Preface
Genome-wide association studies (GWAS) are an effective approach for identifying genetic variants associated to disease risk. GWAS can be confounded by population stratification—systematic ancestry differences between cases and controls—which has previously been addressed by methods that infer genetic ancestry. Those methods perform well in data sets in which population structure is the only kind of structure present, but are inadequate in data sets that also contain family structure or cryptic relatedness. Here, we review recent progress on methods that correct for stratification while accounting for these additional complexities.
GWAS have identified hundreds of common variants associated to disease risk or related traits1 (see Web Resources). These studies have overcome the dangers of population stratification, which can produce spurious associations if not properly corrected2–3. However, accounting for population structure is more challenging when family structure or cryptic relatedness is also present, motivating the development of new methods. Because the spurious associations that have been reported primarily occur at markers with unusual allele frequency differences between subpopulations2, 4, it is critical for new methods aiming to correct for stratification to be evaluated by considering unusually differentiated markers.
The prevailing paradigm in recent years has been to use Genomic Control to measure the extent of inflation due to population stratification or other confounders, and to correct for stratification (if necessary) using methods that infer genetic ancestry, such as Structured Association or Principal Components Analysis. A limitation of this strategy is that it fails to account for other types of sample structure, such as family structure or cryptic relatedness5–6. Modeling family structure is a necessity in studies with family-based sample ascertainment, and there is increasing evidence that cryptic relatedness may occur in a wide range of data sets (see below). Family-Based Association Tests offer one potential solution for dealing with family structure. More recently, approaches using Mixed Models that incorporate the full covariance structure across individuals have been proposed.
Below, we review each of these methods, conduct simulations to evaluate their performance, discuss stratification in the specific context of low-frequency or rare variants, and conclude with guidelines and recommendations.
Detecting Stratification
A widely used approach to evaluate whether confounding due to population stratification exists is to compute the Genomic Control λ (λGC), defined as the median χ2(1 dof) association statistic across SNPs divided by its theoretical median under the null distribution7–9. A value of λGC≈1 indicates no stratification, whereas λGC>1 indicates stratification or other confounders such as family structure or cryptic relatedness (see below), or differential bias10. P-P plots are a standard tool for visualization of test statistics (Figure 1). Values of λGC<1.05 are generally considered benign, although inflation in λGC is proportional to sample size.
Figure 1. P-P plots for visualization of stratification or other confounders.
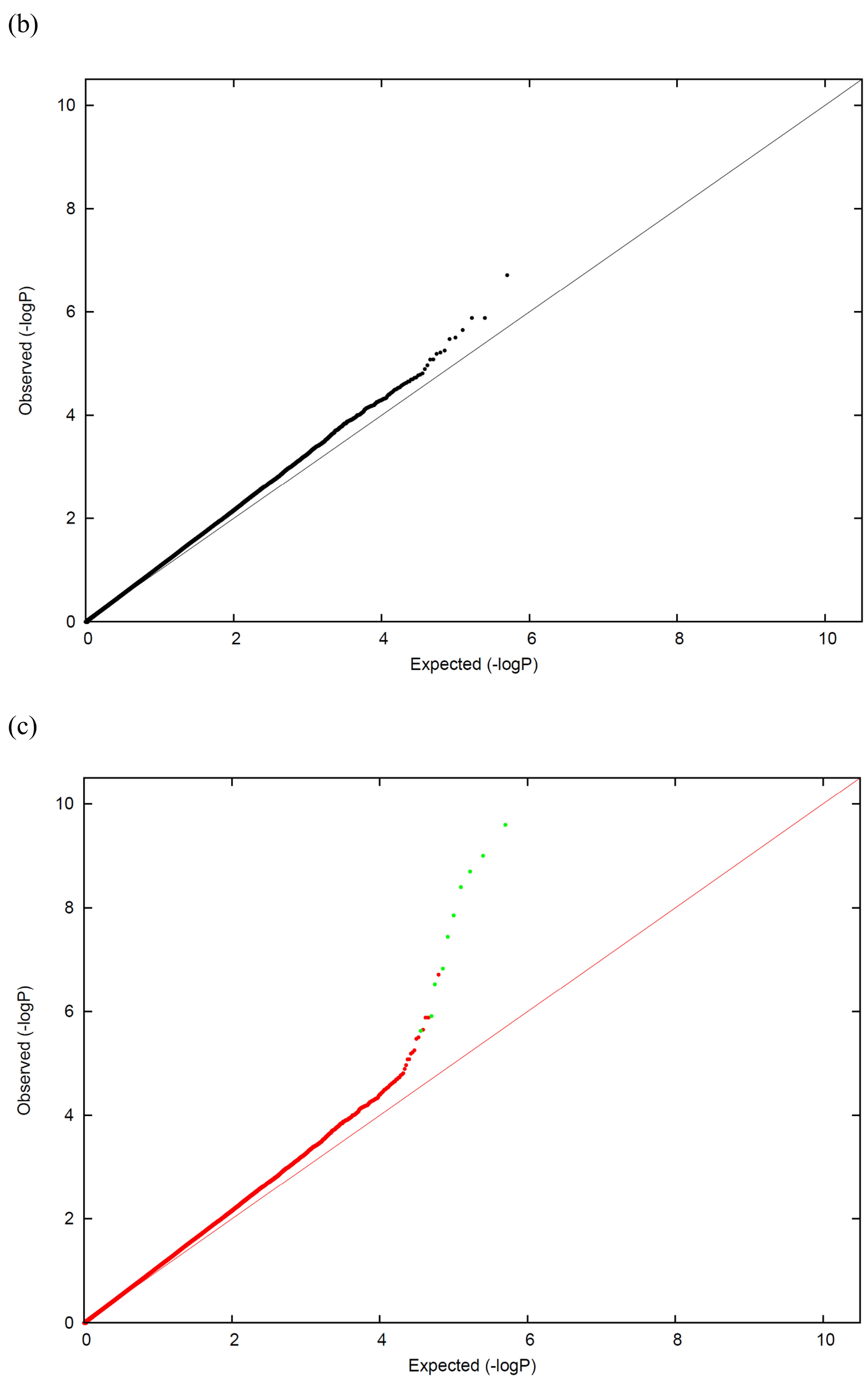
