Abstract
Chronic alcohol exposure affects the central nervous system, influences behavior, and induces neuroadaptive changes in vertebrate species including our own. The molecular mechanisms responsible for chronic alcohol effects have not been fully elucidated due to the complexity of alcohol’s actions. Here we use zebrafish, a novel tool in alcohol research, to reveal a large number of genes that respond to chronic alcohol treatment. We demonstrate differential gene expression in response to chronic alcohol treatment using full genome DNA microarrays and find a total of 1914 genes to show a minimum of 2-fold and significant expression level change (1127 were up- and 787 were down-regulated). Approximately two-thirds of these genes had no known previous functional annotation. The results of the microarray analyses correlated well with those obtained on a selected subset of genes analyzed by quantitative real-time RT-PCR. Analyses of the differentially expressed genes with known annotations were enriched for a variety of molecular functions. Only a fraction of these known genes has been reported in the literature to be alcohol related. We conclude that the zebrafish is an excellent tool for the analysis of genes associated with alcohol’s actions in vertebrates, one which may facilitate the discovery and better understanding of the mechanisms of alcohol abuse.
Keywords: chronic alcohol exposure, DNA Microarray, gene chip, gene expression, gene function, zebrafish brain, alcohol adaptation
INTRODUCTION
Chronic alcohol exposure induces neuroadaptive changes leading to tolerance, adaptation, and alcohol seeking in humans and other non-human animals [19, 13, 26, 51]. Modification of gene expression may underlie the altered brain function resulting from chronic alcohol exposure [56, 45, 50, 22, 39, 16] and has been observed in cultured neurons [8] and in the brain of the rat [9, 49, 70], the mouse [2, 3, 70] and humans (postmortem brain tissue from alcoholics, e.g. [41, 46, 63, 43].
Zebrafish has also been utilized in alcohol research (e.g. [29, 10,23]). The first DNA microarray analysis of the effects of alcohol has been published using zebrafish [39]. However, this latter study employed a complex mixed alcohol dosing regimen that involved repeated short exposure to alcohol and repeated withdrawal from the substance followed by a long term withdrawal before the gene expression analysis. Briefly, the effect of continuous chronic alcohol exposure has not been analyzed at the gene expression level. Nevertheless, significant behavioral adaptation to the substance in zebrafish has been demonstratedafter such chronic exposure using a social behavior paradigm [28]. The goal of the current study is to investigate the gene expression changes that accompany chronic alcohol exposure using a genome wide DNA microarray system in zebrafish.
The rationale for conducting this work in zebrafish is several fold. Most importantly, this small and prolific vertebrate offers unprecedented efficiency for high throughput mutagenesis and drug screens, a major advantage because the complexity of alcohol’s actions requires global analyses. Our work is the first to attempt comprehensive gene expression profiling of the effects of chronic alcohol exposure alone. With this analysis we hope to identify individual molecular targets, cluster of targets, and/or key biochemical interactions associated with functional changes induced by chronic alcohol in the brain. These putative targets then may serve as candidate genes in future follow up forward and reverse genetic studies or drug screens using zebrafish.
MATERIALS AND METHODS
Zebrafish: The rationale for its use and the methods of its maintenance
Although rodents have been successfully utilized in alcohol research, a recent upsurge of alcohol studies with zebrafish suggests that this species also has some utility in this endeavor. First, analysis of zebrafish responses to alcohol may reveal evolutionarily conserved mechanisms common to vertebrates. Second, zebrafish may allow perhaps the most efficient discovery of novel mechanisms associated with alcohol exposure among vertebrate laboratory organisms. Alcohol administration in zebrafish is simple: the subject is immersed in the alcohol solution and its blood-brain alcohol reaches steady levels within 40 min [20, 28] allowing precise control of the duration and dose of alcohol exposure. This dosing method may be superior to drug self-administration paradigms or passive alcohol exposure methods employed with mammalian laboratory species. For example, passive alcohol administration paradigms including alcohol vapor and/or invasive injection methods induce potential stress or anxiety whereas active alcohol administration paradigms (self administration) depend upon motivational characteristics as well as homeostatic processes associated with fluid and/or food intake (see e.g. [47]). It is also important to note that zebrafish as a model of functional changes in behavior induced by alcohol has not only face validity (e.g. [29, 30, 31, 20, 21] but also construct validity [4, 44, 59, 39]. Last, this small (4 cm long) fish is housed in large numbers in small tanks (a shoaling fish), and its prolific nature lends it to both forward and reverse genetic studies that require testing of a large number of subjects.
Adult long-fin zebrafish (six month old) bred and raised in our Vivarium (University of Toronto Mississauga, Mississauga ON, Canada) were randomly selected for this experiment. The fish originated from founders that were purchased from a local pet store (Big Al’s Aquarium Warehouse, Mississauga, ON, Canada) and were of the second filial generation bred in our facility. The rationale for the choice of this “pet store” variety fish was as follows. Genetically well defined strains of zebrafish may enhance reproducibility within and across laboratories. Nevertheless, these strains having been bred for several generations and having lost numerous alleles (due to inbreeding) are expected to possess idiosyncratic features. A genetically heterogeneous population, such as the long fin wild type (these fish originate from a Singapore breeding facility where the effective population size approaches tens of thousands of fish) may be closer to what one may consider the “prototypical” zebrafish and we felt such a population would be a better starting point for the first comprehensive DNA microarray-based gene expression analysis [28]. All fish were raised and housed in a standard manner as described previously [28]. In all experiments approximately 50–50% males and females were included each treatment group.
Alcohol treatment
Our treatment employed for the chronic alcohol group was a continuous exposure to alcohol without repeated and/or prolonged withdrawal from the substance. We choose this simple chronic paradigm to avoid the potential complications arising from mixing alcohol adaptation related processes with withdrawal induced or sensitization induced mechanisms [11]. Although in the human clinic chronic alcohol exposure almost certainly has several components associated not only with adaptation to the substance but also with multiple withdrawal episodes, we wanted to focus on the mechanisms of adaptation alone, a reductionist approach we feel will allow us to disentangle the complexities of alcohol effects. Another reason why we decided to employ this continuous alcohol exposure dosing regimen is that we utilized the exact same procedure in our prior, behavioral, studies [28] and this way the behavioral and gene expression results will be comparable. Two groups of fish were compared: chronic alcohol exposed and chronic freshwater exposed. Fish were randomly assigned to these groups and sample sizes were 30 for both the Chronic freshwater (control), and the Chronic 0.50% alcohol exposed fish. All fish were maintained under identical conditions in the same vivarium room as described before [28]. The chronic alcohol concentration of 0.50% was achieved in fish of both the behavioral and gene expression studies by increasing the alcohol concentration in the holding tank by 0.125% in a stepwise manner once every four days as described before [28]. Once the 0.50% concentration was reached, it was maintained for an additional 9 days (total of 21 days with continuous alcohol exposure). The alcohol dose was chosen based upon previous studies with zebrafish (for references see [28] and so that the brain alcohol levels would approximate what is observed in the human clinic [58, 67]. Increased mortality or morbidity was not observed in any group. To ascertain consistent and continuous alcohol exposure, the alcohol concentration of the holding tanks was monitored using the AM1 Alcohol Analyzer (Analox Instruments, London, UK) and the water (or alcohol solution) of exposure tanks was changed daily as described before [28].
Analysis of alcohol levels in the brain
In order to properly interpret the alcohol induced gene expression changes we have analyzed the amount of alcohol in the brain of our fish as described before [28]. Briefly, a small subset of fish exposed to the 0.5% EtOH concentration or the freshwater (0.0% EtOH concentration) were analyzed: immediately after the alcohol treatment period, fish were decapitated, their brains were quickly removed and homogenized. The homogenate (whole brain extract) was analyzed using the AM1 Alcohol Analyser (Analox Instruments, London, UK). The instrument works on the principle that alcohol-oxygen oxidoreductase (AOD) catalyzes the enzymatic oxidation of ethyl alcohol to acetaldehyde and the oxygen consumption by the enzyme reduces the oxygen content of the solution, which is quantified by an oxygen sensitive sensor. The instrument has been successfully utilized to measure small alcohol amounts from liquid solution or homogenized solid tissue samples and is expected to be one of the most precise methods (see [28] and references therein).
Total RNA Preparation
At end of the chronic treatment period zebrafish were killed by decapitation and the heads were placed on dry-ice. The brain of the fish was removed and immediately frozen in liquid nitrogen and was subsequently stored at −80°C until RNA extraction. Brain samples were homogenized in TRIzol reagent (100mg/ml) and total RNA was extracted according to the manufacturer's protocol (Invitrogen Corporation, Carlsbad, CA). In order to minimize between-animal variability, samples were pooled for fish kept in the same aquarium (n=10 per aquarium). We had 3 such aquaria for alcohol treated and 3 for control fish. That is, a total of 6 RNA sample pools were prepared and separately analyzed (i.e. 6 DNA microarray chips were used). The total RNA was purified using the RNeasy Mini Kit (QIAGEN Inc. Mississauga, CA). The RNA concentrations were determined; the A260/A280 and A260/A230 ratios were calculated as indices of protein and volatile compound contamination, respectively, using a spectrophotometer (NanoDrop ND-1000; NanoDrop Technologies, Inc. Wilmington, DE, USA). The integrity of the total RNA was determined by electrophoresis on a 1.5% denatured agarose formaldehyde gel stained with ethidium bromide. Total RNA was then used for microarray analysis and quantitative RT-PCR experiments.
