Abstract
Our knowledge of Anopheles gambiae molecular biology has mainly been based on studies using inbred laboratory strains. Differences in the environmental exposure of these and natural field mosquitoes have inevitably led to physiological divergences. We have used global transcript abundance analyses to probe into this divergence, and identified transcript abundance patterns of genes that provide insight on specific adaptations of caged and field mosquitoes. We also compared the gene transcript abundance profiles of field mosquitoes belonging to the two morphologically indistinguishable but reproductively isolated sympatric molecular forms, M and S, from two different locations in the Yaoundé area of Cameroon. This analysis suggested that environmental exposure has a greater influence on the transcriptome than does the mosquito’s molecular form-specific genetic background.
Keywords: Anopheles gambiae, field mosquitoes, laboratory strains, transcriptomic divergence, M and S molecular forms
INTRODUCTION
The mosquito Anopheles gambiae is the primary vector of Plasmodium falciparum, the causative agent of human malaria in sub-Saharan Africa. This mosquito species transmits malaria between humans across a wide variety of ecological settings throughout Africa (Gillies & de Meillon, 1968; Coetzee et al., 2000). This ecological plasticity is reflected, at the genetic level, by large numbers of molecular and chromosomal polymorphisms that provide a great evolutionary potential as a reservoir of genetic variability (Coluzzi et al., 2002; Pombi et al., 2008; Costantini et al., 2009). The recent development of genomic and functional genomic approaches in malaria vector research, together with the feasibility of rearing this mosquito species in the laboratory, have fostered the use of this model system to explore the physiology, genetics and evolution of anopheline vectors of malaria. Several major breakthroughs in our understanding of various facets of mosquito biology such as insecticide resistance, immunity and vector-parasite interactions have resulted from these studies (reviewed in Enayati et al., 2005; Michel and Kafatos, 2005; Barillas-Mury and Kumar, 2005; Chen et al., 2008; Yassine and Osta, 2010). The mosquito immune system plays a key role in its interaction with the malaria parasite Plasmodium, and a variety of mosquito effector molecules have been shown to either facilitate or prevent infection in the mosquito (Dong et al., 2006; Mendes et al., 2008; Volohonsky et al., 2010; Mitri et al., 2009; Jaramillo-Gutierrez et al., 2009; Dong et al., 2009; Garver et al., 2009; Dong and Dimopoulos, 2009). However, our current knowledge of the general molecular physiology and immune system of A. gambiae is almost exclusively based on studies performed using laboratory strains that have been selected and maintained under laboratory conditions for the past 15–30 years (Aguilar et al., 2005; Cohuet et al., 2006; Tripet, 2009). These laboratory colonies are likely to have diverged significantly from field populations from which they originated, as a result of strong founder effect at the onset of the colony, enhanced genetic drift and inbreeding in small caged populations and/or selection for some specific trait such as the ability to feed on artificial blood sources (Tchuinkam et al., 1993), susceptibility/refractoriness to parasite infection (Vernick et al., 1995; Collins et al., 1986) and/or susceptibility/resistance to insecticides (Müller et al., 2007). The colonization process together with limited environmental variance under laboratory conditions and standardized rearing methods have lead to limited genetic variation which in turn results in a dramatically reduced phenotypic variance in lab-reared colonies (Tripet et al., 2008, Tripet 2009). For example, studies in Drosophila have indicated that inbred populations have lower fitness (or stress resistance) and are less adaptable than are the outbred populations in the field from which they were derived (Frankham et al., 2000; Hoffmann et al., 2001; Woodworth et al., 2002; Reed et al., 2003a,b). It has also been shown that microsatellite DNA polymorphisms in A. gambiae are dramatically reduced in laboratory colonies when compared to field populations (Norris et al., 2001). Hence, inferences generated from studies using non-natural model systems and/or inbred colonies of vectors (and parasites) may be limited and only reflect some basic characteristics of the natural systems, prompting for subsequent validation at real-life situations. Here, we compared gene transcript abundance profiles between one laboratory colony of A. gambiae and two field populations from the vicinity of Yaoundé, Cameroon, to explore the divergence of their global transcriptomes.
Anopheles gambiae is the nominal member of a species complex that groups together 7 named species of relatively recent and rapid origin, all of which are closely related, sharing considerable genetic variation and being morphologically indistinguishable from one another (Powell et al., 1999; Coluzzi et al., 2002; Besansky et al., 2003). Diversification and further radiation of most species within the An. gambiae complex has been suggested to be driven by divergent ecological selection, allowing colonization of specific larval development sites, thus fostering assortative mating and speciation (Coluzzi, 1982, Ayala & Coluzzi, 2005). Within An. gambiae, the process of ecological divergence and lineage splitting is ongoing (Lehmann & Diabaté, 2008; Manoukis et al., 2008; Costantini et al., 2009; Simard et al., 2009) and recent studies have subdivided natural populations of A. gambiae into two morphologically identical and broadly sympatric molecular forms, called M-form and S-form, which are reproductively isolated and genetically different at several DNA loci, reflecting barriers to gene flow (della Torre et al., 2001, 2002, 2005; Turner et al., 2005; Lehmann & Diabate, 2008; White et al., 2010). Although interbreeding between the M and S forms yields fertile progeny (Diabaté et al. 2008), MS hybrids are rarely observed in nature; when these forms overlap in time and space, the rate of heterogamous insemination is only ~1% (Tripet et al., 2001), clearly demonstrating the existence of a pre-mating barrier. Thus, both indirect and direct genetic evidence indicate incomplete but substantial barriers to gene flow between the two molecular forms of A. gambiae s.s., pointing to the very earliest stages of speciation (della Torre et al., 2002; Costantini et al., 2009). A significant body of knowledge on the genetic differences and geographic distribution of the M and S forms has been established (della Torre et al., 2005; Lehmann & Diabate, 2008; Costantini et al., 2009; Simard et al., 2009; White et al., 2010), but the variations and differences in their physiology and the genes and pathways involved in local adaptation and speciation are still largely unidentified. In Central Africa, both molecular forms are sympatric and can be found together in the same larval development sites. However, the M molecular form is more prevalent in urbanized/polluted areas, habitats of marginal quality, whereas the S form seems to predominate in more rural settings (della Torre et al., 2005; Kristan et al., 2003; Carrara et al., 2004; Wondji et al., 2005, Simard et al., 2009). Here, both molecular forms are chromosomally homosequential at the cytogenetic level, showing mainly standard chromosomes without inversions (Pombi et al., 2008; Simard et al., 2009), offering opportunities to explore genetic and physiological differences between molecular forms without the confounding effect of chromosomal inversions.
