SUMMARY
We present structural and biochemical evidence for a redox switch in the archaeal transcriptional regulator SurR of Pyrococcus furiosus, a hyperthermophilic anaerobe. P. furiosus produces H2 during fermentation, but undergoes a metabolic shift to produce H2S when elemental sulfur (S0) becomes available. Changes in gene expression occur within minutes of S0 addition, and the majority of these S0-responsive genes are regulatory targets of SurR, a key regulator involved in primary S0 response. SurR was shown in vitro to have dual functionality, activating transcription of some of these genes, notably the hydrogenase operons, and repressing others, including a gene encoding sulfur reductase. This work demonstrates via biochemical and structural evidence that the activity of SurR is modulated by cysteine residues in a CxxC motif that constitute a redox switch. Oxidation of the switch with S0 inhibits sequence-specific DNA binding by SurR, leading to deactivation of genes related to H2 production and derepression of genes involved in S0 metabolism.
Keywords: SurR, Pyrococcus furiosus, regulatory transcription factor, elemental sulfur response, hydrogen production, redox switch
Introduction
Though basal transcription in archaea is most similar to the eukaryotic system, regulation of transcription is typified for the most part by the bacterial system, both in the sequence similarities of the regulators themselves to bacterial families and in their regulatory mechanisms (Bartlett, 2005, Bell, 2005). The most prevalent DNA-binding domain found in prokaryotes is the helix-turn-helix (HTH) fold with its many variations and subclasses (e.g. winged HTH), and sequence analyses have demonstrated that this holds for predicted archaeal regulators (Koonin & Wolf, 2008, Aravind et al., 2005). Most of the characterized archaeal regulators are transcriptional repressors; however, a handful of activators have also been characterized (Geiduschek & Ouhammouch, 2005).
Some archaeal regulatory transcription factors are regulated by small molecule effectors, including Mdr1 of Archaeoglobus fulgidus which is responsive to metal ions (Bell et al., 1999), NrpR of Methanococcus maripaludis to 2-oxoglutarate (Lie et al., 2005), TrmB of Thermococcus litoralis and P. furiosus to various sugars (Lee et al., 2003, Lee et al., 2005), and LysM of Sulfolobus solfataricus to lysine (Brinkman et al., 2002). Until now, however, no archaeal transcriptional regulator has been shown to be redox-responsive. Notable bacterial examples include OxyR of E. coli (Choi et al., 2001, Storz et al., 1990, Kim et al., 2002, Paget & Buttner, 2003) and Spx of B. subtilis (Nakano et al., 2005, Newberry et al., 2005), but archaea do not contain homologs of these.
Here we describe the first example in an archaeon of a redox-activated transcription factor, the sulfur response regulator, SurR, of the hyperthermophilic anaerobe Pyrococcus furiosus. This organism grows optimally at 100°C and can utilize both carbohydrates and peptides as carbon sources, via fermentation to organic acids, CO2 and H2; however, in the presence of elemental sulfur (S0), there is a complete metabolic shift from production of H2 to production of H2S (Fiala & Stetter, 1986). DNA microarray expression analyses identified an immediate response of P. furiosus to S0 that occurs within 10 minutes of S0 addition. This primary S0 response involves a dramatic decrease in expression of all three hydrogenase operons and concurrent increase in expression of genes necessary for S0 metabolism, including nsr, the gene encoding NAD(P)H sulfur reductase (Schut et al., 2007).
Almost all of the 17 genes or operons differentially expressed during the primary S0 response contain the SurR consensus DNA-binding motif GTTn3AAC in their promoter regions, and the SurR binding site at several of these promoter regions has been mapped by DNase footprinting experiments (Lipscomb et al., 2009). Each footprint contains at least one GTTn3AAC motif with what appear to be variable numbers of regularly spaced degenerate consensus motifs, resulting in a variety of footprint lengths among the promoter regions tested. The position of the SurR footprint at a given promoter appears to dictate whether SurR will act as an activator or repressor, with footprints lying either upstream from or overlapping the basal transcription elements.
SurR has been shown in vitro to activate transcription of hydrogenase operons encoding soluble hydrogenase I (SHI) and the membrane-bound hydrogenase (MBH) and repress transcription of S0-metabolism associated genes nsr and pdo (protein disulfide oxidoreductase (Pedone et al., 2004, Ren et al., 1998)). The pdo gene (PF0094) is divergently transcribed from surR (PF0095), and there is only 132 bp of intergenic space between the two genes. SurR exerts dual functionality at this shared promoter region, activating transcription from its own gene while repressing transcription from pdo (Lipscomb et al., 2009).
Genes involved in the primary S0 response are thought to comprise the majority of the SurR regulon, and SurR appears to operate as an activator for genes whose expression decreases in the presence of S0 and a repressor for genes whose expression increases with S0 (Lipscomb et al., 2009). In this work we present structural and biochemical evidence for S0-dependent redox regulation of SurR DNA-binding activity, highlighting the key role that this transcription factor plays in the regulatory pathways of H2 and S0 metabolism in P. furiosus.
Results
The SurR structure contains a DNA binding domain with an internal disulfide bond
SurR is a 232 amino acid protein encoded in the P. furiosus genome (locus PF0095). The crystal structure was determined by the Se-SAD method at 2.3 Å resolution (Table 1 and Fig. S1) from a recombinant form purified under aerobic conditions. SurR has relatively few sequence homologs; however, the crystal structure revealed similarities to characterized transcriptional regulators containing winged HTH DNA binding domains. SurR is a homodimer with 2-fold symmetry, and each monomer is composed of three distinct domains (Fig. 1A). The N-terminal (residues 1–75) and C-terminal (residues 166–219) regions each form a winged HTH DNA-binding fold related to the HTH/ArsR family. The linker region between the N- and C-terminal winged HTH folds of the single chain (residues 76–165) has little structural homology with other known structures in the Protein Data Bank (PDB) and forms the major interface for dimerization. This hydrophobic core in the dimer structure consists of a four-stranded beta sheet, a short two-stranded beta sheet, and two long alpha helices interacting in a manner that resembles a coiled coil.
Table 1.