We display simulated P-P plots for genome-wide scans with no causal markers under three scenarios. (a) No stratification: P-values fit the expected distribution. (b) Stratification without unusually differentiated markers: P-values exhibit modest genome-wide inflation. (c) Stratification with unusually differentiated markers: P-value exhibit modest genome-wide inflation, plus severe inflation at a small number of markers.
If population stratification exists, it is important to distinguish between subpopulation differences that are due to very recent genetic drift, and those that arose from more ancient population divergence11. In the case of ancient population divergence, dividing association statistics by λGC will provide a sufficient correction for stratification. In the latter case, markers with unusual allele frequency differences that lie outside the expected distribution, which could be caused by natural selection, make stratification a much more severe problem, and dividing association statistics by λGC is likely to be inadequate. In the case of family structure or cryptic relatedness, dividing association statistics by λGC will generally produce the approximate null distribution, though a refinement to the method may be needed when there is uncertainty in the estimate of λGC 12. However, even if the appropriate null distribution is obtained, in general this approach will not maximize power to detect true associations. Other approaches to correcting for stratification, including approaches that also account for family structure and cryptic relatedness, are described below.
Inferring Genetic Ancestry
Structured Association
Methods that explicitly infer genetic ancestry generally provide an effective correction for population stratification in data sets where population structure is the only type of sample structure. In the Structured Association approach, samples are assigned to subpopulation clusters (possibly allowing fractional cluster membership) using a model-based clustering program such as STRUCTURE13–14, and association statistics are computed by stratifying by cluster (STRAT; see Web Resources)15. The applicability of this approach to large genome-wide data sets has historically been limited by its high computational cost when allowing fractional cluster membership, but faster model-based approaches for inferring population structure have recently been developed16 (ADMIXTURE; see Web Resources). Thus, applying Structured Association to both infer population structure and compute association statistics in genome-wide data sets is likely to become a practical approach.
Principal Components Analysis
Principal Components Analysis (PCA) is a tool that has been used to infer population structure in genetic data for several decades, long before the GWAS era17–20. It should be noted that top PCs do not always reflect population structure: they may reflect family relatedness19, long-range LD (for example, due to inversion polymorphisms4), or assay artifacts10; these effects can often be eliminated by removing related samples, regions of long-range LD, or low-quality data, respectively, from the data used to compute PCs. In addition, PCA can highlight effects of differential bias that require additional quality control21.