Regulation of transcript abundance plays a key role in determining the fitness of a genome, which is critical for development and adaptation. The ability of a population to adapt to the environment is dependent on its phenotypic diversity, which in turn is a consequence of genetic diversity. Polymorphism in thepattern of gene transcript abundance is a widespread phenomenon and has been observed in bacteria (Cooper et al., 2003; Le Gall et al., 2005), yeast (Townsend et al., 2003), mice (Schadt et al., 2003), and humans (Yan et al., 2002). Although most of this variation might be neutral and reflect genetic distance between taxa/populations, several recent studies have demonstrated that natural selection through environmental pressure is a determinant of at least part of this polymorphismin gene transcript abundance (Cooper et al., 2003; Le Gall et al., 2003; Townsend et al., 2003; Enard et al., 2002; Oleksiak et al., 2002; Ranz et al., 2003; Ogura et al., 2004; Whitehead & Crawford, 2006; Giger et al., 2006). These particular studies suggest that long-term behavioral differences are a result of substantial physiological remodeling which, in turn, is guided by the transcriptome. Natural selection can lead to mutations in cis- and trans-acting transcriptional control elements that promote divergence in transcript abundance (Denver et al., 2005; Rifkin et al., 2005). Consistent with these predictions, at the early stages of speciation, as in the case of the M and S molecular forms of An. gambiae, divergence between incipient species might be more easily detectable at the transcriptome level, before reciprocal monophyly is established at the molecular level.
Despite the profound influence of the transcriptome on the divergence that occurs between individuals and populations exposed to different environmental conditions, most studies have addressed divergence only at the genome sequence level. To date, a single study by Cassone et al. (2008), employing the Affymetrix GeneChip platform, has investigated differences in gene transcript abundance between laboratory colonies of the M and S molecular forms and showed ~1–2% of their genes as differently expressed, with a strong bias toward transcription- and sensory process-related functions. In the present work, we compared the transcriptome within and between the two molecular forms of An. gambiae collected from the same larval development sites in two distinct ecological settings around Yaoundé, in order to assess the degree of variation that can be attributed to the genetic background of molecular forms and to environmental conditions, respectively.
RESULTS AND DISCUSSION
Transcriptomic divergence between laboratory colony and field mosquitoes
To identify consistent transcriptomic differences between laboratory colony and field mosquitoes, we compared the global transcriptomic profiles of 4-day-old females of the A. gambiae Keele laboratory colony strain to those of the field S and M molecular form mosquitoes originating from two different locations in the Yaoundé area (the S molecular form collected at Nkolondom (A in figure 1) and the M molecular form collected at Nkolbisson (B in figure 1) (Fig 1). The study was designed to identify transcriptomic differences that had resulted from a continuous exposure over many generations (through natural selection) of these mosquito groups to different environmental conditions (laboratory vs. field), rather than differences that related to the immediate environmental exposure or life history of the individuals that were used for the assays. For this reason, we reared the field mosquitoes from the larval to the adult stage under insectary conditions identical to those of the laboratory colony mosquitoes. The observed differences in gene transcript abundance are thought to be a result of an accumulation of mutations that have influenced gene transcript abundance over many generations in mosquitoes that have adapted to either the field or the laboratory environment.
Figure 1.
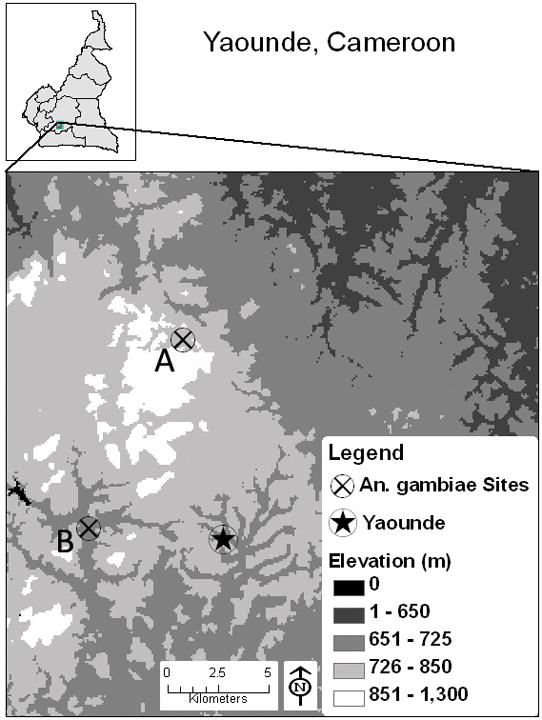
Geographic locations of the two sites from which field Anopheles gambiae were collected. A: Nkolondom, B: Nkolbisson.
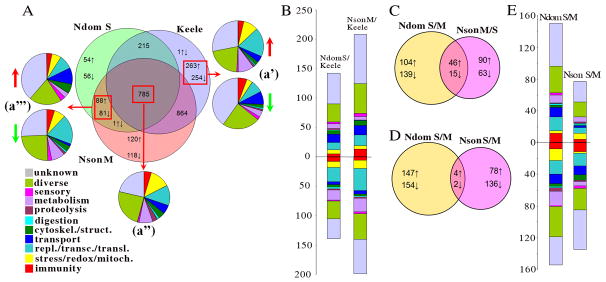
A comparison between the transcriptomes of these laboratory colony and field mosquitoes showed that 518 genes displayed differential transcript abundance: 254 genes showed a higher transcript abundance in the field mosquitoes, and 263 genes showed a higher transcript abundance in the laboratory colony mosquitoes. Microarray-assayed gene transcript abundance was validated with quantitative RT-PCR for eight genes in the two sets of laboratory-versus-field mosquito comparisons (Figure 2). A tyrosine protein kinase (AGAP005763-RA) was the only gene displaying opposite transcript abundance patterns between the Nkolondom and Nkolbisson mosquitoes when compared to the laboratory mosquitoes (Fig 3a′; Table S1). The transcripts showing higher transcript abundance in the field mosquitoes corresponded mainly to genes in the immunity, transport, and cytoskeleton functional classes, with a large proportion belonging to the “diverse” and “unknown” groups (Fig. 3B). The fact that a larger number of immune gene transcripts were more abundant in the field mosquitoes, likely reflects an adaptation of laboratory mosquitoes to a lower amount and diversity of microbes when compared to the microbial exposure of field mosquitoes. Among the transport genes that displayed a higher transcript abundance in the field mosquitoes, five have been putatively linked to insecticide resistance (Roth et al., 2003; Hemingway et al., 2004): a multidrug-resistance P glycoprotein [ENSANGT00000028639], an ATP-binding cassette sub-family A member [ENSANGT00000025474], the secretory protein HlyD [ENSANGT00000000827], a proton-associated sugar transporter A deleted in neuroblastoma 5 [AGAP010856-RA], and a multidrug resistance-associated protein [ENSANGT00000028591] (Table S1).
Figure 2.