Data collection and refinement statistics for SurR and AxxA-SurR structures.
| Data collection | SeMet-SurR | AxxA-SurR |
|---|---|---|
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 52.13, 174.70, 52.25 | 39.75, 93.48, 71.06 |
| α=γ, β (°) | 90, 93.11 | 90, 102.44 |
| Resolution (Å)a | 50–2.50 (2.59–2.50) | 50–2.80 (2.90–2.80) |
| Rsyma | 0.064 (0.185) | 0.055 (0.374) |
| I / σI | 14.8/7.5 | 61.1/11.0 |
| Completeness (%)a | 96.7 (99.2) | 99.3 (100.0) |
| Redundancya | 3.8 (3.8) | 7.2 (7.4) |
| Refinement | ||
| Resolution (Å)a | 19.43–2.5 (2.56–2.5) | 32.53–2.80 (2.87–2.80) |
| No. reflections | 28,832 /1,484 | 11,712 /608 |
| Rwork / Rfree | 22.6 /27.1 | 24.4 /28.1 |
| No. atoms | ||
| Protein | 6,959 | 3,324 |
| Ligand/ion | - | - |
| Water | 100 | 33 |
| B-factors | ||
| Protein | 28.7 | 82.3 |
| Ligand/ion | - | - |
| Water | 23.1 | 67.3 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.007 |
| Bond angles (°) | 1.250 | 1.058 |
| MolProbityb Scores | ||
| Favored (%) | 95.4 | 92.5 |
| Allowed (%) | 99.6 | 99.0 |
| Outliers (%) (No. of residues) | 0.4 (3/846) | 1 (4/416) |
Values in parentheses are for highest-resolution shell.
Figure 1.
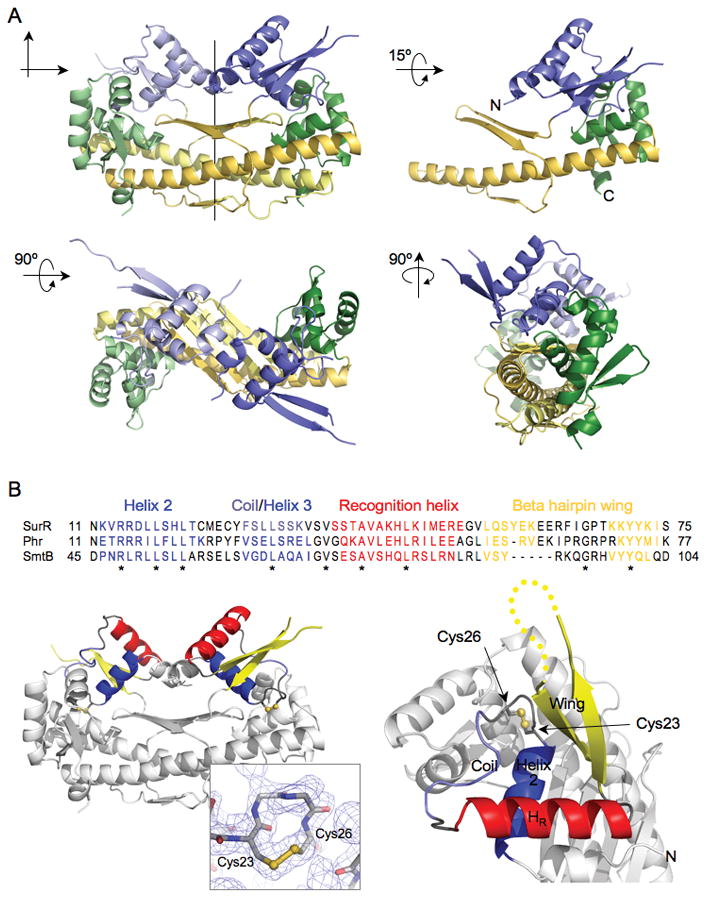
The SurR structure contains winged HTH domains and a disulfide bond.
A. Structure views showing the dimer with 2-fold axis of symmetry, chain A rotated 15o on the horizontal axis, the dimer rotated 90o on the horizontal axis, and the dimer rotated 90o on the vertical axis. Domains are color-coded as follows: N-terminal winged HTH domains are shaded in blue, the middle linker domains are shaded in yellow, and the C-terminal winged HTH domains are shaded in green.
B. Sequence alignment of SurR N-terminal winged HTH domain with those of Phr and SmtB, color-coded according to secondary structure elements shown in structure views of dimer from side (left) and chain A from top (right), with a dotted line representing five missing residues from the beta hairpin wing. Asterisks below alignment denote conserved residues. The internal disulfide bonds are shown as ball-and-stick representation in both structure views, and the electron density map for the disulfide bond is also shown (inset). Sequences were aligned with ClustalW (Thompson et al., 1994). Structure representations in this and all subsequent figures were created with Pymol (www.pymol.org).
Three helices are visible in the N-terminal winged HTH domain; the first two N-terminal helices are joined at a sharp turn, and these are connected with the third helix via a random coil. Several residues of the β-strand hairpin that comprise the wing in the N-terminal winged HTH fold have no interpretable electron density (five amino acids in chain A and four amino acids in chain B), suggesting flexibility in this region of the protein. The third helix is the recognition helix as determined by sequence alignment with structures related to the HTH/ArsR protein family that have the highest sequence identity with the N-terminal region of SurR (residues 1–75) (Fig. 1B). SmtB is one of the best characterized members of the HTH/ArsR protein family (Busenlehner et al., 2003), and Phr is a heat shock regulator from P. furiosus (Vierke et al., 2003) with 32% overall sequence identity to SurR. This comparison illustrates a unique feature of the SurR structure in that it contains a random coil instead of a helix immediately preceding the recognition helix within the HTH motif. The HTH domain of SurR therefore differs significantly from the canonical tri-helical bundle of the HTH domain. Interestingly, immediately adjacent to the N-terminal side of the coil is a CxxC motif with a disulfide bond (Fig. 1B). The presence of the disulfide bond in the HTH region led to the hypothesis that oxidation or reduction of the cysteine residues comprising the CxxC motif might affect the conformation of the DNA-binding domain and in turn modulate the DNA-binding activity of SurR.
Oxidation of the SurR CxxC motif abolishes sequence-specific DNA binding
We investigated the redox state of the cysteine residues in the aerobically-purified protein preparation of SurR used in the previously reported study (Lipscomb et al., 2009) using DTNB (Ellman’s reagent). Surprisingly, aerobically purified SurR was found to be approximately 80% in the reduced state (Table S1); therefore, the crystal structure of SurR containing the disulfide bond did not necessarily represent the form of the protein which had been shown to bind to DNA with sequence specificity in EMSA and footprinting experiments (Lipscomb et al., 2009). The reason for this is not clear, but crystallization conditions or prolonged exposure to oxygen during crystallization may have induced disulfide-bond formation.
Since the form of the protein known to bind DNA and regulate transcription was shown to be predominantly in the reduced state, it was necessary to determine the DNA-binding affinity of the oxidized form represented in the crystal structure. The thiol-specific oxidant diamide (diazenedicarboxylic acid (Kosower & Kosower, 1995)) was used to promote disulfide formation between the cysteine residues in the SurR CxxC motif. After incubation of SurR protein with diamide, no free thiols could be detected in the SurR protein, showing that all had been converted to intramolecular disulfide bonds (Table S1).