Using top PCs as covariates corrects for stratification in GWAS21–22 (EIGENSTRAT; see Web Resources). Like Structured Association, PCA will appropriately apply a greater correction to markers with large differences in allele frequency across ancestral populations. Unlike initial implementations of Structured Association, PCA is computationally tractable in large genome-wide data sets. Related approaches such as Multi-Dimensional Scaling (MDS) and Genetic Matching have also proven useful23–24 (PLINK; see Web Resources). When genome-wide data are not available (for example, in replication studies), Structured Association or PCA can infer genetic ancestry, and hence correct for stratification, using Ancestry-Informative Markers (AIMs)25. A common misconception is that AIMs should be used to infer genetic ancestry even when genome-wide data is available, but in fact the best ancestry estimates are obtained using a very large number of random markers.
A limitation of the above methods is that they do not model family structure or cryptic relatedness. These factors may lead to inflation in test statistics if not explicitly modeled, because samples that are correlated are assumed to be uncorrelated. Although correcting for genetic ancestry and then dividing by the residual λGC will restore an appropriate null distribution, association statistics that explicitly account for family structure or cryptic relatedness are likely to achieve higher power, due to improved weighting of the data.
Family-Based Association Tests
Family-based studies, in which individuals are ascertained from family pedigrees, offer a unique solution to population stratification. Family-Based Association Tests that focus on within-family information (generalizing the Transmission Disequilibrium Test26) are immune to stratification, since transmitted and untransmitted alleles have the same genetic ancestry27–29 (FBAT and QTDT; see Web Resources). However, fully powered statistics for family-based studies will need to incorporate between-family information, which is still susceptible to stratification. A recent suggestion is to transform between-family information into a rank statistic before combining within-family and between-family information, guaranteeing that both sources of information are immune to stratification30–31. This approach performs favorably compared to previous family-based approaches30–31, but places an upper bound on the statistical power that can be extracted from the between-family component of the overall signal, because the transformed rank statistic cannot be more statistically significant than one divided by the number of samples.
Mixed Models
Mixed models can model population structure, family structure and cryptic relatedness32. The basic approach is to model phenotypes using a mixture of fixed effects and random effects. Fixed effects include the candidate SNP and optional covariates such as gender or age, while random effects are based on a phenotypic covariance matrix, which is modeled as a sum of heritable and non-heritable random variation (see Box 1 for details). Mixed models have historically been a theoretically appealing but computationally intensive approach; however, very recent computational advances have now made it possible to apply them to GWAS33–34 (EMMAX and TASSEL; see Web Resources). Methods that explicitly model population structure, family structure and cryptic relatedness are expected to perform better in the presence of these complexities than methods that do not, and this has now been confirmed33–34. For example, in an analysis of seven Wellcome Trust Case Control Consortium phenotypes, the application of mixed models consistently yielded values of λGC that were less than 1.01, in contrast to other approaches33.
Box 1. Mixed models
Simple linear models
Simple linear models represent the phenotype Y as function of fixed effects X :
Here X denotes the genotype at the candidate marker as well as optional covariates such as gender or age, B denotes coefficients of fixed effects, and ε is a normally distributed noise term that accounts for unexplained variation in Y.
PCA addresses the issue of population substructure by including PC covariates in X to explicitly model the ancestry of each individual. If genotype is not causally related to phenotype but genotype and phenotype are both correlated to ancestry, test statistics will be inflated. Using PCA to explicitly model genetic ancestry removes this confounding effect. However, PCA only accounts for fixed effects of genetic ancestry; it does not account for relatedness between individuals, which may also cause inflation in test statistics.
Linear mixed models
Linear mixed models represent the phenotype Y as a function of fixed effects X plus random effects u:
Here u denotes a component of the overall noise variance u + ε that is distributed according to a kinship matrix K. Thus, u represents the heritable component of random variation and ε represents the non-heritable component of random variation.