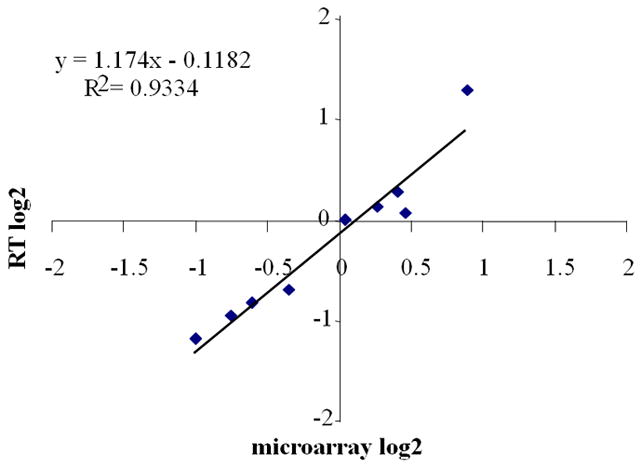
Validation of microarray-assayed gene transcript abundance by real-time quantitative RT-PCR. Log2-transformed microarray transcript abundance data (Ndom-S vs. Keele and Nson-M vs. Keele ratio) for eight genes were plotted against the log2-transformed transcript abundance data obtained by real-time quantitative RT-PCR. The Pearson correlation coefficient (r=0.96), best-fit linear-regression analysis (R2 = 0.93), and slope of the regression line (m = 1.174) showed a high degree of correlation between the two assays in terms of the magnitude of the regulation. The individual values for all these genes are presented in Table S7.
Figure 3.
(A) Venn diagrams showing co-regulation and differential transcript abundance patterns for field and laboratory colony mosquitoes. (a′) 518 genes showed differential transcript abundance between laboratory mosquitoes and at least one of the two field strains (Table S1). Pie charts indicate relative proportions of functional gene groups that were represented by transcripts at higher and lower levels in laboratory colony mosquitoes than in field mosquitoes. The gene with arrows in both directions is a tyrosine protein kinase that showed higher transcript abundance in the Nson M strain and lower transcript abundance in the Ndom S strain when compared to the Keele strain. (a″) 785 genes showed similar transcript levels in field mosquitoes and laboratory colony mosquitoes (Table S8). The pie chart represents the proportions of the functional gene groups. (a‴) Those genes from section (a′) showing similar transcript abundance profiles in both field strains and differential abundance in laboratory mosquitoes are shown in the overlapping section: 88 genes showed higher levels of transcripts in the field mosquitoes, 81 had higher transcript levels in the laboratory colony mosquitoes, and one gene had higher transcript level in the Nson M strain and lower level in the Ndom S strain when compared to laboratory colony mosquitoes (Table S9). The pie charts indicate the relative proportions of functional gene groups that showed transcript abundnace at higher and lower levels in field mosquitoes than in laboratory colony mosquitoes. (B) Proportions of functional gene groups that displayed higher (above the horizontal line) or lower (below the horizontal line) transcript levels in field mosquitoes when compared to the laboratory colony Keele strain (Table S1). (C) Proportions of genes with differential transcript levels between the predominant molecular form at each location and the less abundant form. The numbers of genes with higher and lower transcript abundance are indicated by arrows pointing up and down, respectively (Tables S3 and S4). (D) The proportions of genes with differential transcript abundance between the S and M molecular forms in each location. The numbers of genes with higher and lower transcript abundance are indicated by arrows pointing up and down, respectively. Equivalent differences in transcript abundance in the two locations are indicated in the overlapping section (Tables 1 and S5). (E) Bars indicate the number of functional gene groups showing higher (upper bars) and lower transcript abundance (lower bars) (Tables S3, S4 and S5).
The field mosquitoes also displayed elevated transcript abundance of a larger number of structural and cytoskeletal components (Figure 3B; Table S1). Eight sensory-related genes displayed differential transcript abundance between field and laboratory colony mosquitoes, presumably reflecting a certain degree of olfactory adaptation: Laboratory A. gambiae strains are routinely maintained on either mice or artificial membrane feeding systems, whereas wild A. gambiae are adapted to feed primarily on humans.
The laboratory colony mosquitoes showed elevated transcript abundance of several genes that are involved in oxidoreductive stress-related, mitochondrial, metabolic, and replication-transcription-translation processes (Fig. 3B; Table S1). This increased transcript abundance may be linked to a generally elevated metabolic activity. Higher rates of protein synthesis require higher rates of anabolism and catabolism, which in turn require an elevated mitochondrial activity (Hochachka et al., 1996). This higher translational activity could also explain the generally lower amount of other transcripts or could be a consequence of a shift in resource allocation (Fig. 3B). An organism’s protein pool is in a continual state of flux, with new proteins entering the pool as a result of protein synthesis andnew proteinsbeing removed as a result of protein degradation. This process is energetically expensive, but laboratory colony mosquitoes have adapted to live under optimal conditions without limiting factors (they are provided with food and mates ad libitum, and are largely free of pathogens), and it is possible that these optimal conditions have produced mosquitoes with higher metabolic rates and a stronger biosynthetic machinery.
Transcriptomic comparison between the A. gambiae M and S molecular forms
The global transcriptomic profiles of 4-day-old females of the predominant molecular form from each location (M form in Nkolbisson, S form in Nkolondom) were compared to those of the other molecular form from the same location (Fig. 3C–E; Tables S2–S5). The similarity of gene transcript abundance was 10 times higher between the predominant forms from each location (61 transcripts in common between the M form from Nkolbisson and the S form from Nkolondom) (Fig. 3C, Table S2) than within each molecular form (6 transcripts in common between the M form from Nkolbisson and the M form from Nkolondom) (Fig 3D; Tables 1 and S5), suggesting that adaptation leading to a dominant form under differing environmental conditions has a larger impact on the mosquito’s physiology than does the molecular form itself. A similar pattern has been observed in two other studies: one on transcriptomic differentiation between sedentary and migratory trout (Giger et al., 2006), and another on transcriptome variation affected by natural selection in the fish Fundulus heteroclitus (Whitehead and Crawford, 2006). The results of these studies suggested that the interaction of the genotype with the environment accounts for more transcriptional variation than does genetic ancestry.
Table 1. EXPRESSION LEVELS SPECIFIC OF THE MOLECULAR FORM.