EMSA was used to test the DNA-binding affinity of oxidized SurR. As a control, a SurR variant that lacked the ability to form a disulfide bond was constructed by replacing both cysteine residues in the CxxC motif with alanines (C23A and C26A). This recombinant SurR variant will be referred to as AxxA-SurR. EMSA with pdo-surR intergenic DNA comparing SurR with AxxA-SurR in the presence of either dithiothreitol (DTT) or diamide revealed that oxidation of the SurR cysteine residues reduces its DNA-binding ability (Fig. 2). Several protein-DNA complexes form as protein concentration is increased due to successive binding of SurR proteins at multiple consensus and degenerate consensus sequences within the ~85 bp SurR footprint at the pdo-surR promoter region (Lipscomb et al., 2009). While both proteins shifted the DNA almost equally in the presence of DTT, diamide only affected the wild-type SurR protein. At higher concentrations of SurR, where the lowest mobility protein-DNA complex predominates, the apparent DNA affinity remains the same since in the presence of either DTT or diamide, a complete shift occurred at 1.6 μM protein, although in the presence of diamide, the shift was of an undefined nature and characteristic of non-specific DNA binding. The AxxA-SurR variant displayed a slightly higher DNA affinity than SurR even in the presence of DTT, almost completely shifting the DNA at 0.8 μM protein (compare top and bottom panels in Fig. 2). If oxidation of SurR reduces its DNA-binding affinity, then it would be expected that the AxxA-SurR variant, which cannot be oxidized, would display a higher overall DNA affinity than the aerobically purified SurR which is ~80% in the reduced state. The addition of 10 mM DTT may not be sufficient to reduce 100% of the portion of oxidized SurR protein such that its DNA binding affinity matches the AxxA-SurR variant.
Figure 2.
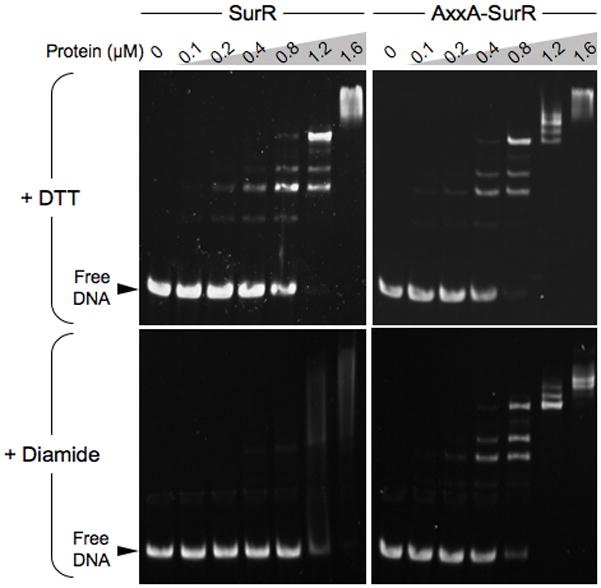
Oxidation of the SurR CxxC motif affects its DNA binding ability. EMSA showing the effect of 10 mM DTT versus 10 mM diamide on the DNA-binding ability of SurR versus AxxA-SurR with pdo-surR intergenic DNA probe (50 nM).
The CxxC motif constitutes a reversible redox switch inducible by S0
The dependence of the DNA-binding ability of SurR upon its redox state suggested that this might represent an in vivo mechanism for regulation of SurR activity. The obvious oxidants present within P. furiosus cells after S0 addition are S0-derived species. Since S0 is not water-soluble, a freshly precipitated colloidal S0 suspension was used to test the effect of S0 species on SurR in EMSA. A comparison of its effect on SurR and AxxA-SurR in EMSA with pdo-surR intergenic DNA is dramatic. While no effect was observed on the shifting pattern of AxxA-SurR, colloidal S0 completely eliminates the specific distinct banding pattern for SurR (Fig. 3A). A nearly complete shift of the free DNA is observed at the highest protein concentration tested; however, the smeared shift appears to be a result of non-specific DNA binding. Colloidal S0 concentrations between 100 and 500 μM are sufficient to convert the specific banding pattern to a smear and reduce DNA binding affinity (using 1.2 μM SurR; Fig. 3B).
Figure 3.
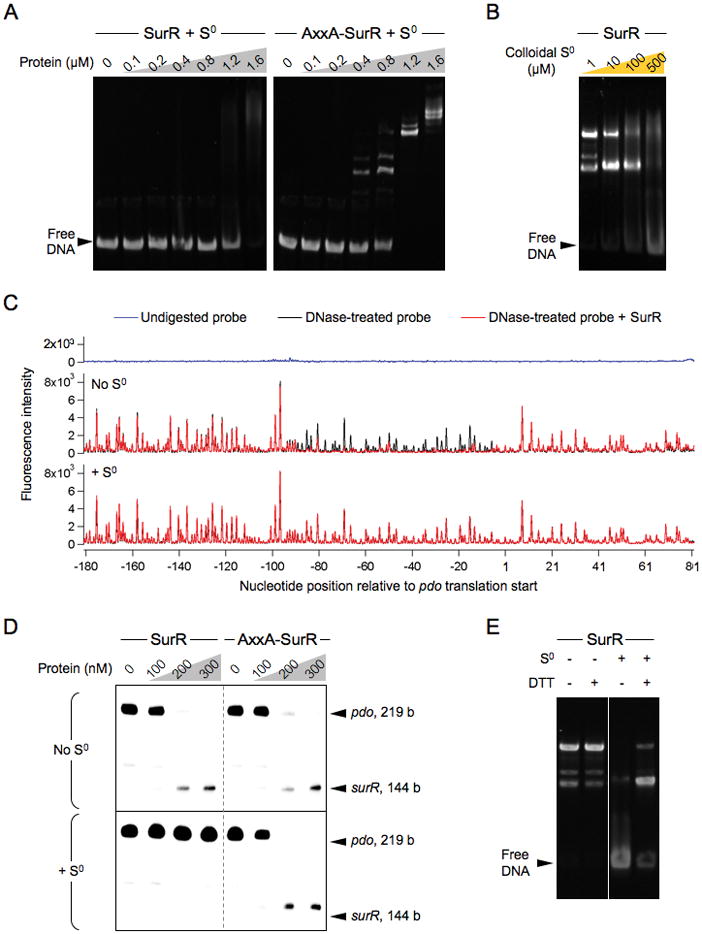
Colloidal S0 reversibly oxidizes SurR.
A. EMSA showing the effect of 10 mM colloidal S0 on the DNA-binding ability of SurR versus AxxA-SurR with pdo-surR intergenic DNA probe (50 nM).
B. EMSA showing the effect of increasing concentrations of colloidal S0 on SurR DNA binding affinity to pdo-surR intergenic DNA probe (25 nM) at a protein concentration of 1.2 μM.