The kinship matrix K is defined according to the pairwise genotypic similarity of individuals, and so its structure is influenced by population structure, family structure and cryptic relatedness. The parameter relates this structure to the phenotype Y : σg2 captures the extent to which genetically similar individuals are phenotypically similar, thus removing confounding effects. The optimal formulation of K, the importance of including PC covariates in fixed effects X, and the effects of these choices have not yet been fully explored.
Population structure: a fixed or random effect?
An important and unanswered question is whether population structure should be modeled as part of the set of random effects together with family structure and cryptic relatedness, or as a separate fixed effect requiring PC covariates and additional model parameters33–34 (see Box 1). Inclusion in random effects is much simpler, and has been shown to provide a sufficient correction for stratification in Finnish and UK data sets33.
However, population structure is actually a fixed effect (i.e. its effect as a function of genetic ancestry is the same for all samples), and spurious associations might result if it is modeled as a random effect based on overall covariance, particularly in the case of unusually differentiated markers. Modeling population structure as a fixed effect provides a higher level of certainty in correcting for stratification, but requires running PCA (or a similar method) to infer the genetic ancestry of each sample34. If family structure is present, inferring genetic ancestry via PCA is a challenge, because family relatedness may lead to artifactual PCs19. A possible solution is to compute PCs using SNP loadings inferred from a set of unrelated samples, either using a different set of samples than those in the disease study or using an unrelated subset of samples from the disease study35. This is likely to be sufficient when the set of unrelated samples used is very large relative to the magnitude of population structure effects. However, unless sample sizes are very large, PCs computed from external SNP loadings will be biased towards zero due to statistical noise in the SNP loadings11, 36. This motivates further work on PCA in related samples.
Modeling phenotypes as fixed
Mixed models view phenotypes as modeled using a fixed set of genotypes. However, as an alternative to mixed models, genotypes can be modeled using a fixed set of phenotypes, a theoretically appealing approach that makes fewer assumptions about phenotypic covariance structure37–38. Simulations in the absence of unusually differentiated markers have shown that using the genotypic covariance matrix to account for both population and family structure can effectively control spurious associations under a variety of settings37 (ROADTRIPS; see Web Resources). However, in the case of unusually differentiated markers, normality assumptions (about genotype distributions) underlying the test statistics will be violated, and stratification may lead to confounding unless PC covariates are used. The question of whether to model random effects only or to include PC covariates as fixed effects is analogous to the mixed model framework. When viewing phenotypes as fixed, PC covariates may be particularly essential since modeling only random effects leads to a uniform correction factor in the absence of missing data37.
Simulations
To illustrate the properties of the above methods in correcting for stratification at normally differentiated or unusually differentiated markers, in the presence or absence of family structure, we carried out two simulations. We considered a case-control study with two subpopulations POP1 and POP2, with 300 cases and 200 controls from POP1 and 200 cases and 300 controls from POP2. We simulated 99,900 normally differentiated markers based on FST(POP1,POP2)=0.01,39 and 100 unusually differentiated markers based on allele frequency difference equal to 0.6 with both minor allele frequencies uniformly distributed on [0.0,0.4]21. In simulation 1, all individuals were unrelated. In simulation 2, all individuals from POP1 were unrelated and individuals from POP2 included 80 case-case sibling pairs, 40 case-control sibling pairs and 130 control-control sibling pairs. We computed λGC for each of the following methods: uncorrected Armitage trend test, EIGENSTRAT21, EMMAX without PC covariates33, EMMAX with PC covariates33, and ROADTRIPS37 (see Web Resources). All PC runs used only one PC, but the additional inclusion of random PCs has little effect on results21. Power to detect causal variants may vary between methods, but our focus here was on correcting false positive associations. We did not simulate the approach described in ref. 30 as this method is completely immune from stratification, ensuring a value of 1.00 in all entries of the table; this approach has appealing properties, but may have reduced power in some instances (see above). We note that the method of ref. 37 with PC covariates incorporated is an approach of potentially high interest, but not currently implemented in ROADTRIPS software.