Genes displaying consistent differential transcript abundance (≥ 1.7-fold) between the A. gambiae S and M molecular forms at both sampling locations (thus molecular form specific traanscriptome patterns). Transcript abundance is indicated as the normalized intensity ratio in fold regulation. Minus values indicate the –fold lower transcript abundance in the experimental sample.
| Gene ID | Gene ID | Gene name | Ndom S vs M | Nson M vs S | Nson S vs M |
|---|---|---|---|---|---|
| Immunity | |||||
| ENSANGT00000008707 | AGAP011952-RA | caspase short class CASPS3 | −1,361 | 1,599 | −1,599 |
| Transport | |||||
| ENSANGT00000010009 | AGAP006518-RA | Charged multivesicular body 1B chromatin modifying | 2,513 | −1,675 | 1,675 |
| Diverse | |||||
| ENSANGT00000013925 | AGAP001215-RA | TPRrepeat containing | −1,343 | 1,506 | −1,506 |
| Unknown | |||||
| ENSANGT00000029116 | Unknown | 1,869 | −1,365 | 1,365 | |
| ENSANGT00000025814 | Unknown | 1,852 | −1,452 | 1,452 | |
| ENSANGT00000010799 | AGAP007620-RA | Unknown | 1,703 | −1,396 | 1,396 |
The number of genes that was found to present different transcript abundance between the M and S molecular forms was 1.4 times higher in Nkolondom than in Nkolbisson (Fig. 3E, Tables S3 and S4), suggesting that the environmental conditions in Nkolondom have a stronger differential effect on the physiology of each molecular form. A similar number of genes showed a higher level of transcript abundance in the two molecular forms from the Nkolondom location, while 63% of the differentially expressed genes showed a higher transcript abundance in the M molecular form than in the S form in the Nkolbisson area. In both locations, mosquitoes of the predominant molecular form displayed an elevated transcript abundance of genes that generally belonged to the immunity and chemosensory functional groups, while the less abundant molecular form displayed an elevated transcript abundance of genes that largely belonged to the stress-redox-mitochondrial functional group.
The higher transcript abundance of stress-related genes and the larger number of repressed genes in the less abundant form is likely to reflect general conditions of higher stress that might result from the environmental conditions or from inter-form competition. Studies in yeast have shown that stressful conditions can result in a twofold larger number of repressed genes; presumably, this response represents a strategy to protect critical functions. A reduced synthesis of irrelevant transcripts and their products may help to conserve energy while the organism tries to adapt to the suboptimal conditions (Gasch & Werner-Washburne, 2002). Similarly, our data suggest that the S molecular form in Nkolbisson, which represents only 17% of the population and showed a 1.7-fold higher number of repressed genes than did the M form, is in a disadvantageous situation with respect to its tolerance of the environment when compared to the M molecular form, with which it competes for resources.
In order to identify potential transcription signatures that are molecular form-specific and may underlie some of the consistent physiological differences between the two forms, we looked at genes that displayed a consistent differential transcript abundance between the two molecular forms in both geographic locations (Fig. 3D; Table 1). The fact that only six such genes were identified underscores the strong effect of environmental adaptation on the mosquitoes’ physiology (Fig. 3D) (Giger et al., 2006). Genes that displayed higher transcript abundance in the M molecular form included a caspase (CASPS3), which is likely to be involved in apoptosis (Adrain et al., 2004; Martin & Baehrecke, 2004), and a tetratrico-peptide repeat containing protein (TRP); this type of repeat has been found to act as a scaffold for a broad range of protein-protein interactions and is involved in the regulation of RNA synthesis or mitosis (Ben-Yehuda et al., 2000). Genes that displayed high transcript abundance in the S molecular form included a charged multivesicular body chromatin modifying protein, which is located in the nuclear matrix and plays a role in the formation of vesicle-filled endosomes that target proteins to the interior of lysosomes, affecting chromatin structure and cell cycle progression (Stauffer et al., 2001). Another three genes with unknown functions [ENSANGT00000029116, ENSANGT00000025814, AGAP007620-RA] also showed an elevated transcript abundance in the S form. None of these six genes displaying differential transcript abundance could be linked to a potential mechanism related to the speciation process on the basis of its predicted function. It is possible that the speciation mechanisms act on mating behavior and that the genes involved are expressed or repressed at the time of the male-female mating interaction, as has been seen for the morphs Cosmopolitan and Zimbabwe of Drosophila melanogaster, which are in an incipient speciation process (Michalak et al., 2007). In spite of our observation, a study comparing gene transcript profiles between laboratory colony A. gambiae M and S forms at different life stages (fourth instar larvae, virgin females, and gravid females) has shown 164 transcripts differently expressed between virgin females of the two molecular forms; among these genes were several with putative implication in olfaction and mate recognition (Cassone et al., 2008). Although transcriptome comparison was done between laboratory colonies, the transcript abundance patterns of a subset of genes were validated by RT-PCR on wild mosquitoes from Cameroon and Burkina Faso. The differences in results between Cassone’s study and ours are likely be related to differences in the experimental design and the field sites from where mosquitoes were collected, as well as the largely different gene transcript abundance assay platforms. Cassone et al used the Affymetrix GeneChip and we used an oligonucleotide glass slide array. Furthermore, the two platforms had been based on different A. gambiae genome annotations, and hence a large number of genes represented on one platform was lacking on the other. In our study, the proportions of the two molecular forms in each location were different, suggesting a strong and different effect of both environments on their physiology. This different effect is reflected in their transcriptomes, and is likely to be masking potential transcriptome signatures that are specific of each molecular form. On the other hand, field mosquitoes used in Cassone’s study were collected from different localities and breeding sites and pooled according to their molecular form: this procedure would result in masking the effect of environmental variation towards detection of specific transcriptomic attributes of each molecular form.
CONCLUDING REMARKS
Our study has shown a substantial divergence, at the transcriptome level, between laboratory colony and field mosquitoes, suggesting a significant impact of environmental conditions on the evolution of the mosquito transcriptome. Gene transcript abundance evolves at a much more greater rate than do DNA sequences, by at least an order of magnitude. For instance, at least 10% of the nematode transcriptome is differentially expressed in organisms separated by 280 generations (Gibson, 2005). Mutations in a variety of different loci can affect transcript abundance, since gene regulation is affected by both cis- and trans-acting elements and transcription factors; furthermore, a single mutation in a single regulatory locus can affect the transcript abundance of dozens of target genes (Brem et al. 2002; Rockman & Kruglyak, 2006; Wray GA, 2007).
Our comparison between the M and S molecular forms of A. gambiae from the Nkolondom and Nkolbisson locations has indicated that the similarity of transcript abundance is larger between the predominant forms from each location than between populations of the same molecular forms in the two locations, suggesting a strong effect of the environmental conditions on the mosquitoes’ physiology. The similarity in the transcript abundance profile between the dominant forms in the two locations is likely to reflect a better adaptation, and hence a less-stressed physiology. Differences in the ability of the mosquitoes to adapt to the environment and to utilize the available resources could result in a “competitive exclusion” between the two forms at each location, the outcome of which might be directly linked to the relative frequencies of each form at the time of colonization of the site.
