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscharke D.C., Woo W.P., Sakala I.G., Sidney J., Sette A., Moss D.J., Bennink J.R., Karupiah G., Yewdell J.W. Poxvirus CD8+ T-cell determinants and cross-reactivity in BALB/c mice. J. Virol. 2006;80(13):6318–6323. doi: 10.1128/JVI.00427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeshappa C.S., Huang H., Xie Y., Wei Y., Mulligan S.J., Deng Y., Xiang J. CD4+ Th-APC with acquired peptide/MHC class I and II complexes stimulate type 1 helper CD4+ and central memory CD8+ T cell responses. J. Immunol. 2009;182(1):193–206. doi: 10.4049/jimmunol.182.1.193. [DOI] [PubMed] [Google Scholar]
- Urbani S., Amadei B., Fisicaro P., Pilli M., Missale G., Bertoletti A., Ferrari C. Heterologous T cell immunity in severe hepatitis C virus infection. J. Exp. Med. 2005;201(5):675–680. doi: 10.1084/jem.20041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh S.R., Gillis J., Peters B., Mothe B.R., Sidney J., Sette A., Johnson R.P. Diverse recognition of conserved orthopoxvirus CD8+ T cell epitopes in vaccinated rhesus macaques. Vaccine. 2009;27(36):4990–5000. doi: 10.1016/j.vaccine.2009.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tang S.T., Lund O., Dziegiel M.H., Buus S., Claesson M.H. High-affinity human leucocyte antigen class I binding variola-derived peptides induce CD4+ T cell responses more than 30 years post-vaccinia virus vaccination. Clin. Exp. Immunol. 2009;155(3):441–446. doi: 10.1111/j.1365-2249.2008.03856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe E.J., Vijaya S., Moss B. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology. 1995;211(1):53–63. doi: 10.1006/viro.1995.1378. [DOI] [PubMed] [Google Scholar]