C. Fluorescence DNase footprinting of SurR (0.3 μM) on pdo-surR probe DNA (10 nM) showing footprint for pdo coding strand in the absence and presence of 1 mM colloidal S0, with the trace for the undigested probe shown at top.
D. In vitro transcription with SurR and AxxA-SurR on pdo-surR template DNA (16 nM) in the absence and presence of 1 mM colloidal S0.
E. EMSA of SurR (1.2 μM) with pdo-surR intergenic DNA probe (50 nM), incubated with or without 1 mM colloidal S0, followed by further incubation in the presence or absence of 2 mM DTT.
In order to establish that the sequence specificity of SurR DNA binding was indeed eliminated as a result of its oxidation by colloidal S0, fluorescence-detected DNase I footprinting of SurR on probe DNA containing the pdo-surR promoter region was performed in the presence and absence of colloidal S0. For the pdo-surR promoter region, the SurR footprint that is readily visible at a protein concentration of 10 nM under non-oxidizing conditions completely disappears with the addition of colloidal S0 (Fig. 3C). This result confirms that, while in EMSA the DNA is still completely shifted by SurR at higher protein concentrations in the presence of colloidal S0 (Fig. 3A, left panel), the affinity of SurR for DNA is reduced to non-specific association since SurR can no longer bind with sequence specificity to its DNA recognition sites. This suggests that oxidation of the cysteine residues in the CxxC motif causes a conformational change in the DNA-binding domain which prevents SurR from binding to DNA in a sequence-specific manner.
Concomitant with its loss of sequence specificity in DNA binding is the necessary loss of regulatory capacity in in vitro transcription assays. This was shown to be the case for the regulation of both pdo and surR. Under standard in vitro transcription conditions, SurR represses the transcription of pdo while activating surR, and the AxxA-SurR variant displays the same activity. Addition of colloidal S0 abolishes the regulatory effect of SurR on transcription of these genes, but it has no effect on the regulation of transcription by AxxA-SurR (Fig. 3D). Repression of the nsr gene by SurR is likewise eliminated when colloidal S0 is present (Fig. S2).
EMSA was used to test the reversibility of SurR oxidation by colloidal S0 by addition of excess reductant. DTT almost completely reverses the effect of oxidation by colloidal S0 (Fig. 3E), but the reductants cysteine, sodium dithionite, and sodium sulfide had no effect (Fig. S3). This result shows that oxidation of SurR by colloidal S0 is reversible, demonstrating that the CxxC motif represents a redox switch capable of modulating the DNA-binding activity of SurR in vitro. Whether the switch is reversible in vivo via chemical or enzymatic means or whether there is a degradation-mediated pathway for disposal of oxidized SurR is not known at this point.
DNA binding by SurR is regulated by a conformational change
Since oxidation of SurR eliminates sequence-specific DNA-binding, the possibility that the quaternary structure of SurR might change upon oxidation was investigated. Analytical gel filtration showed that there was no difference in the elution behavior of as-purified SurR, SurR oxidized with either diamide or colloidal S0, and the AxxA-SurR variant. Each gave rise to only one predominant peak corresponding to a molecular weight of approximately 62 kDa (data not shown), comparable to the calculated molecular weight of a SurR dimer (54 kDa), which is the configuration observed in the SurR crystal structure. The oxidation state of SurR therefore does not affect its quaternary structure.
Since the structure of oxidized SurR does not represent the form capable of sequence-specific DNA binding, crystallization and structure determination of the AxxA-SurR variant was undertaken. The AxxA-SurR structure was solved at 2.6 Å resolution using molecular replacement with the wild-type SurR structure as the searching model (Table 1). Since all of our data show that AxxA-SurR binds DNA and regulates transcription in the same manner as the reduced wild-type protein (Figs. 2 and 3A,D), the structure of the variant should be a good representation of SurR in the reduced state.
The structure of AxxA-SurR is nearly identical to that of oxidized SurR (overall Cα RMS deviation of 1.41 Å) except in the N-terminal HTH region of chain A. The RMS deviation for Cα atoms in the N-terminal HTH domains of the two structures (residues 1–75) is 3.12 Å, excluding the missing residues in the wing, compared to 0.75 Å for residues 76–232. The backbones of the two forms (chain A in both structures) align nicely except in the region beginning at the CxxC motif and extending through to the start of the recognition helix (Fig. 4A). Chain A of the AxxA-SurR structure, however, adopts a more canonical HTH fold: the coil separating helix 2 from the recognition helix in the oxidized structure forms a helix in the AxxA-SurR structure. Interestingly, the HTH region of chain B in the AxxA-SurR structure adopts the same conformation as the oxidized form of the protein (Fig. 4B).
Figure 4.
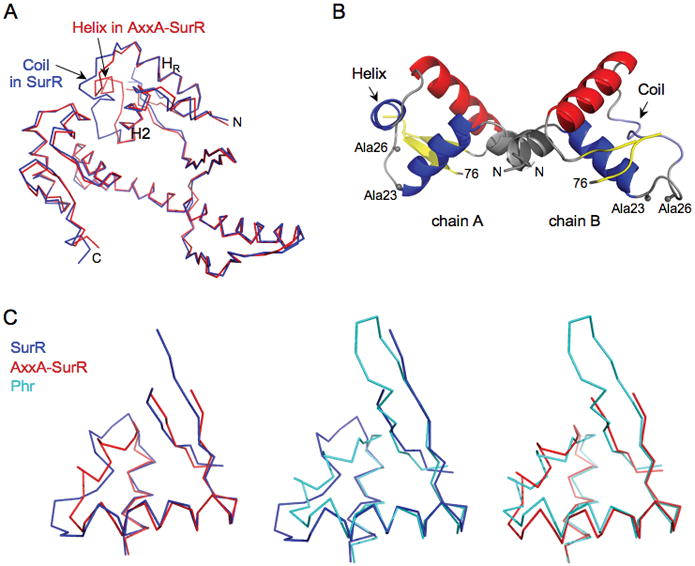
The AxxA-SurR structure adopts a more canonical HTH fold.
A. Chain A of oxidized SurR and ‘reduced’ AxxA-SurR backbone structures overlaid to show the change from coil to helix in the region between helix 2 (H2) and the recognition helix (HR).
B. N-terminal winged HTH region of AxxA-SurR dimer showing structural differences between chains A and B in the helix/coil regions.
C. Backbone overlays of N-terminal winged HTH domains of SurR (residues 11–75), AxxA-SurR (residues 11–75), and Phr (residues 15–77) structures.