The results of the simulations are displayed in Table 1. EIGENSTRAT is effective in correcting for population stratification at both normally and unusually differentiated markers (Simulation 1), but does not control for family structure (Simulation 2). EMMAX corrects for both stratification and population structure except for a modest residual inflation at unusually differentiated markers, which is completely removed by EMMAX with PC covariates; if the number of unusually differentiated markers is small, modest inflation at such markers may not be a major concern. ROADTRIPS corrects for family structure but not for population stratification at unusually differentiated markers, though incorporation of PC covariates could potentially address this. We note that for each method, dividing association statistics by residual λGC is guaranteed to produce statistics with λGC=1, but this approach may be inadequate for spurious associations at unusually differentiated markers, and/or may not maximize power if family structure (or cryptic relatedness) is not fully modeled.
Table 1. Effectiveness of different approaches in correcting for stratification.
We list the λGC (Genomic Control lambda) of each method for normally differentiated (FST = 0.01) and unusually differentiated (Δ = 0.6) markers in Simulation 1 and Simulation 2. In each case, λGC was computed as the median χ2(1 dof) statistic (restricting to the subclass of markers tested) divided by 0.455. EIGENSTRAT corrects for population structure (Simulation 1), EMMAX and ROADTRIPS correct for family structure and for population structure at normally differentiated markers (FST = 0.01), and EMMAX+PCs corrects for family structure and for population structure at normally or highly differentiated markers (FST = 0.01 or Δ = 0.6). We note that the approach of ref. 30 is immune to all of these confounders, implying a value of λGC=1.00 for each column of the table.
| Simulation 1, FST = 0.01 |
Simulation 1, Δ = 0.6 |
Simulation 2, FST = 0.01 |
Simulation 2, Δ = 0.6 |
|
|---|---|---|---|---|
| Armitage trend | 1.40 | 48.4 | 1.57 | 48.3 |
| EIGENSTRAT | 1.00 | 1.00 | 1.17 | 1.14 |
| EMMAX* | 1.00 | 2.05 | 1.01 | 1.62 |
| EMMAX* + PCs | 1.00 | 1.02 | 1.01 | 1.01 |
| ROADTRIPS | 1.00 | 48.4 | 1.00 | 48.3 |
EMMAX can use either the IBS or Balding-Nichols estimate of the kinship matrix33. Results for IBS are displayed in the table, and results for Balding-Nichols are 1.00, 1.91, 1.00, 1.28 for EMMAX and 1.00, 1.03, 1.00, 0.99 for EMMAX + PCs.
Low-frequency and Rare Variants
GWAS have largely focused on common variants, but because most genetic heritability remains unexplained, future work will increasingly focus on variants of low minor-allele frequency (0.5%<MAF<5%) or rare variants (MAF<0.5%)40. First, new low-frequency variants will be identified by the 1000 Genomes Project (see Web Resources) and included in next-generation genotyping arrays. Here, the issues are generally similar to those involving common variants, except that deviation from model specification is more likely, for example if normality assumptions are violated or the genotypic variance of a SNP varies across subpopulations41. Second, exome resequencing projects will aim to identify genes in which individuals with extreme phenotypes have an aggregate excess or deficiency of rare nonsynonymous variants42. Differences in allele frequency spectrum across ancestral populations make stratification a potential concern, but genetic ancestry can be inferred from genotyping array data from the same samples, if available, and included as a covariate. Finally, the advent of whole-exome or whole-genome resequencing raises the question of whether rare variants can be used to infer genetic ancestry with greater precision, perhaps using different methods than the methods currently applied to common variants.
Conclusion
Many different methods of correcting for stratification have been developed, and all of these methods have important advantages. Although mixed models are relatively new and untested, they appear to offer a practical and comprehensive approach for simultaneously addressing confounding due to population stratification, family structure and cryptic relatedness.
In studies where stratification is not a very serious concern, an appealing and simple approach is to use mixed models without including PC covariates. This may include (i) studies in populations of homogeneous ancestry, (ii) studies in structured populations where structure is due to very recent genetic drift, and (iii) studies in any population in which PCA or related methods, applied either to the entire sample or to a subset of unrelated samples, indicate that there is no substantial stratification, i.e. phenotypes are not highly correlated with any of the top PCs.
For studies that do not meet any of the above criteria, an appealing approach is to use mixed models with PC covariates. In family-based studies in which the within-family component contributes much of the overall statistical power, the approach of ref. 30 may also prove useful. In data sets that do not contain family structure or cryptic relatedness, simpler association tests (with or without PC correction, based on above criteria) will probably be sufficient21, 23.