DNA microarrays, cDNA Probe Synthesis, Hybridization and Scanning
The NimbleGen Zv7 Zebrafish Gene Expression 385K microarray used in the study contained 37,157 probe sets with up to 12 probes of 60mer oligonucleotides per gene. The arrays represented at least 24,000 genes plus additional ESTs from multiple Zebrafish tissues. The design of the array relied on gene and EST information from several sources; Ensembl 46 (August 2007, Zv7), RefSeq (September 2007), TIGR (Release 14.0), UniGene (Build 54), Vega 27, and ZGC (August 2007).
10 µg total RNA of each pooled sample was converted to double stranded cDNA using the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen). The synthesized cDNAs were then incubated with 1µl of 4mg/ml RNase A at 37°C for 10 min, precipitated and the pellet dissolved with 20µl of VWR water. Each cDNA sample was quantified and verified to meet the following requirements: (concentration ≥ 100ng/µl; A260/A280 ≥ 1.8, A260/A230 ≥ 1.8). 1 µg cDNA of each sample was synthesized as probe and labeled with Cy3-conjugated random nonamers (TriLink Biotechnologies, San Diego, CA) and hybridized to a NimbleGen Zv7 Zebrafish Gene Expression 385K microarray following the protocol by Roche NimbleGen, Inc (Madison, USA). The microarrays were incubated on the NimbleGen Hybridization System 4 (Roche NimbleGen) for 16 hrs at 42°C. The pools of RNA (see above) from alcohol treated and control zebrafish brains were used to generate cDNA probes, which were then hybridized to microarrays separately. The gene expression level and folds changes (alcohol treated VS control) were calculated from three separate samples.
The hybridized slides were washed at 10X wash buffer I, II and III (Roche NimbleGen), dried by nitrogen gas at room temperature, and scanned with an Axon GenePix Pro 4200A microarray scanner at 5 microns resolution, 532 nm wavelength (Molecular Devices, Sunnyvale, CA) associated software GenePix Pro software (Axon, Union city, CA, USA).
Global Gene Expression Analysis
The scanned images of the arrays were quantified using NimbleScan software (Roche NimbleGen). The expression data were normalized by quantile normalization across replicate on arrays as described previously [6]. The gene expression values were generated by RMA (Robust Multichip Average) analysis [36]. Subsequent microarray data analysis was performed using ArrayStar software (DNASTAR, Inc. Madison, USA).Average ratios of expression values of chronic alcohol treated vs. control fish were calculated from three experiments. Genes were considered differentially expressed when the level of expression change in brain of the alcohol treated fish was at least two fold (up-regulation or down-regulation) as compared to the control fish brains and the difference between the alcohol treated and control groups was significant (p < 0.05, moderated T test, in which the false discovery rate was controlled by the Benjamini Hochberg correction method).
Functional Classification of Differentially Expressed Genes
The UniGene ID numbers of genes that showed significant differential expression were uploaded into the Database for Annotation, Visualization and Integrated Discovery [35, 18] (http://david.abcc.ncifcrf.gov) to obtain DAVID gene ID conversion and functional annotation. The genes were classified into functional groups using ‘GO TERM Molecular Function at all levels’.
Real-time RT-PCR Analysis
Real-time RT-PCR is often employed as a follow up validation procedure subsequent to DNA microarray analysis. While real-time PCR and the microarray analysis may be associated with different technical subtleties and issues, the former is considered more quantitative and more precise. Ten genes were selected as targets from different functional groups and fold change ranges for follow-up investigation. β-actin was used as the control gene for normalization. Primers were designed online with Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi/). The primer pairs were selected from different exons and checked with OligoAnalyzer (Integrated DNA Technologies, Coraville, IA) for the secondary structure and dimer formation. Each primer and amplicon sequence was tested using BLAST to ensure minimal similarity with any other sequence. All primer sequences and gene names used for validation are reported in Table 1.
Table 1.
Primers designed for quantitative RT-PCR
| Gene Product Name | Gene ID | Primer used for qRT-PCR |
|---|---|---|
| Dopamine receptor3 (D3) | NM_183067 | 5’-3’ CCTGTGTGCCATCAGTATCG 5’-3’ ACGGCAAGTAAAACGACACC |
| N-methyl D-aspartate receptor1 (NMDA-R1) |
ENSDART00000034849 | 5’-3’ AAGCCCAAGTGTGGTTTGAC 5’-3’ AGCAGAGCCGTCAGATTTGT |
| P450, Family 11, Subfamily A, Polypeptide1 |
OTTDART00000026902 | 5’-3’ TCCCGAAACCAGAGCAATAC 5’-3’ GCTCAAACTTGCTCCTGACC |
| Neuromedin B Receptor | OTTDART00000006562 | 5’-3’ TTGGTCACTTGTGTGCCAGT 5’-3’ CCTTCAAGCAGGTCCAGAAC |
| Novel Protein Similar To Solute Carrier Family 5 (SLC5A1) |
OTTDART00000028378 | 5’-3’ TCAGAGTGGTCAGCTCTTCG 5’-3’ CTGTTATCATACGCGCCAGA |
| Novel Protein Similar To Opioid Growth Factor Receptor (OGFR) |
OTTDART00000001831 | 5’-3’ AGCCAAGGACATGCAAGACT 5’-3’ ACCCCACCATTCTTTGTGAA |
| Apolipoprotein A-IV (APOA4) |
TC237267 | 5’-3’ AAGTGCTGAGGGAGCGTCT 5’-3’ AAGAGTGGCCTTCAGGGTTT |
| Carboxypeptidase B1 (CPB1) | OTTDART00000027072 | 5’-3’ ATGAGGGAAGAACCATGCAC 5’-3’ CCAGAAGGTTGGTCATGTCA |
| Alcohol Dehydrogenase 8A (ADH8A) |
OTTDART00000010244 | 5’-3’ GCAAAGCCAATTCAGCTCAT 5’-3’ CGAATGCCTTGTCCAGTTTT |
| Solute Carrier Family 6, (neurotransmitter transporter, glycine), member 9 (SLC6A9) |
OTTDART00000032714 | 5’-3’GCCTGCATACTGAAGGTTCC 5’-3’CCCCACTGCATAGCCTACAC |
| β-actin | ZV700S00000303 | 5’-3’ GCTACAGCTTCACCACCACA 5’-3’ AGGAAGGAAGGCTGGAAGAG |
A two-step approach was taken in which the initial reverse transcription was followed by the quantitative PCR amplification. After DNase I (Invitrogen Corporation, CA) treatment, DNA-free total RNA was reverse transcribed using a dT20VN primer (Sigma, Oakville, ON) with SuperScript II. Each RNA reaction had a control reaction without reverse transcriptase to evaluate any genomic DNA contamination. Two µl of the diluted reaction was used as template for each 25 µL real-time PCR amplification. Reactions were assembled using 2X Kapa-SYBR Green Mix (Kapa Biosystems, Inc, Woburn, MA). The M×4000 Multiplex Quantitative PCR System (Stratagene, La Jolla, CA) was used for sample processing and analysis according to the manufacturer’s instructions. Samples were incubated at 95°C for 10 min prior to thermal cycling (40 cycles of: 95°C for 15 s, 57°C for 25 s, and 72°C for 30 s). In order to confirm the amplification specificity and the identity of the PCR products, a melting curve analysis between 55 °C and 95 °C in 1°C increments was also carried out using the M×4000 software. After exporting the ROX-normalized fluorescence measurements to Microsoft Excel, the program LinRegPCR was used to determine the efficiency of each reaction [57]. These efficiencies were used in the final calculation of fold induction from the ΔCt values and the expression of each test gene was normalized to the level of beta-actin within each sample prior to comparisons between samples. To compare the DNA Microarray and qRT-PCR results we calculated the expression fold change seen in the alcohol treated samples relative to the control samples as follows RF = (A/βA)/(Σ(C/βC)/3). Where RF is the relative fold change, A is the mRNA expression level of the alcohol treated samples, C is the mRNA expression level for the control samples, βA is the β actin housekeeping gene expression level in the alcohol treated samples, and βC is the β actin housekeeping gene expression level in the control treated samples. It is notable that although we used the β actin results corresponding to the appropriate sample, the β actin levels were not significantly different between alcohol treated and control samples, confirming that this gene may be considered a housekeeping gene under the current circumstances. The relative fold change results were compared between the DNA microarray and qRT-PCR methods using paired t-tests with Bonferroni correction for type 1 error. Furthermore, we also analyzed the correlation for the results generated by these two methods and calculated a Pearson product moment correlation coefficient based upon the means of fold change values obtained for the 10 selected genes.
RESULTS
Alcohol levels in the brain
The brain of zebrafish (whole brain extract) that were exposed to 0.5% alcohol was found to contain 0.33% (vol/vol %) alcohol (SEM = 0.0335). That is, the concentration of alcohol in the brain of these fish was approximately 66% of the external concentration, a value that corresponds well to the one found by others using a different alcohol quantification method but the same dose and zebrafish population (long fin wild type) [20]. Quantification of alcohol concentration in the brain of fish exposed only to freshwater gave a mean of 0.02 % alcohol (with SEM = 0.018). The latter value being apparently above zero may be due to experimental error variation and to the fact that the AMI alcohol analyzer quantifies alcohol concentration based upon an enzymatic reaction that reduces oxygen content. Thus fluctuation of oxygen levels due to error variation itself could result in the obtained value. Indeed, the alcohol concentration obtained for the freshwater control fish is not significantly different from zero (t = 1.314, df = 4, p > 0.25), whereas the brain alcohol concentration obtained for the alcohol treated fish was significantly above zero (t = 9.863, df = 2, p < 0.01).