EXPERIMENTAL PROCEDURES
Field sites and mosquito collection
Anopheles gambiae larvae and pupae were collected from several pools of water in two market gardening areas (Nkolondom and Nkolbisson) located in the outskirts of Yaoundé, Cameroon, and transferred to the insectary, where they were reared to the adult stage. The village of Nkolondom (11°30′56″E 3°58′20″ N) (A in figure 1) is about 9.5 km northwest of Yaoundé and outside the city limits. Nkolbisson (11°27′15″E 3°52′21″ N) (A in figure 1) is about 6.5 km west of the center of the city, within an urbanized neighborhood. Nkolondom overlooks the city at an elevation of 805 m, while Nkolbisson lies in a valley at 702 m. Both sites are 10 km apart and are separated from one another by a steep hill with its peak at 1,200 m (Fig. 1). They experience a typical four-season equatorial climate, with a mean annual rainfall of about 1,500 mm and mean temperature of 24°C. Although rains are recorded every month, the long dry season extends from late November to early March (with 10–30 mm rainfall/month), and the short dry season includes July and August (80–100 mm rainfall/month). The rainfall peak is in October (250–350 mm/month). Larval collections were conducted at both sites in November and December, 2004.
Mosquito rearing and identification
Eggs of the A. gambiae Keele laboratory colony strain (a mixture of the M and S molecular forms) were brought to the insectary at the OCEAC Research Station in Yaoundé, and larvae and adults were raised there for several generations under standard temperature and humidity conditions of 28±1°C, 70–80% RH with a 12h:12h light-dark cycle. Larvae were fed on baby fish food (TetraMin® Baby) and adults were provided with 10% sugar solution ad libitum. Fourth instars larvae and pupae were collected from natural breeding sites using dippers and transported to the laboratory where they were reared to adults in their own breeding site water, and maintained under temperature, humidity, and feeding conditions identical to those of the laboratory colony mosquitoes. Emerging adults from field collections were readily identified on morphological grounds using reference keys (Gillies & De Meillon, 1968; Gillies & Coetzee, 1987), and only female A. gambiae s.l. specimens were included in the study. They were identified to species and molecular form using the PCR-RFLP method (Fanello et al., 2002). One or two legs of each specimen were directly used as a DNA template source in the PCR reaction mixture.
In agreement with the known geographic distribution of species within the A. gambiae complex in this area (Wondji et al., 2005; Ayala et al., 2009; Simard et al., 2009), only A. gambiae s.s. was present in our collections. Both, the M and S molecular forms were found at both sites, and no hybrids were observed. In the more rural Nkolondom area, 90% (408/453) of the mosquitoes were of the S molecular form and 10% (45/453) of the M form, whereas in the more urbanized and polluted area of Nkolbisson, the M form was predominant (96/115=83%) over the S form (19/115=17%).
Mosquito dissections and RNA extraction
Four-day-old sugar-fed females were used for gene transcript abundance analyses. After the mosquito was anesthetized with CO2, the head was removed to avoid eye pigment contamination, which can inhibit probe labeling reactions, and the legs were dissected out for use in molecular identification (above). The remaining body was maintained in RNAlater for subsequent RNA extraction using the RNeasy® Mini Kit (QIAGEN).
Microarray -based transcription analysis
In each of the microarray –based transcription assays two pools of 20 mosquitoes were compared, except for the Nkolbisson S molecular form from which the pool was of 19 mosquitoes.
Fluorochrome-labeled cRNA probes were synthesized from 2–3 μg RNA extracted from the pools of mosquitoes using the Agilent Technologies low-input RNA labeling kit according to the manufacturer’s instructions and previously described methodology (Dong et al., 2006). Probe quantity was estimated with a Beckman DU640 spectrophotometer. Hybridizations were carried out on the previously described Agilent Technologies -based A. gambiae full genome glass slide microarray with the Agilent Technologies in situ hybridization kit according to manufacturer’s instructions and previously described methods (Dong et al., 2006).
Microarray scanning was done with an Axon GenePix 4200AL scanner, and scan images were analyzed with Genepix Pro 6.0 software (Axon Instruments, Union City, CA). Images were analyzed with Genepix 6.0 software to determine spot size, location and quality, and potentially confounding spots were manually removed from the analysis. The minimum signal intensity was set to 150 fluorescent units, and the signal-to-background ratio cutoff was set to 2.0 for both the Cy5 and Cy3 channels. For the transcriptomic comparison between laboratory colony and field mosquitoes, at least three biological replicates were performed for each experimental set. For the transcriptomic comparison between the M and S molecular forms, the three biological replicates were performed using 3 pools of mosquitoes of the predominant molecular form against one pool of mosquitoes of the less abundant form due to the low numbers of mosquitoes of the latter at each collection site. In all experiments, technical replicates were performed when the hybridization quality was considered insufficient based on signal to background ratio. The background-subtracted median fluorescent values were normalized according to a LOWESS normalization method, and Cy5/Cy3 ratios from replicate assays were subjected to t-tests with a 0.01 P-value using the TIGR MIDAS and MEV software (Dudoit et al., 2003). Transcript abundance values for genes were included when significant P-values were found among replicates of an experimental set. Transcript abundance ratios were averaged with the GEPAS microarray preprocessing software prior to logarithmic (base 2) transformation (Herrero et al., 2003). All the transcript abundance values presented exhibited reproducible regulation trends (up- or down-regulation) in the replicate assays.
Validation of microarray –based transcription data by real-time quantitative PCR
Microarray-assayed transcript abundance was further validated with quantitative RT-PCR for eight genes in the two sets of laboratory-versus-field mosquito comparisons. RNA samples were reverse-transcribed using Superscript II (Invitrogen) with random hexamers. Real-time quantification was performed using the QuantiTect SYBR Green PCR Kit (Qiagen) and ABI Detection System ABI Prism 7300. All PCR reactions were performed in triplicate. The specificity of the PCR reactions was assessed by analysis of melting curves for each data point. The ribosomal protein S7 gene was used for normalization of cDNA templates. The gene names, IDs, and primer sequences used are listed in Table S6. Microarray- and real-time quantitative PCR-generated data showed a high degree of correlation (Pearson correlation coefficient, r = 0.96; best-fit linear-regression, R2 = 0.93; slope of the regression line, m = 1.174) (Fig. 2; Table S7).
Supplementary Material
Table S1. Genes displaying constitutively and significantly (≥ 1.7-fold difference) higher or lower transcript levels in field mosquitoes (Nkolondom-S and Nkolbisson-M mosquitoes), as compared to laboratory strain mosquitoes (Keele strain). For each gene, the fold higher or lower transcript abundance is indicated as the normalized experimental/controlintensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases AGAP and ENSEMBL. Ndom S, Nkolondom S molecular form; Nson M, Nkolbisson M molecular form.
Table S2. Genes that showed differential transcript abundance (≥ 1.7-fold) between the A. gambiae S and M molecular forms at both geographic locations (thus specific for the predominant form). The fold higher or lower transcript abundance is indicated as the normalized experimental/controlintensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.