The HTH region of chain A in the AxxA-SurR structure aligns better with the Phr structure than with oxidized SurR (Fig. 4C). Since Phr contains a canonical HTH motif with a helix at the position where oxidized SurR has a random coil, the conformational change in SurR from oxidized to the reduced state, represented by AxxA-SurR, could indicate a shift toward a protein conformation that more readily binds to DNA in a sequence-specific manner. Moreover, in the AxxA-SurR form, the N- and C-terminal winged HTH motifs in chain A form a pseudodimer (Fig. S4A). Although there is less than 15% sequence identity between the N- and C-terminal HTH domains, they align with an RMSD of 2.55 Å (Residues 13–75 and 166–219, Fig. S4B). A major difference is in the length of the hairpin loop in the wing.
Along with the shift to a more canonical HTH fold in one monomer, the amino acid side-chains that shift in position from the oxidized to the ‘reduced’ state likely play important roles in DNA recognition and binding. The side chains of residues 23 through 36 in the coil region move significantly, with several becoming more solvent-exposed in the ‘reduced’ AxxA-SurR form (Fig. 5). Four residues in particular appear to be more solvent-exposed and positioned more closely to the recognition helix: Ser32, Ser29, Phe28, and Tyr27. Both Ser29 and Ser32 may contribute to sequence-specific DNA binding as the amino acid serine has a somewhat higher propensity to be involved in protein-DNA contacts, particularly with the DNA backbone (Jones et al., 1999, Coulocheri et al., 2007). Phe28 flips toward the recognition helix and the wing of the winged HTH motif, and Tyr27 moves out from a position close to the protein core to a region near the wing. Perhaps the shift in the position of the Phe and Tyr residues could change or stabilize the position of the wing so that it can make necessary contacts with the DNA. For Phr, the wing appears to play an important role in sequence-specific DNA recognition (Liu et al., 2007); however, DNA binding involves three arginine residues in the hairpin loop, none of which are conserved in SurR. Nevertheless, there are some charged residues, including one Arg, in the wing hairpin of SurR which could play important roles in DNA binding, as charged residues in this region in particular can be important in DNA binding by winged HTH proteins (Aravind et al., 2005).
Figure 5.
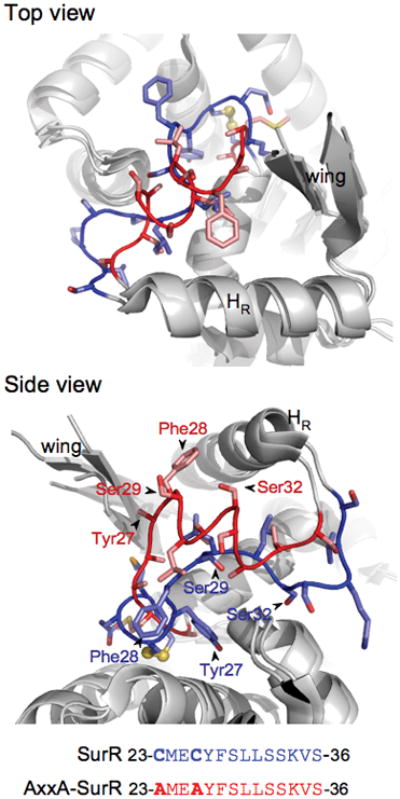
The structures suggest a mechanism for the SurR redox switch. The SurR and AxxA-SurR structures are overlaid, showing the shift of side chain positions in the coil/helix area of the SurR N-terminal HTH domain, with views shown from top and side. Key residues that may be important to DNA binding are labeled in the side view, and the amino acid sequence of the coil/helix region for each structure is shown below. Electron density for the Tyr27 side chain was missing in the AxxA-SurR structure.
Discussion
SurR has a novel overall structure and domain organization (see Fig. S5 for stereo diagrams), even though the N- and C-terminal winged HTH domains follow the general pattern of the HTH/ArsR superfamily. The main dimerization interface occurs in the linker region between these two HTH domains, between two long alpha helices and two sets of beta strands (Fig. 1A). This linker region has few structural homologs and therefore represents a novel fold among known protein structures. A superficially similar fold exists in the related Phr structure, which also has a long alpha-helix coil beneath a four-stranded beta sheet; however, the secondary structure organization within the domain differs from that of SurR.
The structural differences between the oxidized and ‘reduced’ forms of SurR validate that oxidation of the cysteine residues in the CxxC motif plays an important role in modulating the DNA-binding activity of SurR. The SurR CxxC motif represents a regulatory redox switch comparable to those described for OxyR (Choi et al., 2001, Kim et al., 2002) and Spx (Nakano et al., 2005, Newberry et al., 2005) whose transcriptional regulatory activity is governed by direct sensing of redox conditions. The structure of the ‘reduced’ form of SurR (deduced from the AxxA-SurR variant) is suggestive of the mechanism of the redox switch: oxidation of the CxxC motif to a disulfide bond results in an unwinding of the helix N-terminal to the recognition helix in the canonical HTH motif (see Movie S1), causing a reduction in DNA-binding affinity and a loss in DNA-binding specificity. The CxxC motif is just N-terminal to this helix, and therefore formation of a disulfide bond in this region apparently places a constraint on the helix, forcing it into a coil. In fact, for redox-sensitive proteins with CxxC motifs, it is common to find an α-helix immediately following the CxxC motif (Fomenko & Gladyshev, 2003). Disulfide bond formation at the SurR CxxC motif might also be facilitated by the presence of a histidine residue (His171) near the N-terminal cysteine (Cys23) that could stabilize the cysteine thiolate, giving it a higher propensity to be oxidized (Declercq et al., 2001, Fomenko & Gladyshev, 2003).
The slightly different conformations adopted by the HTH regions in each monomer of the ’reduced’ AxxA-SurR structure could be due to the crystal packing; however, it is also suggestive of an allosteric mechanism for SurR. Perhaps the binding of SurR to its cognate DNA begins with one monomer binding to a palindromic half-site, inducing a conformational change which registers at the other monomer HTH fold, and in turn allows it to bind specifically to the other palindrome half-site. There is also a possibility that the alanine mutations in the AxxA-SurR variant are contributing to the disparity between the two chains, and the structure is not entirely representative of the reduced form of SurR, even though there appears to be no functional difference between reduced SurR and the AxxA-SurR variant.
Possible intracellular redox effectors of SurR
Sulfur in its elemental state is not water-soluble, but many S0-dependent organisms are able to internalize solubilized S0 for metabolic purposes (Hedderich et al., 1998). This process begins by natural diffusion of polysulfide from solid sulfur and is probably accelerated by S0-solubilizing agents such as sulfide produced abiotically and from P. furiosus S0 reduction (Blumentals et al., 1990, Hedderich et al., 1998). Colloidal S0 in the form of short chains of S0 and stable S8 rings is formed upon polysulfide oxidation (Kleinjan et al., 2003, Kleinjan et al., 2005). Colloidal S0 or polysulfide are possible in vivo effectors for SurR, as these S0 species are likely present within the cell. Colloidal S0 in particular is a likely candidate considering that one of the regulatory targets of SurR is nsr, encoding a cytosolic protein which appears to utilize colloidal S0 as a substrate for coenzyme A-dependent evolution of sulfide in vitro (Schut et al., 2007, Lipscomb et al., 2009). An increase in the availability of colloidal S0 could be the driving force in the oxidation of SurR, leading to derepression of nsr so that the available S0 can be metabolized.