Gene expression profile
A total of 6 RNA samples from zebrafish brains (i.e. 3 separate pooled samples from alcohol treated and 3 from control fish) were individually hybridized to DNA microarrays. Gene expression levels across different microarrays within a treatment group (i.e. within the control group and within the alcohol treated group) were highly reproducible as reflected by the high correlation coefficients obtained for pairs of microarrays in all possible combinations within each treatment condition (r > 0.9), a result which is in line with recent studies in which pooling of brain samples (zebrafish or human) reduced error variation and increased reliability of results [42, 46, 68, 69, 39]. Notably, vertebrate genomes contain several genes that may be highly similar in terms of nucleotide sequence and also in terms of their function. These genes are expected to respond similarly to alcohol treatment. One of the largest families of structurally and functionally similar proteins are the G-protein coupled olfactory receptors, or odorant receptors. Here we randomly chose six of these receptors whose genes are represented on our microarray with gene ID ENSDART00000014683; ENSDART00000076967; ENSDART00000100123; ENSDART00000100136; ENSDART00000100142 and ENSDART00000103138. Importantly, mRNA expression of all of these genes showed similar levels of down-regulation in chronic alcohol treated fish, with fold changes 4.697; 3.042; 3.584; 3.777; 4.928, respectively. A similar conclusion may be drawn from the analysis of another set of genes, in this case three sequences corresponding to the human opioid growth factor receptor complex (OGFR, with gene ID ENSDART00000080681; OTTDART00000001830 and OTTDART00000001831). All three genes were up-regulated in the chronic alcohol exposed fish with similar fold changes: 4.090, 4.205 and 3.854, respectively. Last, it is also notable that each of the 37157 probe sets on the microarray contains a maximum of 12 different probes corresponding to distinct sequences of the gene represented by that given probe set. We calculated the standard error for each of these probe sets and found minimal variation within a set. Compared to the average fluorescence intensity (expressed in arbitrary units) obtained for the six microarrays (3 arrays for control fish: 2639, 3342, 2721; and 3 arrays for chronic alcohol treated fish: 3117, 4111, 3209) the average variability within the probe sets (expressed as SEM) was negligible (3 arrays for control fish: 0.748, 0.780, 0.754; 3 arrays for chronic alcohol treated fish: 0.741, 0.770, 0.759). The above results demonstrate excellent reliability of the microarray to detect gene expression changes consistently.
The microarray analysis also allowed us to check what proportion of genes was expressed in the brain in the control as well as in the alcohol treated fish as compared to all the genes in the genome. In the past, the estimates of brain specific gene expression showed that about 40% of the genes are expressed in the vertebrate brain [27]. Here with the more extensive genome representation in our microarray we found 56.4% of known genes with Entrez ID to be expressed in the control, and 58.0% of known genes with Entrez ID to be expressed in the chronic alcohol exposed fish brain.
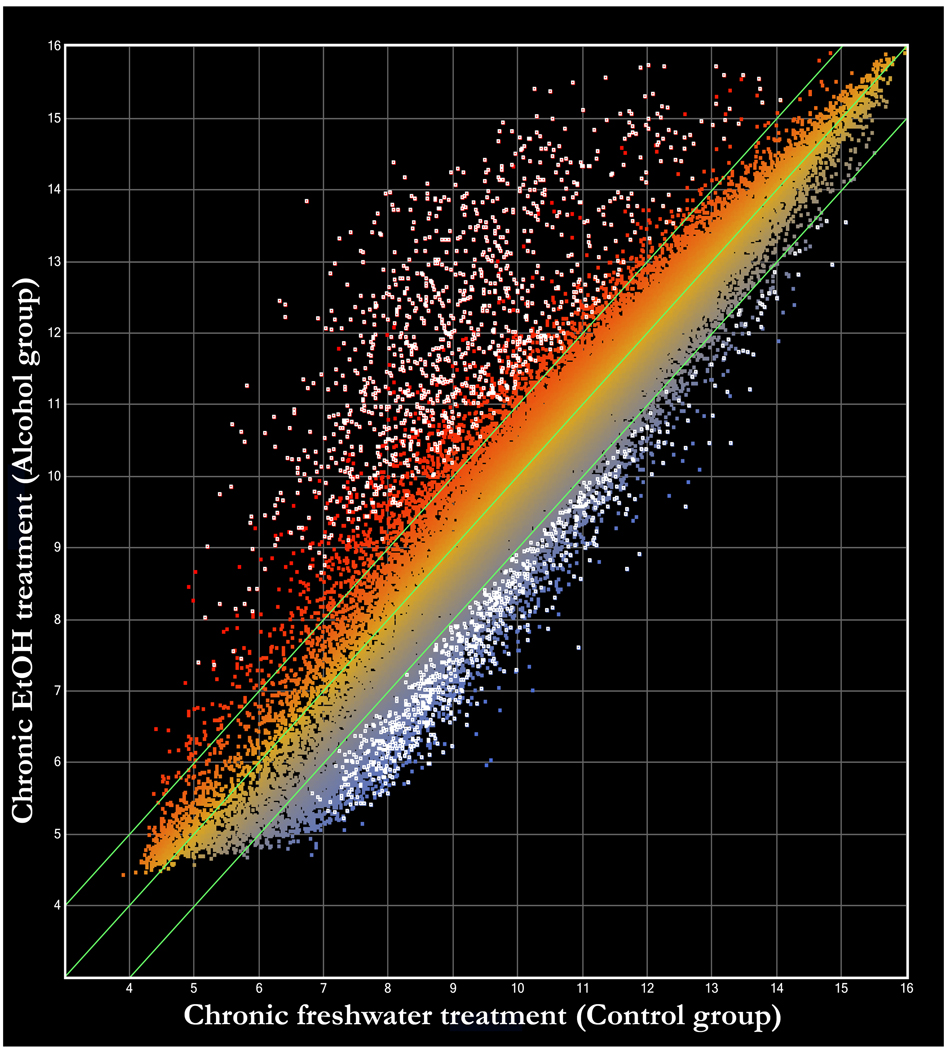
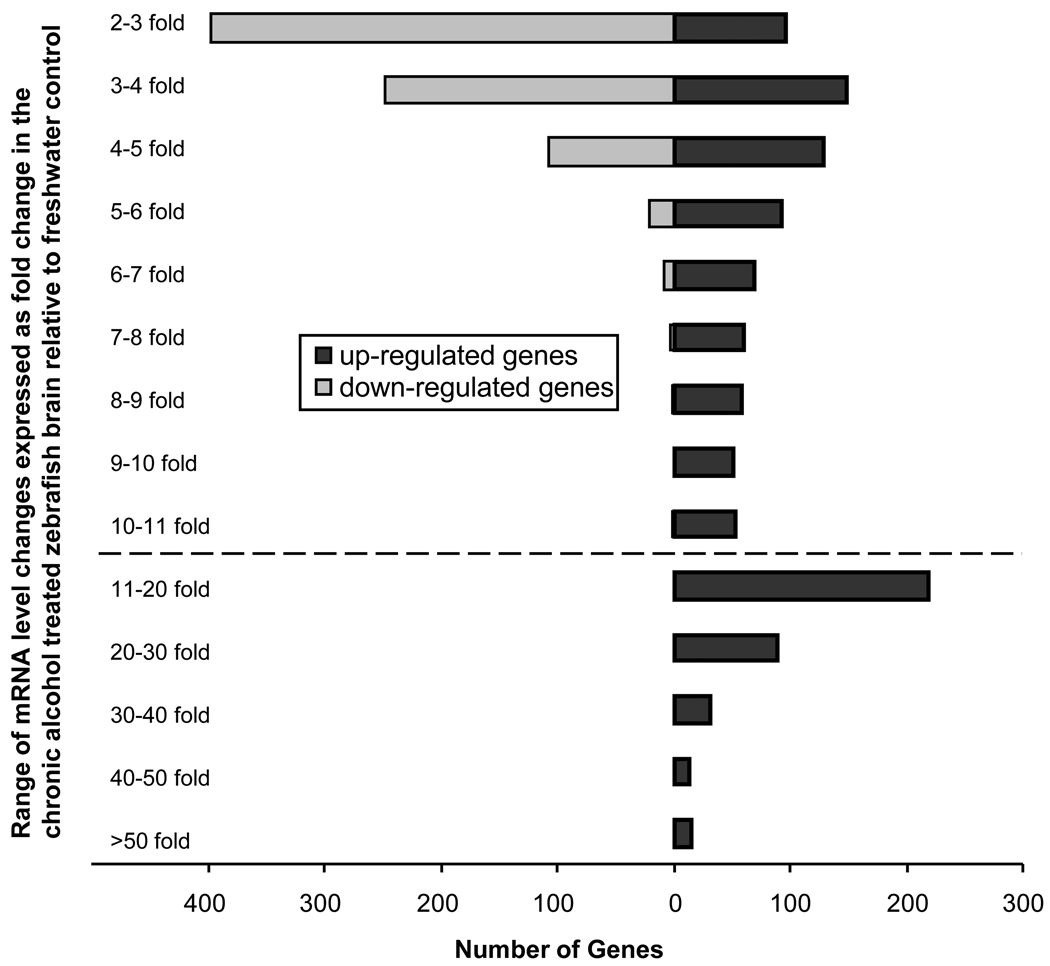
Figure 1 shows the expression level of all genes we tested. 1914 sequences/genes changed their expression level at least two fold and significantly (p < 0.05) between control and alcohol treated zebrafish, which represents more than 5% of the approximately 37,157 probe sets present on the microarray. Of these differentially expressed genes, 696 have known functions (36.4%), and 1218 have no known functions (63.6%). 1127 genes were significantly up-regulated in chronic alcohol treated fish and these included 506 functionally known and 621 unknown genes. The fold change of significantly up-regulated genes ranged from the minimum fold value set by us, i.e. 2 (TC264763) to 79, the maximum fold change we detected that corresponded to a gene encoding a functionally uncharacterized protein (SEQ_IDOTTDART00000017839). 878 genes were significantly and at least two-fold down-regulated, including 597 functionally unknown and 190 known genes. The fold change ranged from the set value of 2 (ENSDART0000089366) to 8, the maximum fold change representing a down regulated gene (SEQ_IDZV700S00004250) whose function is unknown. Figure 2 summarizes the distribution of genes according to up or down-regulation and fold expression change, and shows that a substantial number of genes were up-regulated several fold (more than 10 fold) whereas most down-regulated genes were down-regulated only 2–4 fold. In summary, chronic alcohol treatment induced differential gene expression in the brain of zebrafish in a large number of genes, and up-regulation was observed in almost 30% more genes than down-regulation. Furthermore, more than 60% of the differentially expressed genes we identified represent functionally uncharacterized genes.