Glossary
- Population structure
Sample structure due to differences in genetic ancestry among samples.
- Family structure
Sample structure due to familial relatedness among samples.
- Cryptic relatedness
Sample structure due to distant relatedness among samples with no known family relationships.
- Genomic Control
A method of detecting (or detecting and correcting for) stratification based on the genome-wide inflation of association statistics.
- Structured Association
A method of correcting for stratification in which samples are assigned to subpopulation clusters and evidence of association is stratified by cluster.
- Principal Components Analysis
A dimensionality reduction technique used to infer continuous axes of variation in genetic data, often representing genetic ancestry.
- Family-Based Association Tests
A class of association tests that uses families with one or more affected children as the observations rather than unrelated cases or controls. The analysis treats the allele that is transmitted to (one or more) affected children from each parent as “cases” and the untransmitted alleles as “controls”, to avoid the effects of population structure.
- Mixed Models
A class of models in which phenotypes are modeled using both fixed effects (candidate SNP and fixed covariates) and random effects (phenotypic covariance matrix).
- Differential bias
Spurious differences in allele frequencies between cases and controls due to differences in sample collection, sample preparation and/or genotyping assay procedures.
- Genetic drift
Random fluctuations in allele frequencies over time due to sampling effects, particularly in small populations.
- Multi-Dimensional Scaling
A dimensionality reduction technique, similar to PCA, in which points in a high-dimensional space are projected into a lower-dimensional space while approximately preserving the distance between points.
- Genetic Matching
A method of association testing in which cases and controls are matched for genetic ancestry, as inferred by PCA or other methods.
- Ancestry-Informative Markers
Genetic markers ascertained for large differences in allele frequency between subpopulations that are genotyped to infer genetic ancestry in new samples.
- Transmission Disequilibrium Test
A family-based association test involving case–parent trios in which alleles transmitted from parents to child are compared to untransmitted alleles.
- Rank statistic
a statistic describing the rank, across markers, of association of each marker. Rank statistics can be transformed into quantiles of a standard normal distribution that can be combined with other statistics.
- SNP loadings
The correlations of each SNP to a given PC in PCA. The PC coordinates of each sample are proportional to the sum of normalized genotypes weighted by SNP loadings.
- FST
A measure of the genetic distance between two populations that describes the proportion of overall genetic variation that is due to differences between populations.
- Armitage Trend Test
A standard χ2(1 dof) association test computed as the number of samples times the squared correlation between genotype and phenotype.
- Genetic heritability
The proportion of the total phenotypic variation in a given characteristic that can be attributed to additive genetic effects. In the broad sense, heritability involves all additive and non-additive genetic variance, whereas in the narrow sense, it involves only additive genetic variance.
- Exome resequencing
A study design in which exon capture technologies are used to obtain resequencing data covering all exonic regions for each individual in the study.
Biographies
Alkes L. Price is an Assistant Professor of Statistical Genetics at the Harvard School of Public Health and a 2010–2012 Alfred P. Sloan Research Fellow. His research interests include disease mapping in admixed populations, deconstructing the heritable components of common disease and gene expression traits, and statistical methods for mapping rare variants.
Noah Zaitlen received his Ph.D. from the University of California San Diego under the supervision of Dr. Eleazar Eskin. He is currently a postdoctoral fellow at the Harvard School of Public Health in the laboratory of Dr. Alkes Price. His research focuses on understanding the genetic basis of complex human phenotypes.
David Reich is an Associate Professor in the Harvard Medical School Department of Genetics and an Associate Member of the Broad Institute of Harvard and MIT. His group focuses on developing methods for understanding human population structure and history, and applying this knowledge to help in the search for disease genes.
Nick Patterson is a senior staff scientist at the Broad Institute in Cambridge, Massachusetts. He received his doctorate in Mathematics from Cambridge University, and has worked both in defense (for the U.K. and U.S. governments) and in finance. His current research interests include the genetics of admixed populations and human genetic history.