Primary S0 response and transcriptional regulation
The ability of P. furiosus to sense and respond to S0 within minutes of S0 addition demonstrates the rapidity of the cellular response to this environmental change (Schut et al., 2007). SurR has been shown to bind to the promoter regions of several of the genes whose expression is either significantly increased or decreased during the primary S0 response, and furthermore, the SurR DNA-binding motif elucidated by SELEX was found in 13 out of 17 of these gene/operon promoters (Lipscomb et al., 2009). The relationship of SurR to the intracellular response of P. furiosus to S0 can now be understood from the data presented herein. In its reduced state, SurR is thought to activate transcription of the hydrogenase operons and related genes and at the same time repress transcription from at least four genes/operons that encode proteins involved in S0 metabolism, most notably, NSR. The DNA microarray data mapping S0 response show exactly the opposite regulation, with hydrogenase and related genes being down-regulated and S0-response genes being up-regulated (Schut et al., 2007), but the SurR redox swich now helps to explain this observation. When S0 is present, the CxxC motif presumably becomes oxidized, causing SurR to release from its DNA recognition sites; therefore, the effect that is observed at the transcriptional level is at least in part a result of deactivation of hydrogenase genes and derepression of S0-metabolizing genes when SurR is no longer exerting any transcriptional control due to the loss of its sequence specific DNA-binding ability. This explanation for the physiological role of SurR is summarized in Fig. 6, which now forms our working model for future studies of the mechanisms by which P. furiosus responds to the presence of S0, an insoluble electron acceptor that has a dramatic effect on its primary metabolism.
Figure 6.
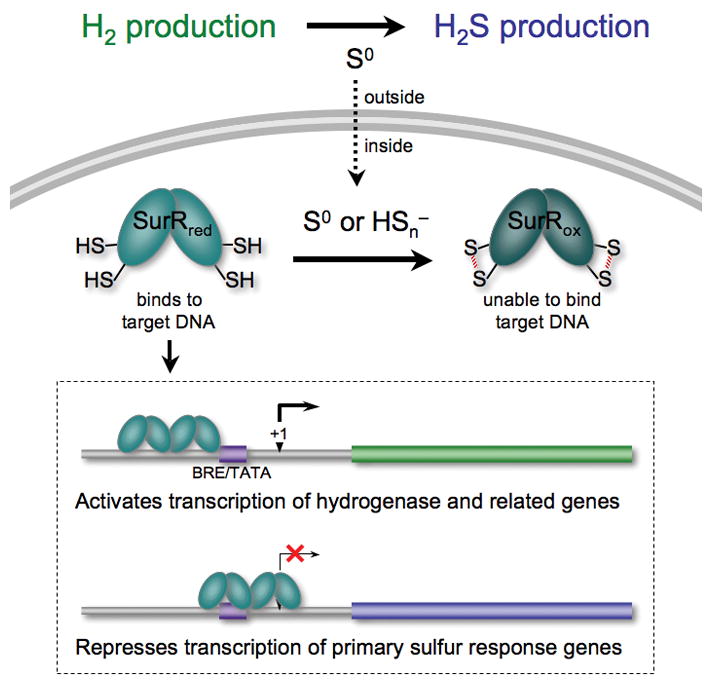
Cartoon of physiological implications of SurR redox switch. In the absence of S0, SurR remains in the reduced state (SurRred), in which it binds DNA and elicits transcriptional control: activating hydrogenase and related genes, presumably by recruiting the basal transcription apparatus to the promoter, while repressing genes of the primary S0 response, most likely by blocking access to the promoter. Under these conditions, P. furiosus produces H2. However, when S0 is available, cells shift from producing H2 to H2S, due at least in part to the deactivation of SurR (SurRox) resulting from oxidation by S0 or polysulfide inside the cell. Once oxidizing S0 species are depleted inside the cell, SurR is thought to be converted back to its reduced active state to resume transcriptional regulation of target genes, thereby promoting the production of H2 by the cell.
Experimental procedures
Cloning of SurR and construction of AxxA-SurR variant
SurR was cloned as described previously (Lipscomb et al., 2009) using a modified pET24d vector that incorporated a cleavable his-tag into the protein N-terminus. This vector was used to create the AxxA-SurR expression vector in which both cysteine codons were mutated to alanine codons (C23A and C26A) using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and the primer 5'-ggatttgttatctcacctaactgctatggaagcctactttagccttttaagtagc with its complement.
Expression and purification of SurR and AxxA-SurR
The auto-induction protein expression protocol established by Studier (Studier, 2005) was followed for expression of his-tag cleavable SurR and AxxA-SurR, as well as selenomethionine-labeled SurR and AxxA-SurR. Protein purification was performed as described previously (Lipscomb et al., 2009). Protein samples intended for crystallization trials underwent a final gel-filtration polishing step using a HiPrep 26/60 Sephacryl S-100 High Resolution (GE Healthcare) gel filtration column equilibrated with 20 mM HEPES, 200 mM NaCl, pH 7.6. Protein-containing fractions were pooled and concentrated to 8–12 mg/mL using an Amicon Ultra-15 centrifugal filter device (Millipore). Protein concentration was determined using a Bio-Rad DC Protein Assay kit.
Crystallization, optimization and data collection
Both SeM-labeled SurR and AxxA-SurR were screened against 384 crystallization conditions from Crystal Screen, Crystal Screen 2, MembFac, PEG/ION Screen, and Crystal Screen Cryo from Hampton Research, Wizard I and II from Emerald BioSystems, and MemSys from Molecular Dimensions with the modified microbatch under oil method (Darcy et al., 1996) using the Oryx 6 crystallization robot from Douglas Instruments. All the screens were stored at 18oC. Optimization was performed after the initial hits from screening using the same method. Crystals were flash-frozen and stored in liquid nitrogen. SeM-SurR crystals were obtained from 20 mM CaCl2, 25% (vol/vol) MPD, and 100 mM sodium acetate, pH 4.4. AxxA-SurR crystals were obtained from 70 mM sodium acetate, pH 4.8, with 30% (vol/vol) glycerol. The datasets of SurR and AxxA-SurR were collected at Advance Photon Source (APS) beamlines 22ID and 22BM, respectively at 100 K. The wavelength was 0.9724 Å for the single SAD dataset of SeMet-SurR, whereas AxxA-SurR’s data was collected using 1.00 Å wavelength.