Figure 1.
The expression level of 1914 genes in the zebrafish brain shows at least a two fold and significant (p < 0.05) difference between chronic alcohol (EtOH) treated and freshwater control fish (white dots). The 37,157 dots on the figure represent single genes or expressed sequence tags (EST’s). The ordinate shows the gene expression of the alcohol treated fish and the abscissa shows the gene expression results for the control fish (expressed in relative arbitrary units of fluorescence intensity on a logarithm scale). Genes whose expression level is identical between the two groups lie on the diagonal line. Genes whose expression level is at least two fold different between the groups lie outside of the two lines parallel to the central diagonal line. The dots positioned above the upper diagonal line represent genes that are upregulated and the dots appearing below the lower diagonal line represent genes that are downregulated in the alcohol treated group as compared to the freshwater control group. White dots represent those genes whose expression level differs between the alcohol treated and freshwater control groups at least two fold AND the difference is significant (p < 0.05).
Figure 2.
Distribution of genes according to the change of their expression level in response to chronic alcohol treatment. Black bars represent up-regulated and gray bars down regulated genes. The distribution of genes is shown using 1-fold bins from the set threshold (2 fold) up to 11 fold and using 10-fold bins from 11 fold up (below the dashed line). Note the substantial number of genes that show overexpression (up-regulation) in the range between 11 to 50 fold. Also note that the majority of down-regulated genes fall in the category of 2–3 fold reduction of mRNA expression.
qRT-PCR verification
Quantitative reverse transcriptase polymerase chain reaction, or qRT-PCR, is perhaps the most precise method available for the quantification of gene expression. Here, we analyzed the expression of 10 genes and investigated whether the results correlate with those of the DNA microarray analysis. We chose these genes to include ones with different fold expression changes between control and alcohol treated samples. For example, we chose 6 up-regulated genes with fold changes ranging from about 4 to 20 folds and 2 down-regulated genes with fold changes about 2 to 6 fold downregulation (0.45 and 0.165 fold change, respectively). These differentially expressed genes were also chosen so that they would represent a variety of functional groups. We also included 2 genes whose expression level, according to our Microarray results, did not significantly change as a result of alcohol treatment. Table 2 summarizes these genes and shows both the microarray and the qRT-PCR fold change results along with a statistical comparison of the data generated by these two methods. Briefly, the qRT-PCR analysis of the expression level of these genes generally confirmed the microarray results. No significant differences were found in the fold change values quantified using the two methods. We also investigated the correlation between the results obtained by the DNA Microarray and the qRT-PCR and calculated a Spearman correlation coefficient. We chose this non-parametric statistic for two reasons. One, the distribution of data was not normal and two, we were interested in the ability of the two methods to detect relative differences between genes in mRNA expression and not the absolute fold change value. The correlation coefficient we obtained was high and significant (r = 0.881, p < 0.001).
Table 2.
Comparison of qRT-PCR and DNA Microarray results
| Gene Product Name | Gene ID | qRT-PCR fold change** (mean ± SEM) |
DNA Microarray fold change (mean ± SEM) |
Statistical comparison PCR vs Microarray* |
|---|---|---|---|---|
| Genes detected as significantly upregulated by Microarray | ||||
| P450, Family 11, Subfamily A, Polypeptide1 |
OTTDART00000026902 | 7.073 ± 1.286 | 12.705 ± 3.593 | t = 1.322 df = 2 p > 0.05 |
| Novel Protein Similar To Solute Carrier Family 5 (SLC5A1) |
OTTDART00000028378 | 79.34 ±10.004 | 31.78 ± 11.81 | t = 4.154 df = 2 p > 0.05 |
| Novel Protein Opioid Growth Factor-like Receptor (OGFR) |
OTTDART00000001831 | 9.826 ± 2.929 | 4.253 ± 0.900 | t = 1.73 df = 2 p > 0.05 |
| Apolipoprotein A-IV (APOA4) |
TC237267 | 53.32 ± 14.683 | 50.70 ± 3.416 | t = 0.861 df = 2 p > 0.05 |
| Carboxypeptidase B1 (CPB1) |
OTTDART00000027072 | 15.81 ± 10.585 | 19.45 ± 8.341 | t = −0.278 df = 2 p > 0.05 |
| Alcohol Dehydrogenase 8A (ADH8A) |
OTTDART00000010244 | 4.038 ± 0.368 | 3.800 ± 0.994 | t = 0.240 df = 2 p > 0.05 |
| Genes detected as significantly downregulated by Microarray | ||||
| Neuromedin B Receptor |
OTTDART00000006562 | 0.400 ±0.069 | 0.450 ± 0.11 | t = 0.204 df = 2 p > 0.85 |
| Solute Carrier Family 6, (glycine transporter) member 9 (SLC6A9) |
OTTDART00000032714 | 0.450 ± 0.111 | 0.165 ± 0.047 | t = 1.836 df = 2 p > 0.05 |
| Genes detected as not significantly changed by Microarray | ||||
| Dopamine receptor3 (D3) |
NM_183067 | 0.918 ± 0.048 | 0.871 ± 0.116 | t = 0.629 df = 2 p > 0.05 |
| N-methyl D-aspartate receptor1 (NMDA-R1) |
ENSDART00000034849 | 1.025 ± 0.106 | 1.008 ± 0.088 | t = 0.200 df = 2 p > 0.05 |
statistical comparison was performed using paired t-test with Bonferroni correction for multiple comparisons.
fold changes are expressed as the ratio between alcohol treated and control samples, a ratio larger than 1 represents up and a ratio smaller than 1 represents down-regulation of gene expression.
Functional classification of differentially expressed genes
Chronic alcohol treatment responsive genes (the differentially expressed genes) that had UniGene IDs were classified into functional groups using the DAVID Bioinformatics Functional Group Analysis application. The analysis was conducted based upon molecular functions. Functional information of chronic alcohol responsive genes was obtained for as many genes as possible from public domain databases. The analysis demonstrated that the chronic alcohol responsive genes were implicated in a wide variety of Gene Ontology (GO) functions, including enzyme regulator activity, transporter activity, transcription regulator activity, hydrolase activity, receptor activity, oxidation activity, nucleotide binding, transferase activity, ion binding, and molecular transducer activity (Table 3). Most functional groups contained both up and down regulated genes. All of the genes classified under the terms “binding and oxidation” were increased in expression level in the alcohol treated fish brain. Genes classified by the functional groups enzyme regulator activity, enzyme inhibitor activity; ion binding, transporter activity, transferase activity, hydrolase activity were predominantly up-regualted. The down-regulated genes were mainly represented in the functional groups gated-channel activity, cation channel activity, and transcription regulator activity (Table 3).
Table 3.