Table S3. Genes displaying differential transcript abundance (≥ 1.7-fold) between the A. gambiae S and M molecular forms in Nkolondom. The fold higher or lower transcript abundance is indicated as the normalized experimental/control intensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to gene annotation data from the mosquito genome databases AGAP and ENSEMBL.
Table S4. Genes displaying differential transcript abundance (≥ 1.7-fold) between the A. gambiae S and M molecular forms at the Nkolbisson location. The fold higher or lower transcript abundance is indicated as the normalized experimental/controlintensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.
Table S5. Genes that showed differential transcript abundance in each location for the same molecular form. The fold higher or lower transcript abundance is indicated as the normalized experimental/control intensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.
Table S6. PCR primers used for microarray data validation by real-time QRT-PCR.
Table S7. Individual gene transcript abundance values (log2 ratio) obtained by microarray and real-time RT-PCR to validate microarray gene transcript abundance data.
Table S8. Genes with similar transcript abundance in the field mosquitoes (S from Nkolondom and M from Nkolbisson) and in the Keele laboratory colony. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.
Table S9. Genes with similarly transcript abundance in the field S form mosquitoes from Nkolondom and M form mosquitoes from Nkolbisson and with differential transcript abundance when compared to the Keele laboratory colony. The fold higher or lower transcript abundance is indicated as the normalized experimental/controlintensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.
Acknowledgments
We thank the insectary and genome core facility personnel at the Johns Hopkins Malaria Research Institute and OCEAC for assistance with mosquito rearing. We also thank Dr. Deborah McClellan at the Editing Referral Service, William H. Welch Medical Library, at the Johns Hopkins University School of Medicine. This work has been supported by the National Institutes of Health/National Institute of Allergy and Infectious Disease 1R01AI061576, the Ellison Medical Foundation, the World Health Organization/TDR, and a Johns Hopkins Malaria Research Institute Pilot Grant.
Footnotes
AUTHORS’ CONTRIBUTIONS
Ruth Aguilar: performed experiments, analyzed data and wrote the manuscript.
Frederic Simard: designed experiments and wrote the manuscript.
Colince Kamdem: performed experiments.
Tim Shields: analyzed data.
Gregory E. Glass: analyzed data.
Lindsey Garver: performed experiments and analyzed data.
George Dimopoulos: coordinated the project, designed experiments, analyzed data and wrote the manuscript.
Contributor Information
Ruth Aguilar, Email: ruth.aguilar@cresib.cat.
Frederic Simard, Email: Frederic.Simard@ird.fr.
Colince Kamdem, Email: kamdem_d@yahoo.com.
Tim Shields, Email: tshields@jhsph.edu.
Gregory E. Glass, Email: ggurrigl@jhsph.edu.
Lindsey S. Garver, Email: lgarver@jhsph.edu.
References
- Adrain C, Creagh EM, Cullen SP, Martin SJ. Caspase-dependent inactivation of proteasome function during programmed cell death in Drosophila and man. J Biol Chem. 2004;279(35):36923–30. doi: 10.1074/jbc.M402638200. [DOI] [PubMed] [Google Scholar]
- Aguilar R, Dong Y, Warr E, Dimopoulos G. Anopheles infection responses; laboratory models versus field malaria transmission systems. Acta Trop. 2005;95(3):285–91. doi: 10.1016/j.actatropica.2005.06.005. Review. [DOI] [PubMed] [Google Scholar]
- Ayala FJ, Coluzzi M. Chromosome speciation: humans, Drosophila, and mosquitoes. Proc Natl Acad Sci U S A. 2005;102(Suppl 1):6535–42. doi: 10.1073/pnas.0501847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D, Costantini C, Ose K, Kamdem GC, Antonio-Nkondjio C, Agbor JP, Awono-Ambene P, Fontenille D, Simard F. Habitat suitability and ecological niche profile of major malaria vectors in Cameroon. Malar J. 2009;8(1):307. doi: 10.1186/1475-2875-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillas-Mury C, Kumar S. Plasmodium-mosquito interactions: a tale of dangerous liaisons. Cell Microbiol. 2005;7(11):1539–45. doi: 10.1111/j.1462-5822.2005.00615.x. Review. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Dix I, Russell CS, McGarvey M, Beggs JD, Kupiec M. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics. 2000;156(4):1503–17. doi: 10.1093/genetics/156.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayire O, Fontenille D, Touré Y, Sagnon N. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc Natl Acad Sci U S A. 2003;16;100(19):10818–23. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Carrara GC, Calzetta M, Santolamazza F, Di Deco MA, della Torre A, Petrarca V. Preliminary data on the distribution of species and forms of the Anopheles gambiae complex (Diptera: Culicidae) at sites of Angola. Parassitologia. 2004;46 (Suppl 1):85. [Google Scholar]
- Cassone BJ, Mouline K, Hahn MW, White BJ, Pombi M, Simard F, Constantin C, Besansky NJ. Differential gene expression in incipient species of Anopheles gambiae. Molecular Ecology. 2008;17(10):2491–504. doi: 10.1111/j.1365-294X.2008.03774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XG, Mathur G, James AA. Gene expression studies in mosquitoes. Adv Genet. 2008;64:19–50. doi: 10.1016/S0065-2660(08)00802-X. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M, Craig M, le Sueur D. Distribution of African malaria mosquitoes belonging to the Anopheles gambiae complex. Parasitol Today. 2000;16(2):74–7. doi: 10.1016/s0169-4758(99)01563-x. [DOI] [PubMed] [Google Scholar]
- Cohuet A, Osta MA, Morlais I, Awono-Ambene PH, Michel K, Simard F, Christophides GK, Collins FH, Sakai RK, Vernick KD, Paskewitz S, Seeley DC, Miller LH, Collins WE, Campbell CC, Gwadz RW. Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science. 1986;234(4776):607–10. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- Coluzzi M. Spatial distribution of chromosomal inversions and speciation in Anopheline mosquitoes. Prog Clin Biol Res. 1982;96:143–53. [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298(5597):1415–8. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Cooper TF, Rozen DE, Lenski RE. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci. 2003;100:1072–1077. 17. doi: 10.1073/pnas.0334340100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini C, Ayala D, Guelbeogo WM, Pombi M, Some CY, Bassole IH, Ose K, Fotsing JM, Sagnon N, Fontenille D, Besansky NJ, Simard F. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Torre A, Costantini C, Besansky NJ, Caccone A, Petrarca V, Powell JR, Coluzzi M. Speciation within Anopheles gambiae--the glass is half full. Science. 2002;298(5591):115–7. doi: 10.1126/science.1078170. [DOI] [PubMed] [Google Scholar]