Structure solution and refinement
Data were indexed, integrated, and scaled using the HKL2000 software suite (Otwinowski & Minor, 1997); the statistics are in Table 1. Phasing of SeM-SurR was done through the SECSG web-based structure solution pipeline (Liu et al., 2005). The pipeline uses an array of separate programs to screen parameter space to achieve the best solution. The program package SOLVE/RESOLVE (Terwilliger, 2000, Terwilliger, 2002, Terwilliger & Berendzen, 1999) was used to identify and refine the heavy atom positions and calculate the initial electron density map. The ARP/wARP software traced the initial model (Perrakis et al., 1999). Four SurR monomers make two non-crystallographic dimers related by a non-crystallographic 2-fold axis of symmetry and occupy the asymmetric unit with an estimated solvent content of 49% based on a Matthews’ coefficient (Vm) of 2.4 Å3/Da. Then the experimental phases were improved using non-crystallographic symmetry averaging with DM (Cowtan & Zhang, 1999). The ARP/wARP traced about 40% of the model automatically. The final model was completed using Coot (Emsley & Cowtan, 2004), and the restraint refinement was performed using REFMAC5 (Murshudov et al., 1997). NCS restraints were employed up to the last stage of the refinement. The AxxA-SurR structure was determined by Phaser (McCoy, 2007) from the CCP4 package (Potterton et al., 2003) using SurR as the model structure, and the final model was completed using Coot (Emsley & Cowtan, 2004). Validation of both structures was performed using MOLPROBITY (Lovell et al., 2003).
Structural comparison and alignment
Comparison searches against all known protein structures were performed with Dali (Holm & Sander, 1996) and VAST (Gibrat et al., 1996, Madej et al., 1995). Pairwise alignments were made with DaliLite (Holm & Park, 2000) and Pymol (www.pymol.org).
Free thiol quantification
A protocol for colorimetric quantitation of free sulfhydryls with Ellman’s reagent (Coligan, 1996) was adapted for use in a 96-well microplate. DTT standard solutions were prepared in degassed 0.1 M sodium phosphate buffer (pH 8.0). Protein samples of 10, 25 and 50 μM were prepared in degassed 20 mM HEPES, 200 mM NaCl (pH 7.6). A second set of samples was prepared in the presence of 10 mM diamide, and all samples were incubated at room temperature for 15 min prior to assaying.
Electromobility shift assay
EMSAs were performed essentially as described previously (Lipscomb et al., 2009). Protein-DNA incubations were carried out in the presence of 10 mM diamide, colloidal S0, or DTT in 1x EMSA buffer (20 mM HEPES, 200 mM KCl, 5% (vol/vol) glycerol, 1 mM EDTA, pH 7.5). EMSA binding reactions were incubated at 70°C for 20 min. The colloidal S0 solution was made immediately prior to use by diluting an anaerobic solution of polysulfide made from S0 and Na2S (4:1 mole ratio) with aerobic EMSA buffer to form the colloidal S0 suspension. For oxidation reversibility experiments, after incubation of the EMSA reactions, a fresh solution of DTT was added to a final concentration of 20 mM. A second incubation was carried out for 5 min at 70°C before gel electrophoresis. Gels were stained with SYBR Green I nucleic acid gel stain.
Fluorescence DNase I footprinting
Modified fluorescence footprinting (Wilson et al., 2001) was performed based on the DNase I footprinting method (Galas & Schmitz, 1978) essentially as described previously (Lipscomb et al., 2009) except that for some samples, colloidal S0 was added to the protein-DNA binding reaction as described for EMSA. The pdo-surR probe was PCR-amplified from pUC18-cloned promoter-ORF DNA (previously described (Lipscomb et al., 2009)) using 5' HEX (hexachlorofluorescein) labeled primers. Sample preparation, data collection, and analysis were performed as described previously (Lipscomb et al., 2009).
In vitro transcription
In vitro transcription experiments (Hethke et al., 1996) were carried out as previously described (Lipscomb et al., 2009) with PCR-amplified pdo-surR template DNA (16 nM) and different amounts of SurR and AxxA-SurR (0–300 nM), in the absence and presence of 1 mM colloidal S0. The transcription products were analysed on an 8% polyacrylamide (vol/vol) urea gel in 1xTBE buffer and visualized by a PhosphorImager (FLA 5000, Fuji).
Analytical gel filtration
A Superdex 75 10/300 GL size exclusion column (GE Healthcare) was used to determine the quaternary structure of four SurR protein samples (in 20 mM HEPES, 200 mM NaCl, pH 7.6): untreated SurR, untreated AxxA-SurR, SurR treated with 10 mM diamide for 10 min at room temperature, and SurR treated with 2 mM colloidal S0 for 10 min at room temperature. Molecular weight standards used were from Sigma.
Supplementary Material
Acknowledgments
We would like to thank F. Sugar for helpful advice in protein purification and W. Lanzilotta for consultation regarding the structures and the supplementary movie. This work was supported by the National Institutes of Health (GM62407 to B.C.W. and GM042025 to R.A.S.), the Georgia Research Alliance (to B.C.W.), the University of Georgia Research Foundation (to B.C.W.) the National Science Foundation (MCB-9631093 to R.A.S.), the Department of Energy (FG05-95ER20175 to M.W.W.A. and FG02-08ER64690 to R.A.S.), and by the priority program of the Deutsche Forschungsgemeinschaft for “Regulation of genome function and gene regulation in Archaea” (to M.T.). Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
Accession codes
The structures were deposited into the Protein Data Bank (PDB), with PDB IDs 2QLZ for SurR and 2QUF for AxxA-SurR.