Functional clustering of alcohol responsive genes
| Molecular Function |
GO terms | Number of upregulated genes |
Number of downregulated genes |
|---|---|---|---|
| Enzyme regulator activity |
protease inhibitor activity | 21 | 2 |
| endopeptidase inhibitor activity | 20 | 2 | |
| enzyme inhibitor activity | 21 | 2 | |
| serine-type endopeptidase inhibitor activity | 8 | 1 | |
| enzyme regulator activity | 24 | 3 | |
| peptidase activity | 11 | 2 | |
| Binding | vitamin binding | 8 | 0 |
| pyridoxal phosphate binding | 6 | 0 | |
| cofactor binding | 10 | 0 | |
| Transmembrane transporter activity |
secondary active transmembrane transporter activity | 4 | 3 |
| active transmembrane transporter activity | 5 | 4 | |
| Transcription regulator activity |
transcription factor activity | 10 | 18 |
| transcription regulator activity | 10 | 22 | |
| sequence-specific DNA binding | 8 | 14 | |
| DNA binding | 20 | 21 | |
| nucleic acid binding | 28 | 24 | |
| Hydrolase activity | serine-type peptidase activity | 6 | 1 |
| serine-type endopeptidase activity | 6 | 0 | |
| endopeptidase activity | 9 | 0 | |
| peptidase activity | 11 | 2 | |
| Receptor activity | transmembrane receptor activity | 4 | 7 |
| G-protein coupled receptor activity | 2 | 4 | |
| Oxidoreductase activity, tetrapyrrole binding |
monooxygenase activity | 6 | 0 |
| heme binding | 5 | 0 | |
| tetrapyrrole binding | 5 | 0 | |
| iron ion binding | 6 | 0 | |
| auxiliary transport protein activity, transporter activity |
transporter activity | 24 | 18 |
| active transmembrane transporter activity | 5 | 4 | |
| substrate-specific transporter activity | 14 | 15 | |
| ion transmembrane transporter activity | 5 | 15 | |
| transmembrane transporter activity | 7 | 15 | |
| substrate-specific transmembrane transporter activity | 6 | 15 | |
| cation transmembrane transporter activity | 3 | 13 | |
| cation channel activity | 2 | 9 | |
| metal ion transmembrane transporter activity | 2 | 10 | |
| auxiliary transport protein activity | 2 | 7 | |
| calcium channel inhibitor activity | 2 | 6 | |
| substrate specific channel activity | 2 | 10 | |
| ion channel activity | 2 | 10 | |
| calcium channel activity | 2 | 6 | |
| channel activity | 2 | 10 | |
| passive transmembrane transporter activity | 2 | 10 | |
| Nucleotide binding, transferase activity, transferring phosphorus- containing groups |
protein-tyrosine kinase activity | 1 | 5 |
| transferase activity | 32 | 11 | |
| nucleoside-triphosphatase activity | 11 | 2 | |
| protein kinase activity | 8 | 7 | |
| pyrophosphatase activity | 11 | 2 | |
| hydrolase activity, acting on acid anhydrides, in | |||
| phosphorus-containing anhydrides | 11 | 2 | |
| hydrolase activity, acting on acid anhydrides | 11 | 2 | |
| phosphotransferase activity, alcohol group as acceptor | 8 | 8 | |
| kinase activity | 12 | 8 | |
| protein serine/threonine kinase activity | 7 | 3 | |
| transferase activity, transferring phosphorus-containing | |||
| groups | 13 | 8 | |
| ATP binding | 19 | 9 | |
| adenyl ribonucleotide binding | 19 | 9 | |
| ATPase activity | 5 | 1 | |
| adenyl nucleotide binding | 19 | 9 | |
| ribonucleotide binding | 21 | 11 | |
| purine ribonucleotide binding | 21 | 11 | |
| purine nucleotide binding | 21 | 11 | |
| nucleotide binding | 22 | 13 | |
| Ion binding | cation binding | 33 | 21 |
| metal ion binding | 34 | 21 | |
| ion binding | 34 | 21 | |
| transition metal ion binding | 25 | 7 | |
| zinc ion binding | 16 | 7 | |
| Molecular transducer activity |
transmembrane receptor activity | 4 | 7 |
| receptor activity | 8 | 10 | |
| molecular transducer activity | 10 | 11 | |
| signal transducer activity | 10 | 11 | |
Several of the significantly differentially expressed genes were found to belong to the cytochrome P450 family (Table 4) and solute carrier family (Table 5). These genes are known to be involved in multiple mechanisms and biochemical pathways. Several myelination-related genes were also observed among the significantly up and down-regulated genes responding to chronic alcohol treatment in the zebrafish brain. For example, peripheral myelin protein (OTTDART00000025430), a major component of myelin, was down-regulated in the brain of alcohol treated zebrafish. A number of myelin-related genes that were previously identified as differentially expressed in the brain of patients that suffered from alcoholism [40, 41, 42, 43, 46] also showed comparably altered expression levels in the chronic alcohol treated zebrafish. Furthermore, neuronal acetylcholine receptor subunit alpha-9 (ENSDART00000077335, down-regulated), nicotinic acetylcholine receptor subunit gamma (BC079485.1, down-regulated); cGMP-dependent protein kinase 1, alpha isozyme (AW018995, up-regulated) were observed to change their expression level in the current study, and these genes too have been reported as alcohol responsive genes by others (e.g. [12]).
Table 4.
List of Cytochrome P450 family genes whose expression significantly and at least 2 fold changed in response to chronic alcohol treatment
| Gene Product name | Gene ID | Fold change |
|---|---|---|
| cytochrome P450, family 2, subfamily A, polypeptide 6 | TC256080 ZV700S00005088 |
17.900 up 8.070 up |
| cytochrome P450, family 2, subfamily C, polypeptide 8; | TC249614 | 4.694 up |
| cytochrome P450, family 2, subfamily C, polypeptide19 | ZV700S00000732 | 9.047 up |
| similar to Cytochrome P450 family 2 subfamily G polypeptide 1 |
NM_001020822 | 9.907 up |
| cytochrome P450, family 2, subfamily J | BC081666.1 | 7.904 up |
| cytochrome P450, family 2, subfamily J, polypeptide 2, |
OTTDART00000005960 | 13.506 up |
| similar to cytochrome P450, family 2, subfamily J | OTTDART00000005967 | 9.481 up |
| cytochrome P450, family 2, subfamily K, polypeptide 6 |
OTTDART00000024129 | 29.998 up |
| Similar to cytochrome P450, family 2, subfamily U | ZV700S00000465 | 4.118 up |
| cytochrome P450 family 2 subfamily X polypeptide 12 |
ENSDART00000026629 | 6.875 up |
| cytochrome P450, family 3, subfamily A, polypeptide 5 |
ZV700S00004361 | 6.872 up |
| cytochrome P450, family 4, subfamily V, polypeptide 2 |
NM_001077602 | 21.121 up |
| cytochrome P450 family | NM_001079704 | 5.278 up |
| Cytochrome P450. | ZV700S00004360 | 5.027 up |
| cytochrome P450, family 8, subfamily B, polypeptide 1 |
ZV700S00003386 ZV700S00005260 |
6.399 up 6.365 up |
| cytochrome P450, family 11, subfamily A, polypeptide 1; |
OTTDART00000026902 OTTDART00000026903 OTTDART00000026904 OTTDART00000026905 |
11.007 up 4.898 up 4.414 up 6.185 up |
| similar to Cytochrome P450, family 12, subfamily G, polypeptide1 |
NM_001020822 | 9.907 up |
| cytochrome P450, family 22, subfamily X, polypeptide 12 |
ENSDART00000026629 | 6.875 up |
| Similar to cytochrome P450, family 46, subfamily A, polypeptide 1 |
OTTDART00000029923 | 10.954 up |
| cytochrome P450, family 71, subfamily A, | NM_001044309 | 19.293 up |
Table 5.
list of Solute carries family genes whose expression significantly and at least 2 fold changed in response to chronic alcohol treatment
| Gene Product name | Gene ID | Fold changes |
|---|---|---|
| solute carrier family 2, member 9 (Facilitated glucose transporter) |
TC251933 | 3.484 up |
| solute carrier family 2 (facilitated glucose transporter), member 12 |
OTTDART00000027012 | 2.790 up |
| similar to solute carrier family 5 (sodium/glucose cotransporter), member 1 |
OTTDART00000028378 | 21.546 up |
| solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
TC261898 ZV700S00003520 |
9.688 up 11.041 up |
| solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
OTTDART00000032714 | 6.533 down |
| similar to solute carrier family 6 (neutral amino acid transporter), member 19 |
ZV700S00003698 | 11.517 up |
| solute carrier family 7 (cationic amino acid transporter), member 1 |
ZV700S00004691 | 63.554 up |
| solute carrier family 7 (cationic amino acid transporter), member 7 |
ZV700S00002874 | 5.764 up |
| similar to solute carrier family 7(neutral amino acid transporter) member 10; |
OTTDART00000002042 OTTDART00000002041 |
23.502 up 12.385 up |
| similar to solute carrier family 12 (sodium/chloride transporters), member 3 |
OTTDART00000020878 | 3.566 down |
| solute carrier family 13 (sodium/sulphate symporters), member 1 |
OTTDART00000026911 | 6.920 up |
| solute carrier family 13 member 3 (sodium- dependent dicarboxylate transporter), |
ZV700S00005271 | 15.762 up |
| solute carrier family 15 (Oligopeptide transporter) member 1 |
AW018970 OTTDART00000024429 OTTDART00000030736 |
17.018 up 17.711 up 18.987 up |
| Solute carrier family 16 member 5 (Monocarboxylate transporter) |
ENSDART00000041559 ENSDART00000074436 |
3.333 down 4.363 down |
| solute carrier family 22 (organic cation transporter), member 1 |
TC244476 | 11.718 up |
| solute carrier family 22 (organic anion transporter), member 6" |
NM_207077 | 3.790 up |
| solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
OTTDART00000025053 | 3.240 up |
| "solute carrier family 25 (mitochondrial carrier), member 4 |
OTTDART00000027611 | 2.215 down |
| solute carrier family 25, member 42 | NM_001045453 | 3.573 down |
| similar to solute carrier family 26, member 3 | OTTDART00000009290 OTTDART00000009291 |
16.802 up 13.055 up |
| similar to vertebrate solute carrier family 26 | OTTDART00000019974 | 3.387 down |
| solute carrier family 27 (fatty acid) | ZV700S00005292 | 5.772 up |
| Solute carrier family 29 member 2 (Nucleoside transporter) |
ENSDART00000053572 | 2.353 down |
| Similar to solute carrier family 29 (Nucleoside transporters), member 2 |
ZV700S00006259 | 18.570 up |
| Similar to solute carrier family 30 (zinc transporter), member 2 |
OTTDART00000006662 | 2.716 down |
| solute carrier family 34 (sodium phosphate), member 2 |
BC054940.1 NM_182877 |
9.914 up 7.842 up |
| similar to solute carrier family 35 (UDP- glucuronic acid), member D1 |
OTTDART00000027646 | 3.834 up |
| similar to solute carrier family 43, member 2 | ZV700S00004830 | 8.617 up |
| Mitochondria solute carrier protein (Hypothetical protein) |
ZV700S00005450 | 4.322 up |
| similar to Mitochondria solute carrier protein | ZV700S00003943 | 24.718 up |
In addition, a number of other differentially expressed genes were found to be associated with neuronal communication related processes. For example, genes encoding a protein involved in the regulation of G-protein signaling (OTTDART00000024799), a G-protein coupled receptor (ENSDART00000104325) and different voltage-gated ion channels such as Voltage-dependent L-type calcium channel subunit alpha-1F (ENSDART00000011078) belong to this functional cluster. Genes whose protein products function in synaptic transmission also cluster under the above category. For example, the genes for syntaxin-1A (ENSDART00000081947), and the member of the solute carrier family (SLC6A6, TC261898) were found differentially expressed in the chronic alcohol treated zebrafish. Genes whose products are involved in ethanol metabolism including alcohol dehydrogenases, e.g. alcohol dehydrogenase 5 (AF295407), alcohol dehydrogenase 8a (OTTDART00000010244) and alcohol dehydrogenase 8b (OTTDART00000010244) were found significantly up-regulated. The genes whose products are associated with fatty acid metabolism such as apolipoprotein A-IV (TC237267), Apolipoprotein B (TC238656) were also found up-regulated. A functionally uncharacterized gene similar to ATP synthase H+ transporting, mitochondrial F0 complex, subunit b, isoform 1 (OTTDART00000028541) was up-regulated in alcohol treated zebrafish. ATP is a fundamental molecule involved in energy storage and almost all catalytic processes. But extracellular ATP also plays an important role in synaptic transmission, acting as a neurotransmitter and/or a neuromodulator [14].