Footnotes
Web Resources
http://genome.gov/gwastudies/ (NHGRI catalog of published GWAS)
http://pritch.bsd.uchicago.edu/software.html (STRUCTURE and STRAT software13, 15)
http://www.genetics.ucla.edu/software/admixture/ (ADMIXTURE software16)
http://www.hsph.harvard.edu/faculty/alkes-price/software/ (EIGENSTRAT, implemented in EIGENSOFT software19, 21)
http://pngu.mgh.harvard.edu/~purcell/plink/ (PLINK software23)
http://biosun1.harvard.edu/~fbat/fbat.htm (FBAT software27, 29)
http://www.sph.umich.edu/csg/abecasis/QTDT / (QTDT software28)
http://genetics.cs.ucla.edu/emmax/ (EMMAX software33)
http://www.maizegenetics.net/ (TASSEL software34)
http://www.stat.uchicago.edu/~mcpeek/software/index.html (ROADTRIPS software37)
http://www.1000genomes.org/ (1,000 Genomes Project)
References
- 1.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 2.Campbell CD, et al. Demonstrating stratification in a European American population. Nat Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 3.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voight BF, Pritchard JK. Confounding from cryptic relatedness in case-control association studies. PLoS Genet. 2005;1:e32. doi: 10.1371/journal.pgen.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weir BS, Anderson AD, Hepler AB. Genetic relatedness analysis: modern data and new challenges. Nat Rev Genet. 2006;7:771–780. doi: 10.1038/nrg1960. [DOI] [PubMed] [Google Scholar]
- 7.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 9.Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 10.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 11.Price AL, et al. The impact of divergence time on the nature of population structure: an example from Iceland. PLoS Genet. 2009;5:e1000505. doi: 10.1371/journal.pgen.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devlin B, Bacanu SA, Roeder K. Genomic Control to the extreme. Nat Genet. 2004;36:1129–1130. doi: 10.1038/ng1104-1129. author reply 1131. [DOI] [PubMed] [Google Scholar]
- 13.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 15.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menozzi P, Piazza A, Cavalli-Sforza L. Synthetic maps of human gene frequencies in Europeans. Science. 1978;201:786–792. doi: 10.1126/science.356262. [DOI] [PubMed] [Google Scholar]
- 18.Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton University Press; 1994. [Google Scholar]
- 19.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, Zhang S, Zhao H, Cooper RS. Association mapping, using a mixture model for complex traits. Genet Epidemiol. 2002;23:181–196. doi: 10.1002/gepi.210. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luca D, et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am J Hum Genet. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seldin MF, Price AL. Application of ancestry informative markers to association studies in European Americans. PLoS Genet. 2008;4:e5. doi: 10.1371/journal.pgen.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 27.Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- 28.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange C, DeMeo DL, Laird NM. Power and design considerations for a general class of family-based association tests: quantitative traits. Am J Hum Genet. 2002;71:1330–1341. doi: 10.1086/344696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Won S, et al. On the analysis of genome-wide association studies in family-based designs: a universal, robust analysis approach and an application to four genome-wide association studies. PLoS Genet. 2009;5:e1000741. doi: 10.1371/journal.pgen.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lasky-Su J, et al. On genome-wide association studies for family-based designs: an integrative analysis approach combining ascertained family samples with unselected controls. Am J Hum Genet. 2010;86:573–580. doi: 10.1016/j.ajhg.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 33.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X, Li S, Cooper RS, Elston RC. A unified association analysis approach for family and unrelated samples correcting for stratification. Am J Hum Genet. 2008;82:352–365. doi: 10.1016/j.ajhg.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Zou F, Wright FA. Convergence and prediction of principal components scores in high dimensional settings. Annals of Statistics. doi: 10.1214/10-AOS821. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thornton T, McPeek MS. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am J Hum Genet. 2010;86:172–184. doi: 10.1016/j.ajhg.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rakovski CS, Stram DO. A kinship-based modification of the armitage trend test to address hidden population structure and small differential genotyping errors. PLoS One. 2009;4:e5825. doi: 10.1371/journal.pone.0005825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F(ST) Nat Rev Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abney M, McPeek MS. Association testing with principal-components-based correction for population stratification [Abstract number 58]. Presented at the annual meeting of the American Society of Human Genetics; Philadelphia, PA. 2008. http://www.ashg.org/2008meeting/abstracts/fulltext/ [Google Scholar]
- 42.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]