- della Torre A, Fanello C, Akogbeto M, Dossou-yovo J, Favia G, Petrarca V, Coluzzi M. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001;10(1):9–18. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem Mol Biol. 2005;35(7):755–69. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- Diabaté A, Dabiré RK, Heidenberger K, Crawford J, Lamp WO, Culler LE, Lehmann T. Evidence for divergent selection between the molecular forms of Anopheles gambiae: role of predation. BMC Evol Biol. 2008;11(8):5. doi: 10.1186/1471-2148-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2(6):e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Dimopoulos G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J Biol Chem. 2009;284(15):9835–44. doi: 10.1074/jbc.M807084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5(5):e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques Suppl. 2003:45–51. [PubMed] [Google Scholar]
- Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 2005;14(1):3–8. doi: 10.1111/j.1365-2583.2004.00529.x. Review. [DOI] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16(4):461–4. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- Fontenille D, Cohuet A, Awono-Ambene P, Kengne P, Antonio-Nkondjio C, Wondji C, Simard F. Malaria vectors: from the field to genetics. Research in Africa. Rev Epidemiol Sante Publique. 2005;53(3):283–90. doi: 10.1016/s0398-7620(05)84605-x. [DOI] [PubMed] [Google Scholar]
- Frankham R, Manning H, Margan SH, Briscoe DA. Does equalization of family sizes reduce genetic adaptation to captivity? Anim Conserv. 2000;3:357–363. [Google Scholar]
- Garver LS, Dong Y, Dimopoulos G. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog. 2009;5(3):e1000335. doi: 10.1371/journal.ppat.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics. 2002;(4–5):181–92. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- Gibson G. Mutation accumulation of the transcriptome. Nat Genet. 2005;37(5):458–60. doi: 10.1038/ng0505-458. [DOI] [PubMed] [Google Scholar]
- Giger T, Excoffier L, Day PJR, Champigneulle A, Hansen MM, Powell R, Lagardier CR. Life history shapes gene expression in salmonids. Curr Biol. 2006;16:R281–R282. doi: 10.1016/j.cub.2006.03.053. [DOI] [PubMed] [Google Scholar]
- Gillies MT, Coetzee M. A supplement to the Anophelinae of Africa south of the Sahara (Afrotropical Region). Publications of the South African Institute for Medical Research no. 55. South African Institute for Medical Research; Johannesburg: 1987. [Google Scholar]
- Gillies MT, De Meillon B. The Anophelinae of Africa south of the Sahara (Ethiopian Zoogeographical Region). Publications of the South African Institute for Medical Research no. 54. 2. South African Institute for Medical Research; Johannesburg: 1968. [Google Scholar]
- Hemingway J, Hawkes NJ, McCarroll L, Ranson H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem Mol Biol. 2004;34(7):653–65. doi: 10.1016/j.ibmb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Herrero J, Al-Shahrour F, Diaz-Uriarte R, Mateos A, Vaquerizas JM, Santoyo J, Dopazo J. GEPAS: A web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka W, Buck LT, Doll CJ, Land SC. Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci USA. 1996;93:493–9498. doi: 10.1073/pnas.93.18.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Hallas R, Sinclair C, Partridge L. Rapid loss of stress resistance in Drosophila melanogaster under adaptation to laboratory culture. Evolution. 2001;55:436–438. doi: 10.1111/j.0014-3820.2001.tb01305.x. [DOI] [PubMed] [Google Scholar]
- Jaramillo-Gutierrez G, Rodrigues J, Ndikuyeze G, Povelones M, Molina-Cruz A, Barillas-Mury C. Mosquito immune responses and compatibility between Plasmodium parasites and anopheline mosquitoes. BMC Microbiol. 2009;30(9):154. doi: 10.1186/1471-2180-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristan M, Fleischmann H, della Torre A, Stich A, Curtis CF. Pyrethroid resistance/susceptibility and differential urban/rural distribution of Anopheles arabiensis and An. gambiae s.s. malaria vectors in Nigeria and Ghana. Med Vet Entomol. 2003;17(3):326–32. doi: 10.1046/j.1365-2915.2003.00449.x. [DOI] [PubMed] [Google Scholar]
- Le Gall T, Darlu P, Escobar-Paramo P, Picard B, Denamur E. Selection-driven transcriptome polymorphism in Escherichia coli/Shigella species. Genome Res. 2005;15(2):260–8. doi: 10.1101/gr.2405905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Diabate A. The molecular forms of Anopheles gambiae: a phenotypic perspective. Infect Genet Evol. 2008;8(5):737–46. doi: 10.1016/j.meegid.2008.06.003. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoukis NC, Powell JR, Touré MB, Sacko A, Edillo FE, Coulibaly MB, Traoré SF, Taylor CE, Besansky NJ. A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae. Proc Natl Acad Sci U S A. 2008;26;105(8):2940–5. doi: 10.1073/pnas.0709806105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Baehrecke EH. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131(2):275–84. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- Mendes AM, Schlegelmilch T, Cohuet A, Awono-Ambene P, De Iorio M, Fontenille D, Morlais I, Christophides GK, Kafatos FC, Vlachou D. Conserved mosquito/parasite interactions affect development of Plasmodium falciparum in Africa. PLoS Pathog. 2008;16;4(5):e1000069. doi: 10.1371/journal.ppat.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P, Malone JH, Lee IT, Hoshino D, Ma D. Gene expression polymorphism in Drosophila populations. Mol Ecol. 2007;16(6):1179–89. doi: 10.1111/j.1365-294X.2007.03201.x. [DOI] [PubMed] [Google Scholar]
- Michel K, Kafatos FC. Mosquito immunity against Plasmodium. Insect Biochem Mol Biol. 2005;35(7):677–89. doi: 10.1016/j.ibmb.2005.02.009. Review. [DOI] [PubMed] [Google Scholar]
- Mitri C, Jacques JC, Thiery I, Riehle MM, Xu J, Bischoff E, Morlais I, Nsango SE, Vernick KD, Bourgouin C. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009;5(9):e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Donnelly MJ, Ranson H. Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. BMC Genomics. 2007;29(8):36. doi: 10.1186/1471-2164-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris DE, Shurtleff AC, Touré YT, Lanzaro GC. Microsatellite DNA polymorphism and heterozygosity among field and laboratory populations of Anopheles gambiae s.s (Diptera: Culicidae) J Med Entomol. 2001;38 (2):336–340. doi: 10.1603/0022-2585-38.2.336. [DOI] [PubMed] [Google Scholar]
- Ogura A, Ikeo K, Gojobori T. Comparative analysis of gene expression for convergent evolution of camera eye between octopus and human. Genome Res. 2004;14:555–1561. doi: 10.1101/gr.2268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- Pombi M, Caputo B, Simard F, Di Deco MA, Coluzzi M, della Torre A, Costantini C, Besansky NJ, Petrarca V. Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC Evol Biol. 2008;8:309. doi: 10.1186/1471-2148-8-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Petrarca V, della Torre A, Caccone A, Coluzzi M. Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia. 1999;41(1–3):101–13. Review. [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Reed DH. PhD Dissertation. University of Houston; Houston, TX: 1998. Population size, selection, and mutation accumulation. [Google Scholar]