We have identified several functionally uncharacterized, i.e., not annotated genes, that appear to be similar to already known alcohol responsive genes previously identified in mammals. These include genes encoding opioid receptors [53, 12] and growth factors whose expression has been shown to change in response to various forms of alcohol treatment in mice or in human alcoholic patients [54, 12]. In our microarray analysis these genes are annotated as functionally uncharacterized genes similar to the one that encodes a human opioid receptor (OTTDART00000001830), similar to human insulinlike growth factor receptor (TC244476), and functionally uncharacterized genes similar to those that encode the human insulin-like growth factor binding protein and acid labile subunit (IGFALS, OTTDART00000024228).
Last, we noted some important genes whose expression level did not change as a result of chronic alcohol exposure. Neuroadaptations induced by chronic drug exposure are believed to share mechanisms with the plastic changes that underlie memory formation [50]. Two of the many key players in neuronal plasticity that are believed to be also involved in drug addiction are the NMDA and the AMPA receptors, glutamate gated ion channels [38]. Interestingly, although several sequences corresponding to subunits of these receptors on our microarray did detect significant mRNA expression, the level of expression did not differ between control and alcohol treated zebrafish. It is important to note, however, that posttranscriptional mechanisms, including trafficking the already translated gene product to the cell membrane (e.g. the synaptic density), have been suggested to underlie plastic changes associated with drug addiction as well as memory formation in these receptors [5, 7, 66], processes that cannot be detected by DNA microarrays.
Discussion
Recently, adaptation to alcohol after chronic exposure identical in dosing regimen employed here has been demonstrated in zebrafish [28]. In this latter study the chronic alcohol (or freshwater) exposure was followed by application of acute doses of alcohol and the alcohol effects were quantified by measuring shoaling behavior (social preference) in zebrafish. The results showed that in response to acute alcohol treatment alone (prior “chronic” freshwater treatment) experimental zebrafish reduced their shoaling behavior. The reduction of shoaling response, however, was abolished by prior chronic alcohol treatment. That is, fish that received chronic alcohol before the acute alcohol exposure behaved as freshwater control, a phenomenon that was interpreted as adaptation to alcohol. Chronic alcohol exposure induced changes were also detected in neurochemical analyses [28]. Levels of dopamine, DOPAC (the metabolite of dopamine), serotonin and 5-HIAA (the metabolite of serotonin) all increased in the studied genetically heterogeneous zebrafish population in response to chronic alcohol treatment as compared to freshwater control.
The molecular mechanisms underlying the behavioral adaptation and these neurochemical changes, or in general, the effects of chronic alcohol exposure at the molecular level are largely unknown in zebrafish. We view our comprehensive DNA microarray analysis as the first step in the direction of answering these questions. Due to the complex pharmacological profile of alcohol, identification of underlying mechanisms may be best achieved using comprehensive methods and this was our rationale for the use of whole genome DNA microarrays. Our array was not only the most comprehensive thus far employed for the analysis of chronic alcohol induced gene expression changes, but unlike another recent microarray analysis [39], our alcohol dosing regimen was devoid of complicating factors associated with repeated and short term alcohol exposure induced sensitization and long term withdrawal induced functional changes [11]. Briefly, the gene expression changes we detected could only be the result of long lasting and stable alcohol exposure. Despite this reductionist approach, we identified close to 2000 differentially expressed genes less then 40% of which had known functions. Bioinformatic analysis of these functionally annotated genes suggested that chronic alcohol treatment engaged a broad spectrum of molecular mechanisms in the zebrafish brain. Knowledge of these mechanisms may be used for future targeted mutagenesis and phenotypical functional testing studies and/or for the design of candidate gene based forward genetics.
Our bioinformatic analysis identified several functional groups of differentially expressed genes that corresponded well to those identified in postmortem brains of human patients with alcoholism or in animal models (mice and rats) or in neuronal cell cultures (Table 6, [41, 65, 63, 61, 46, 16]. These results confirm evolutionary conservation of numerous biochemical processes engaged by alcohol and suggest that zebrafish is appropriate for the analysis of these processes.
Table 6.
Comparison of functional gene clusters found to respond with differential gene expression to chronic alcohol treatment in different experimental models
| Molecular function |
Thibault et al., 2000 In vitro neurons |
Lewohl et al., 2000 Human |
Mayfield et al.,2002 Human |
Sokolov et al., 2003 Human |
Daniels and Buck, 2002 Mouse |
Saito et al., 2002 Rat |
Our study Zebrafish |
|---|---|---|---|---|---|---|---|
| Binding activity |
56 | 44 | 30 | 62 | 53 | 15 | 123 |
| Enzyme & Catalytic activity |
34 | 37 | 0 | 0 | 34 | 11 | 41 |
| Signal transducer activity |
24 | 19 | 11 | 18 | 17 | 5 | 22 |
| Transcription regulator activity |
17 | 7 | 0 | 12 | 10 | 0 | 32 |
| Transporter activity |
6 | 18 | 12 | 22 | 9 | 4 | 42 |
| Enzyme regulator activity |
5 | 6 | 0 | 0 | 7 | 0 | 27 |
| Defense/imm unity protein activity |
4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Apoptosis regulator activity |
3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Translation regulator activity |
0 | 0 | 0 | 0 | 3 | 0 | 0 |
| number genes of analysis |
nearly 6000 genes |
Over 4000 genes |
approximat ely 10 000 genes |
expression of 12,626 genes |
7634 unique genes |
over 5000 genes |
approximate ly 32,292 genes |
| Animal model |
human SH- SY5Y neuroblasto ma cells ( neural cells) |
frontal cortex of alcoholics |
frontal and motor cortices of alcoholics |
postmortem brains |
hippocampus of DBA/2 J and C57BL/6 J mice |
rat hippocamp us |
Brain of zebrafish |
The number of differentially expressed genes we identified in chronic alcohol treated zebrafish represents approximately 5% of the genome, a result in agreement with two, less comprehensive, studies conducted recently in humans [22, 24].
The large number of differentially expressed genes confirmed a complex effect profile of alcohol, but in this complexity we did note important functional clusters. For example, differential expression of numerous genes encoding ion channels was notable. This is in good agreement with the increasing amount of evidence suggesting that alcohol interacts directly with ion channels [33, 46, 12]. This interaction may trigger secondary homeostatic or feedback responses at the gene expression level and this is what we may have detected here for the first time. We have found several genes downregulated after chronic alcohol treatment that encoded cation channels including calcium channels (e.g. L type voltage-dependent calcium channel alpha 1F and 1S subunits [ENSDART00000011078, ENSDART00000039686]). Others have proposed that calcium influx via L-type calcium channels is required for dopamine release at dopaminergic synapses, a process that may underlie rewarding properties of drugs of abuse [60, 25]. Accordingly, reduced L-type calcium channel expression may lead to reduction of dopaminergic neurotransmission, which then may explain the anhedonic effects of chronic alcohol use and the need for continued stimulation via this drug (avoidance of withdrawal effects).
One striking finding of our study concerns the large number of differentially expressed genes (mostly showing upregulation in response to chronic alcohol treatment) all encoding solute carrier (SLC) family proteins (Table 5). The solute carrier group of membrane transport proteins includes over 300 members organized into 47 families [34]. These transmembrane proteins play key roles in the transport of small molecules including neurotransmitters across vesicular and plasma membranes [32]. In our study, we found 11 SLC family genes whose expression level was altered after chronic alcohol exposure including SLC2, SLC5, SLC6, SLC7, SLC12, SLC13, SLC15, SLC16, SLC22, SLC25, SLC26. Some genes in this family have unknown functions. But, for example, the SLC6 transporter family includes the transporters that mediate the Na+ -dependent uptake of dopamine, 5-HT, norepinephrine, glycine and GABA [32]. This family of genes has not been previously associated with alcohol related changes in the vertebrate brain. The above mechanisms all relate to neuronal communication and our results clearly indicate that several mechanistic aspects of these processes were altered by chronic alcohol exposure.