References
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Bartlett MS. Determinants of transcription initiation by archaeal RNA polymerase. Curr Opin Microbiol. 2005;8:677–684. doi: 10.1016/j.mib.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Bell SD. Archaeal transcriptional regulation--variation on a bacterial theme? Trends Microbiol. 2005;13:262–265. doi: 10.1016/j.tim.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Bell SD, Cairns SS, Robson RL, Jackson SP. Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol Cell. 1999;4:971–982. doi: 10.1016/s1097-2765(00)80226-9. [DOI] [PubMed] [Google Scholar]
- Blumentals, Itoh M, Olson GJ, Kelly RM. Role of Polysulfides in Reduction of Elemental Sulfur by the Hyperthermophilic Archaebacterium Pyrococcus furiosus. Appl Environ Microbiol. 1990;56:1255–1262. doi: 10.1128/aem.56.5.1255-1262.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Bell SD, Lebbink RJ, de Vos WM, van der Oost J. The Sulfolobus solfataricus Lrp-like protein LysM regulates lysine biosynthesis in response to lysine availability. J Biol Chem. 2002;277:29537–29549. doi: 10.1074/jbc.M203528200. [DOI] [PubMed] [Google Scholar]
- Busenlehner LS, Pennella MA, Giedroc DP. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol Rev. 2003;27:131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, Ryu S. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- Coulocheri SA, Pigis DG, Papavassiliou KA, Papavassiliou AG. Hydrogen bonds in protein-DNA complexes: Where geometry meets plasticity. Biochimie. 2007;89:1291–1303. doi: 10.1016/j.biochi.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- Crankshaw MW, Grant GA. Modification of Cysteine. In: Coligan JE, editor. Current protocols in protein science. Brooklyn, N.Y.: Wiley; 1996. pp. 5.23.1–5.23.18. [DOI] [PubMed] [Google Scholar]
- Darcy A, Elmore C, Stihle M, Johnston JE. A novel approach to crystallising proteins under oil. J Cryst Growth. 1996;168:175–180. [Google Scholar]
- Declercq JP, Evrard C, Clippe A, Stricht DV, Bernard A, Knoops B. Crystal structure of human peroxiredoxin 5, a novel type of mammalian peroxiredoxin at 1.5 A resolution. J Mol Biol. 2001;311:751–759. doi: 10.1006/jmbi.2001.4853. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fiala G, Stetter KO. Pyrococcus-Furiosus Sp-Nov Represents a Novel Genus of Marine Heterotrophic Archaebacteria Growing Optimally at 100-Degrees C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- Fomenko DE, Gladyshev VN. Identity and functions of CxxC-derived motifs. Biochemistry. 2003;42:11214–11225. doi: 10.1021/bi034459s. [DOI] [PubMed] [Google Scholar]
- Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Ouhammouch M. Archaeal transcription and its regulators. Mol Microbiol. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr Op Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- Hedderich R, Klimmek O, Kroger A, Dirmeier R, Keller M, Stetter KO. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev. 1998;22:353–381. [Google Scholar]
- Hethke C, Geerling AC, Hausner W, de Vos WM, Thomm M. A cell-free transcription system for the hyperthermophilic archaeon Pyrococcus furiosus. Nucleic Acids Res. 1996;24:2369–2376. doi: 10.1093/nar/24.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Jones S, van Heyningen P, Berman HM, Thornton JM. Protein-DNA interactions: A structural analysis. J Mol Biol. 1999;287:877–896. doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- Kim SO, Merchant K, Nudelman R, Beyer WF, Jr, Keng T, DeAngelo J, Hausladen A, Stamler JS. OxyR: a molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- Kleinjan WE, de Keizer A, Janssen AJ. Kinetics of the chemical oxidation of polysulfide anions in aqueous solution. Water Res. 2005;39:4093–4100. doi: 10.1016/j.watres.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Kleinjan WE, de Keizer A, Janssen JH. Biologically Produced Sulfur. Top Curr Chem. 2003;230:167–188. [Google Scholar]
- Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosower NS, Kosower EM. Diamide: an oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Engelmann A, Horlacher R, Qu Q, Vierke G, Hebbeln C, Thomm M, Boos W. TrmB, a sugar-specific transcriptional regulator of the trehalose/maltose ABC transporter from the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem. 2003;278:983–990. doi: 10.1074/jbc.M210236200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Moulakakis C, Koning SM, Hausner W, Thomm M, Boos W. TrmB, a sugar sensing regulator of ABC transporter genes in Pyrococcus furiosus exhibits dual promoter specificity and is controlled by different inducers. Mol Micro. 2005;57:1797–1807. doi: 10.1111/j.1365-2958.2005.04804.x. [DOI] [PubMed] [Google Scholar]
- Lie TJ, Wood GE, Leigh JA. Regulation of nif expression in Methanococcus maripaludis - Roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J Biol Chem. 2005;280:5236–5241. doi: 10.1074/jbc.M411778200. [DOI] [PubMed] [Google Scholar]
- Lipscomb GL, Keese AM, Cowart DM, Schut GJ, Thomm M, Adams MW, Scott RA. SurR: a transcriptional activator and repressor controlling hydrogen and elemental sulphur metabolism in Pyrococcus furiosus. Mol Microbiol. 2009;71:332–349. doi: 10.1111/j.1365-2958.2008.06525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Vierke G, Wenke AK, Thomm M, Ladenstein R. Crystal structure of the archaeal heat shock regulator from Pyrococcus furiosus: A molecular chimera representing eukaryal and bacterial features. J Mol Biol. 2007;369:474–488. doi: 10.1016/j.jmb.2007.03.044. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Lin D, Tempel W, Praissman JL, Rose JP, Wang BC. Parameter-space screening: a powerful tool for high-throughput crystal structure determination. Acta Crystallogr D Biol Crystallogr. 2005;61:520–527. doi: 10.1107/S0907444905003239. [DOI] [PubMed] [Google Scholar]
- Lovell SC, I, Davis W, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Madej T, Gibrat JF, Bryant SH. Threading a Database of Protein Cores. Proteins Struct Funct Genet. 1995;23:356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nakano S, Erwin KN, Ralle M, Zuber P. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol. 2005;55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- Newberry KJ, Nakano S, Zuber P, Brennan RG. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc Natl Acad Sci USA. 2005;102:15839–15844. doi: 10.1073/pnas.0506592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Paget MS, Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- Pedone E, Ren B, Ladenstein R, Rossi M, Bartolucci S. Functional properties of the protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus: a member of a novel protein family related to protein disulfide-isomerase. Eur J Biochem. 2004;271:3437–3448. doi: 10.1111/j.0014-2956.2004.04282.x. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Ren B, Tibbelin G, de Pascale D, Rossi M, Bartolucci S, Ladenstein R. A protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus contains two thioredoxin fold units. Nat Struct Biol. 1998;5:602–611. doi: 10.1038/862. [DOI] [PubMed] [Google Scholar]
- Schut GJ, Bridger SL, Adams MW. Insights into the Metabolism of Elemental Sulfur by the Hyperthermophilic Archaeon Pyrococcus furiosus: Characterization of a Coenzyme A- Dependent NAD(P)H Sulfur Oxidoreductase. J Bacteriol. 2007;189:4431–4441. doi: 10.1128/JB.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Tartaglia LA, Ames BN. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. Automated structure solution, density modification and model building. Acta Crystallogr D Biol Crystallogr. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierke G, Engelmann A, Hebbeln C, Thomm M. A novel archaeal transcriptional regulator of heat shock response. J Biol Chem. 2003;278:18–26. doi: 10.1074/jbc.M209250200. [DOI] [PubMed] [Google Scholar]
- Wilson DO, Johnson P, McCord BR. Nonradiochemical DNase I footprinting by capillary electrophoresis. Electrophoresis. 2001;22:1979–1986. doi: 10.1002/1522-2683(200106)22:10<1979::AID-ELPS1979>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