- Reed DH, Frankham R. Correlation between fitness and genetic diversity. Cons Biol. 2003a;17:230–237. [Google Scholar]
- Reed DH, Lowe EH, Briscoe DA, Frankham R. Fitness and adaptation in a novel environment: effect of inbreeding, prior environment, and lineage. Evol Int J Org Evol. 2003b;57(8):1822–1828. doi: 10.1111/j.0014-3820.2003.tb00589.x. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438(7065):220–3. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Genetics of global gene expression. Nature Reviews Genetics. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Roth CW, Holm I, Graille M, Dehoux P, Rzhetsky A, Wincker P, Weissenbach J, Brey PT. Identification of the Anopheles gambiae ATP-binding cassette transporter superfamily genes. Mol Cells. 2003;15(2):150–8. [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Simard F, Ayala D, Kamdem GC, Pombi M, Etouna J, Ose K, Fotsing JM, Fontenille D, Besansky NJ, Costantini C. Ecological niche partitioning between Anopheles gambiae molecular forms in Cameroon: the ecological side of speciation. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J Cell Sci. 2001;114(Pt 13):2383–93. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- Tchuinkam T, Mulder B, Dechering K, Stoffels H, Verhave JP, Cot M, Carnevale P, Meuwissen JH, Robert V. Experimental infections of Anopheles gambiae with Plasmodium falciparum of naturally infected gametocyte carriers in Cameroon: factors influencing the infectivity to mosquitoes. Trop Med Parasitol. 1993;44(4):271–6. [PubMed] [Google Scholar]
- Townsend JP, Cavalieri D, Hartl DL. Population genetic variation in genome-wide gene expression. Mol Biol Evol. 2003;20(6):955–63. doi: 10.1093/molbev/msg106. [DOI] [PubMed] [Google Scholar]
- Tripet F. Ecological immunology of mosquito-malaria interactions: Of non-natural versus natural model systems and their inferences. Parasitology. 2009;136(14):1935–42. doi: 10.1017/S0031182009006234. Review. [DOI] [PubMed] [Google Scholar]
- Tripet F, Aboagye-Antwi F, Hurd H. Ecological immunology of mosquito-malaria interactions. Trends Parasitol. 2008;24(5):219–27. doi: 10.1016/j.pt.2008.02.008. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet F, Touré YT, Taylor CE, Norris DE, Dolo G, Lanzaro GC. DNA analysis of transferred sperm reveals significant levels of gene flow between molecular forms of Anopheles gambiae. Mol Ecol. 2001;10(7):1725–32. doi: 10.1046/j.0962-1083.2001.01301.x. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3(9):e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernick KD, Fujioka H, Seeley DC, Tandler B, Aikawa M, Miller LH. Plasmodium gallinaceum: a refractory mechanism of ookinete killing in the mosquito, Anopheles gambiae. Exp Parasitol. 1995;80(4):583–95. doi: 10.1006/expr.1995.1074. [DOI] [PubMed] [Google Scholar]
- Volohonsky G, Steinert S, Levashina EA. Focusing on complement in the antiparasitic defense of mosquitoes. Trends Parasitol. 2010;26(1):1–3. doi: 10.1016/j.pt.2009.10.003. [DOI] [PubMed] [Google Scholar]
- White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol Ecol. 2010 doi: 10.1111/j.1365-294X.2010.04531.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead, Crawford Neutral and adaptive variation in gene expression. Proc Natl Acad Sci U S A. 2006;103(14):5425–30. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C, Simard F, Petrarca V, Etang J, Santolamazza F, della Torre A, Fontenille D. Species and populations of the Anopheles gambiae complex in Cameroon with special emphasis on chromosomal and molecular forms of Anopheles gambiae s.s. Journal of Medical Entomology. 2005;42(6):998–1005 (8). doi: 10.1093/jmedent/42.6.998. [DOI] [PubMed] [Google Scholar]
- Woodworth LM, Montgomery ME, Briscoe DA, Frankham R. Rapid genetic deterioration in captivity: causes and consequences. Conserv Genet. 2002;3:277–288. [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Yassine H, Osta MA. Anopheles gambiae innate immunity. Cell Microbiol. 2010;12(1):1–9. doi: 10.1111/j.1462-5822.2009.01388.x. Review. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes displaying constitutively and significantly (≥ 1.7-fold difference) higher or lower transcript levels in field mosquitoes (Nkolondom-S and Nkolbisson-M mosquitoes), as compared to laboratory strain mosquitoes (Keele strain). For each gene, the fold higher or lower transcript abundance is indicated as the normalized experimental/controlintensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases AGAP and ENSEMBL. Ndom S, Nkolondom S molecular form; Nson M, Nkolbisson M molecular form.
Table S2. Genes that showed differential transcript abundance (≥ 1.7-fold) between the A. gambiae S and M molecular forms at both geographic locations (thus specific for the predominant form). The fold higher or lower transcript abundance is indicated as the normalized experimental/controlintensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.
Table S3. Genes displaying differential transcript abundance (≥ 1.7-fold) between the A. gambiae S and M molecular forms in Nkolondom. The fold higher or lower transcript abundance is indicated as the normalized experimental/control intensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to gene annotation data from the mosquito genome databases AGAP and ENSEMBL.
Table S4. Genes displaying differential transcript abundance (≥ 1.7-fold) between the A. gambiae S and M molecular forms at the Nkolbisson location. The fold higher or lower transcript abundance is indicated as the normalized experimental/controlintensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.
Table S5. Genes that showed differential transcript abundance in each location for the same molecular form. The fold higher or lower transcript abundance is indicated as the normalized experimental/control intensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.
Table S6. PCR primers used for microarray data validation by real-time QRT-PCR.
Table S7. Individual gene transcript abundance values (log2 ratio) obtained by microarray and real-time RT-PCR to validate microarray gene transcript abundance data.
Table S8. Genes with similar transcript abundance in the field mosquitoes (S from Nkolondom and M from Nkolbisson) and in the Keele laboratory colony. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.
Table S9. Genes with similarly transcript abundance in the field S form mosquitoes from Nkolondom and M form mosquitoes from Nkolbisson and with differential transcript abundance when compared to the Keele laboratory colony. The fold higher or lower transcript abundance is indicated as the normalized experimental/controlintensity ratio. Fold up-regulation is indicated by positive numbers, and fold down-regulation by negative numbers. Putative functions have been assigned to genes according to the gene annotation data from the mosquito genome databases. Transcripts lacking AGAP annotation IDs are indicated based on the ENSEMBL ID.