Alcohol and its metabolite acetaldehyde may be produced physiologically in the organism [55] and vertebrates may encounter alcohol in their natural environment too and may take up the substance passively or via food or drink. Thus not surprisingly alcohol metabolic pathways have been found conserved in several vertebrate species (human, rat, mouse and zebrafish). The major pathway for alcohol metabolism in mammals is a characteristic two-step enzymatic process: alcohol dehydrogenase converts alcohol to acetaldehyde, and subsequently acetaldehyde is metabolized to acetate by aldehyde dehydrogenase [64, 71]. In our current study we found several zebrafish genes encoding alcohol dehydrogenase (AF295407.1; OTTDART00000010244; OTTDART00000010247; OTTDART00000026022) and aldehyde dehydrogenase (BC081581.1) to be up-regulated in the brain of zebrafish after chronic alcohol exposure. This up-regulation may represent a homeostatic compensatory response that could, at least partially, underlie the adaptation to chronic alcohol exposure at the behavioral level in zebrafish we observed previously [28]. We have also identified several other genes that may be involved in the metabolism of alcohol. The gene P4502E1 (CYP2E1) belongs to a large and diverse superfamily [62, 17]. Although its protein product is not abundant in the mammalian brain [48], it has been shown to play a significant role in ethanol oxidation [71]. We also observed a large number of Cytochrome P450 (CYP450) family genes (Table 4) up-regulated in response to chronic alcohol treatment. The most common reaction catalyzed by cytochrome P450 is the monooxygenase reaction. Protein products of these genes are also implicated in steroid and fatty acid metabolism. In summary, based upon our findings with SLC and P450 family genes, we conclude that chronic alcohol treatment induces numerous metabolic processes including hormone metabolism in the brain of zebrafish, a conclusion that supports the hypotheses proposed for mice and men (e.g. [46, 12].
Our study also identified members of the Apolipoprotein family to respond to chronic alcohol treatment. These include Apolipoprotein-A-IV (TC237267) and apolipoprotein B (TC238656; ZV700S00005187). Apolipoproteins are lipid-binding proteins involved in the transport of triglycerides, phospholipids, and cholesterol in the plasma [1]. The gene encoding Apolipoprotein-A1 has been reported to show increased expression after chronic ethanol exposure in several studies [46, 12, 15] but the specific genes we identified for zebrafish have not been found to be alcohol treatment responsive. Notably, the genes coding for apolipoprotein Al and A-IV are closely linked and tandemly organized on the human chromosome 11, and Apolipoprotein-A-IV and Apolipoprotein-A1 genes are believed to have originated from a common evolutionary ancestor [37].
Although more than 70% of zebrafish gene sequences we identified as differentially expressed have not been annotated in the gene ontology (GO) database [52], we could verify a number of genes that were previously reported as alcohol responsive in other vertebrate organisms (rats, mice and humans). These genes involve those whose products are associated with processes including myelination, neurotransmission, hormonal responses, calcium signaling, G-protein mediated signaling and cAMP-dependent responses, results that again confirm evolutionarily conservation of alcohol related mechanisms.
We also identified a number of functionally uncharacterized alcohol responsive genes using zebrafish. These results are also notable as they demonstrate how much work still needs to be done. Appropriate understanding of the complex actions of alcohol will only be possible if most, if not all, of these genes are functionally characterized and their biochemical/biological roles revealed. The functional characterization of these genes is likely to proceed fastest with the use of zebrafish.
Although at the very first stages of discovery, the above suggest that zebrafish is a translationally relevant tool with which the genetics of alcohol induced changes in the vertebrate brain may be studied.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by NIH/NIAAA (1R01AA015325-01A2 grant to RG). Some of the procedures were performed at the Canadian Drosophila Microarray Centre which was supported by an NSERC Canada multi-user facility grant. J.T.W. is supported by an NSERC Canada Discovery Grant. Z.R. is supported by a CIHR Team Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ando S, Tachibana A, Yamada S, Kishimura H. Apolipoprotein complexity in Japanese eel Anguilla japonica: truncated apolipoprotein A–I and apolipoprotein A-I-like protein in plasma lipoproteins. Biosci Biotechnol Biochem. 2005;69:2258–2262. doi: 10.1271/bbb.69.2258. [DOI] [PubMed] [Google Scholar]
- 2.Bachtell RK, Wang YM, Freeman P, Risinger FO, Ryabinin AE. Alcohol drinking produces brain region-selective changes in expression of inducible transcription factors. Brain Res. 1999;847:157–165. doi: 10.1016/s0006-8993(99)02019-3. [DOI] [PubMed] [Google Scholar]
- 3.Bachtell RK, Weitemier AZ, Galvan-Rosas A, Tsivkovskaia NO, Risinger FO, Phillips TJ, Grahame NJ, Ryabinin AE. The Edinger-Westphal-lateral septum urocortin pathway and its relationship to alcohol consumption. J Neurosci. 2003;23:2477–2487. doi: 10.1523/JNEUROSCI.23-06-02477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbazuk WB, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell JA, McPherson JD, Johnson SL. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beitner-Johnson D, Guitart X, Nestler EJ. Common intracellular actions of chronic morphine and cocaine in dopaminergic brain reward regions. Ann N Y Acad Sci. 1992;654:70–87. doi: 10.1111/j.1749-6632.1992.tb25957.x. [DOI] [PubMed] [Google Scholar]
- 6.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 7.Borgland SL, Taha SA, Sarti F, Fields HL, Bonci A. Orexin A in the VTA is critical for the induction of synaptic plasticity and behavioral sensitization to cocaine. Neuron. 2006;49:589–601. doi: 10.1016/j.neuron.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Chandler LJ, Sutton G, Norwood D, Sumners C, Crews FT. Chronic ethanol increases N-methyl-D-aspartate-stimulated nitric oxide formation but not receptor density in cultured cortical neurons. Mol Pharmacol. 1997;51:733–740. doi: 10.1124/mol.51.5.733. [DOI] [PubMed] [Google Scholar]
- 9.Chang SL, Patel NA, Romero AA. Activation and desensitization of Fos immunoreactivity in the rat brain following ethanol administration. Brain Res. 1995;679:89–98. doi: 10.1016/0006-8993(95)00210-h. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee D, Gerlai R. High precision liquid chromatography analysis of dopaminergic and serotoninergic responses to acute alcohol exposure in zebrafish. Behav Brain Res. 2009;200:208–213. doi: 10.1016/j.bbr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemmesen L, Hemmingsen R. Physical dependence on ethanol during multiple intoxication and withdrawal episodes in the rat: evidence of a potentiation. Acta Pharm. Toxicol. 1984;55:345–350. doi: 10.1111/j.1600-0773.1984.tb01993.x. [DOI] [PubMed] [Google Scholar]
- 12.Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- 13.Crews FT, Braun CJ, Hoplight B, Switzer RC, III, Knapp DJ. Binge ethanol consumption causes differential brain damage in young adolescent rats compared with adult rats. Alcohol Clin Exp Res. 2000;24:1712–1723. [PubMed] [Google Scholar]
- 14.Cunha RA, Ribeiro JA. Purinergic modulation of [(3)H]GABA release from rat hippocampal nerve terminals. Neuropharmacology. 2000;39:1156–1167. doi: 10.1016/s0028-3908(99)00237-3. [DOI] [PubMed] [Google Scholar]
- 15.Damodaran S, Dlugos CA, Wood TD, Rabin RA. Effects of chronic ethanol administration on brain protein levels: a proteomic investigation using 2-D DIGE system. Eur J Pharmacol. 2006;547:75–82. doi: 10.1016/j.ejphar.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Daniels GM, Buck KJ. Expression profiling identifies strain-specific changes associated with ethanol withdrawal in mice. Genes Brain and Behavior. 2002;1:35–45. doi: 10.1046/j.1601-1848.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- 17.Danielson P. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3:561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- 18.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 19.Diamond I, Gordon AS. Cellular and molecular neuroscience of alcoholism. Physiol Rev. 1997;77:1–20. doi: 10.1152/physrev.1997.77.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Dlugos CA, Rabin RA. Ethanol effects on three strains of zebrafish: model system for genetic investigations. Pharmacol Biochem Behav. 2003;74:471–480. doi: 10.1016/s0091-3057(02)01026-2. [DOI] [PubMed] [Google Scholar]
- 21.Dooley K, Zon LI. Zebrafish: a model system for the study of human disease. Curr Opi Genet Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 22.Fan L, Bellinger F, Ge YL, Wilce P. Genetic study of alcoholism and novel gene expression in the alcoholic brain. Addict Biol. 2004;9:11–18. doi: 10.1080/13556210410001674040. [DOI] [PubMed] [Google Scholar]
- 23.Fernandes Y, Gerlai R. Long-term behavioral changes in response to early developmental exposure to ethanol in zebrafish. Alcoholism: Clinical and Experimental Research. 2009;33:601–609. doi: 10.1111/j.1530-0277.2008.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- 25.Flatscher-Bader T, van der Brug MP, Landis N, Hwang JW, Harrison E, Wilce PA. Comparative gene expression in brain regions of human alcoholics. Genes Brain Behav. 2006;5:78–84. doi: 10.1111/j.1601-183X.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- 26.Fleming M, Mihic SJ, Harris RA. Ethanol. In: Hardman JG, Limbird LE, editors. The pharmacological basis of therapeutics. 10th ed. New York: McGraw-Hill; 2001. pp. 429–445. [Google Scholar]
- 27.Fritzsch B. Of Mice and Genes: Evolution of Vertebrate Brain Development. Brain Behav Evol. 1998;52:207–217. doi: 10.1159/000006564. [DOI] [PubMed] [Google Scholar]
- 28.Gerlai R, Chatterjee D, Pereira T, Sawashima T, Krishnannair R. Acute and Chronic alcohol dose: Population differences in behavior and neurochemistry of zebrafish. Genes Brain and Behavior. 2009;8:586–599. doi: 10.1111/j.1601-183X.2009.00488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerlai R, Lahav M, Guo S, Rosenthal A. Drinks like a fish: zebra fish (Danio rerio) as a behavior genetic model to study alcohol effects. Pharmacol Biochem Behav. 2000;67:773–782. doi: 10.1016/s0091-3057(00)00422-6. [DOI] [PubMed] [Google Scholar]
- 30.Gerlai R, Lee V, Blaser R. Effects of acute and chronic ethanol exposure on the behavior of adult zebrafish (Danio rerio) Pharmacol Biochem Behav. 2006;85:752–761. doi: 10.1016/j.pbb.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerlai R. Zebra fish: an uncharted behavior genetic model. Behav Genet. 2003;33:461–468. doi: 10.1023/a:1025762314250. [DOI] [PubMed] [Google Scholar]
- 32.Gether U, Andersen PH, Larsson OM, Schousboe A. Neurotransmitter transporters: molecular function of important drug targets. Trends Pharmacol Sci. 2006;27:375–383. doi: 10.1016/j.tips.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Harris RA. Ethanol actions on multiple ion channels: which are important? Alcohol Clin Exp Res. 1999;23:1563–1570. [PubMed] [Google Scholar]
- 34.Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins: Introduction. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 35.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karathanasis SK, Oettgen P, Haddad IA, Antonarakis SE. Structure, evolution, and polymorphisms of the human apolipoprotein A4 gene (APOA4) Proc Natl Acad Sci USA. 1986;83:8457–8461. doi: 10.1073/pnas.83.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessels HW, Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron. 2009;61:340–350. doi: 10.1016/j.neuron.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kily LJ, Cowe YC, Hussain O, Patel S, McElwaine S, Cotter FE, Brennan CH. Gene expression changes in a zebrafish model of drug dependency suggest conservation of neuro-adaptation pathways. J Exp Biol. 2008;211:1623–1634. doi: 10.1242/jeb.014399. [DOI] [PubMed] [Google Scholar]
- 40.Kril JJ, Halliday GM. Brain shrinkage in alcoholics: a decade on and what have we learned? Prog Neurobiol. 1999;58:381–387. doi: 10.1016/s0301-0082(98)00091-4. [DOI] [PubMed] [Google Scholar]
- 41.Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24:1873–1882. [PubMed] [Google Scholar]
- 42.Lewohl JM, Van Dyk DD, Craft GE, Innes DJ, Mayfield RD, Cobon G, Harris RA, Dodd PR. The application of proteomics to the human alcoholic brain. Ann N Y Acad Sci. 2004;1025:14–26. doi: 10.1196/annals.1316.002. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90:1050–1058. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- 44.Loucks E, Carvan MJ., 3rd Strain-dependent effects of developmental ethanol exposure in zebrafish. Neurotoxicol Teratol. 2004;26:745–755. doi: 10.1016/j.ntt.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 45.Matsumoto I, Wilce PA, Buckley T, Dodd P, Puzke J, Spanagel R, Zieglgansberger W, Wolf G, Leng S, Rommelspacher H, Finckh U, Schmidt LG. Ethanol and gene expression in brain. Alcohol Clin Exp Res. 2001;25:82–86. doi: 10.1097/00000374-200105051-00015. [DOI] [PubMed] [Google Scholar]
- 46.Mayfield RD, Lewohl JM, Dodd A, Herlihy PR, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- 47.Miller SS, Goldman ME, Erickson CK, Shorey RL. Induction of physical dependence on and tolerance to ethanol in rats fed a new nutritionally complete and balanced liquid diet. Psychopharmacology (Berl.) 1980;68:55–59. doi: 10.1007/BF00426650. [DOI] [PubMed] [Google Scholar]
- 48.Montoliu C, Sancho-Tello M, Azorin I, Burgal M, Vallés S, Renau-Piquera sJ, Guerri C. Ethanol increases cytochrome P4502E1 and induces oxidative stress in astrocytes. J Neurochem. 1995;65:2561–2570. doi: 10.1046/j.1471-4159.1995.65062561.x. [DOI] [PubMed] [Google Scholar]
- 49.Nakahara T, Hirano M, Uchimura H, Shirali S, Martin CR, Bonner AB, Preedy VR. Chronic alcohol feeding and its influence on c-Fos and heat shock protein-70 gene expression in different brain regions of male and female rats. Metabolism. 2002;51:1562–1568. doi: 10.1053/meta.2002.35595. [DOI] [PubMed] [Google Scholar]
- 50.Nestler EJ. Molecular neurobiology of addiction. Am J Addict. 2001;10:201–217. doi: 10.1080/105504901750532094. [DOI] [PubMed] [Google Scholar]
- 51.Obernier JA, Bouldin TW, Crews FT. Binge ethanol exposure in adult rats causes necrotic cell death. Alcohol Clin Exp Res. 2002;26:547–557. [PubMed] [Google Scholar]
- 52.Olsvik PA, Lie KK, Stavrum AK, Meier S. Gene-expression profiling in gill and liver of zebrafish exposed to produced water. Int J Environ Anal Chem. 2007;87:195–210. [Google Scholar]
- 53.Oswald LM, Wand GS. Opioids and alcoholism. Physiol Behav. 2004;8:339–358. doi: 10.1016/j.physbeh.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Pucilowski O, Ayensu WK, D'Ercole AJ. Insulin-like growth factor I expression alters acute sensitivity and tolerance to ethanol in transgenic mice. Eur J Pharmacol. 1996;305:57–62. doi: 10.1016/0014-2999(96)00177-x. [DOI] [PubMed] [Google Scholar]
- 55.Quertemont E, Didone V. Role of acetaldehyde in mediating the pharmacological and behavioral effects of alcohol. Alcohol Res Health. 2006;29:258–265. [PMC free article] [PubMed] [Google Scholar]
- 56.Rahman S, Miles MF. Identification of novel ethanol-sensitive genes by expression profiling. Pharmacol Ther. 2001;92:123–134. doi: 10.1016/s0163-7258(01)00163-2. [DOI] [PubMed] [Google Scholar]
- 57.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 58.Redmond AD. Blood alcohol concentration and conscious level. Alcohol. Alcohol. 1983;18:89–91. [Google Scholar]
- 59.Reimers MJ, La Du JK, Periera CB, Giovanini J, Tanguay RL. Ethanol-dependent toxicity in zebrafish is partially attenuated by antioxidants. Neurotoxicol Teratol. 2006;28:497–508. doi: 10.1016/j.ntt.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 60.Rossetti ZL, Isola D, De Vry J, Fadda F. Effects of nimodipine on extracellular dopamine levels in the rat nucleus accumbens in ethanol withdrawal. Neuropharmacology. 1999;38:1361–1369. doi: 10.1016/s0028-3908(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 61.Saito M, Smiley J, Toth R, Vadasz C. Microarray analysis of gene expression in rat hippocampus after chronic ethanol treatment. Neurochemistry Research. 2002;27:1221–1229. doi: 10.1023/a:1020937728506. [DOI] [PubMed] [Google Scholar]
- 62.Sigel A, Sigel H, Sigel RKO. The Ubiquitous Roles of Cytochrome450 Proteins. Met Ions Life Sci. 2007;3:1–26. [Google Scholar]
- 63.Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. Alcohol ClinJ Neurosci Res. 2003;72:756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- 64.Swift R. Direct measurement of alcohol and its metabolites. Addiction. 2003;98:73–80. doi: 10.1046/j.1359-6357.2003.00605.x. [DOI] [PubMed] [Google Scholar]
- 65.Thibault C, Lai C, Wilke N, Duong B, Olive MF, Rahman S, Dong H, Hodge CW, Lockhart DJ, Miles MF. Expression profiling of neural cells reveals specific patterns of ethanol-responsive gene expression. Mol Pharmacol. 2000;58:1593–1600. doi: 10.1124/mol.58.6.1593. [DOI] [PubMed] [Google Scholar]
- 66.Ungless MA, Whistler JL, Malenka RC, Bonci A. Single cocaine exposure in vivo induces long-term potentiation in dopamine neurons. Nature. 2001;411 doi: 10.1038/35079077. 583-537. [DOI] [PubMed] [Google Scholar]
- 67.Urso T, Gavaler JS, Van Thiel DH. Blood ethanol levels in sober alcohol users seen in an emergency room. Life Sci. 1981;28:1053–1056. doi: 10.1016/0024-3205(81)90752-9. [DOI] [PubMed] [Google Scholar]
- 68.Van der Ven K, De Wit M, Keil D, Moens L, Van Leemput K, Naudts B, De Coen W. Development and application of a brain-specific cDNA microarray for effect evaluation of neuro-active pharmaceuticals in zebrafish (Danio rerio) Comp Biochem Physiol B Biochem Mol Biol. 2005;141:408–417. doi: 10.1016/j.cbpc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 69.van der Ven K, Keil D, Moens LN, Van Leemput K, van Remortel P, De Coen WM. Neuropharmaceuticals in the environment: mianserin-induced neuroendocrine disruption in zebrafish (Danio rerio) using cDNA microarrays. Environ Toxicol Chem. 2006;25:2645–2652. doi: 10.1897/05-495r.1. [DOI] [PubMed] [Google Scholar]
- 70.Vilpoux C, Warnault V, Pierrefiche O, Daoust M, Naassila M. Ethanol-sensitive brain regions in rat and mouse: a cartographic review, using immediate early gene expression. Alcohol Clin Exp Res. 2009;33:945–969. doi: 10.1111/j.1530-0277.2009.00916.x. [DOI] [PubMed] [Google Scholar]
- 71.Zimatkin SM, Buben AL. Ethanol oxidation in the living brain. Alcohol Alcohol. 2007;42:529–532. doi: 10.1093/alcalc/agm059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.