Abstract
PIWI-interacting RNAs (piRNAs) protect genome integrity from transposons. In Drosophila ovarian somas, primary piRNAs are produced and loaded onto Piwi. Here, we describe roles for the cytoplasmic Yb body components Armitage and Yb in somatic primary piRNA biogenesis. Armitage binds to Piwi and is required for localizing Piwi into Yb bodies. Without Armitage or Yb, Piwi is freed from the piRNAs and does not enter the nucleus. Thus, piRNA loading is required for Piwi nuclear entry. We propose that a functional Piwi–piRNA complex is formed and inspected in Yb bodies before its nuclear entry to exert transposon silencing.
Keywords: Piwi, Armitage, Yb, Zucchini, primary piRNA, Drosophila
In Drosophila, three sets of endogenous small RNAs have been identified so far: microRNAs (miRNAs), endogenous siRNAs (endo-siRNAs/esiRNAs), and PIWI-interacting RNAs (piRNAs) (Ghildiyal and Zamore 2009; Malone and Hannon 2009; Siomi and Siomi 2009). Of these, piRNAs are considered unique because of their germline-specific expression and specific interaction with germline-specific Argonaute proteins, PIWI proteins (Cox et al. 2000; Harris and Macdonald 2001; Saito et al. 2006; Brennecke et al. 2007; Gunawardane et al. 2007; Nishida et al. 2007; Li et al. 2009). The identification of the piRNAs associated with three PIWI proteins (Aubergine [Aub], Argonaute 3 [AGO3], and Piwi) has revealed distinct features of piRNAs associated with each PIWI and has led to two models for piRNA biogenesis: the primary processing pathway and the amplification loop pathway (Aravin et al. 2007; Klattenhoff and Theurkauf 2008; Ghildiyal and Zamore 2009). In the amplification loop model, the Slicer (endonuclease) activity of Aub and AGO3 determines the formation of the 5′ end of piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). Zucchini (Zuc), a putative cytoplasmic nuclease (Pane et al. 2007), is involved in the primary processing pathway (Li et al. 2009; Malone et al. 2009); however, its precise molecular function remains unclear. Furthermore, the factors other than zuc required for primary piRNA biogenesis are unknown.
The ovarian somatic cell (OSC) line consists of ovarian somas only (Saito et al. 2009). The expression of Aub and AGO3 is not detectable in OSCs (Saito et al. 2009) because both proteins are germ cell-specific. This implies that the amplification loop does not operate in OSCs (Saito et al. 2009). However, OSCs express piRNAs and are loaded onto Piwi, indicating that the piRNAs in OSCs are generated specifically through the primary processing pathway (Saito et al. 2009). Thus, OSCs are an ideal tool to elucidate the molecular mechanisms of primary piRNA processing and Piwi function. Loss of zuc function drastically reduced the level of primary piRNAs in the ovaries (Malone et al. 2009). We recapitulated this in OSCs: Zuc depletion by RNAi caused a severe reduction in the piRNA level in OSCs (Saito et al. 2009). This result prompted us to screen for other factors necessary for primary piRNA production using RNAi in OSCs.
Results and Discussion
Armitage is required for primary piRNA biogenesis and accumulates at distinct cytoplasmic foci in OSCs
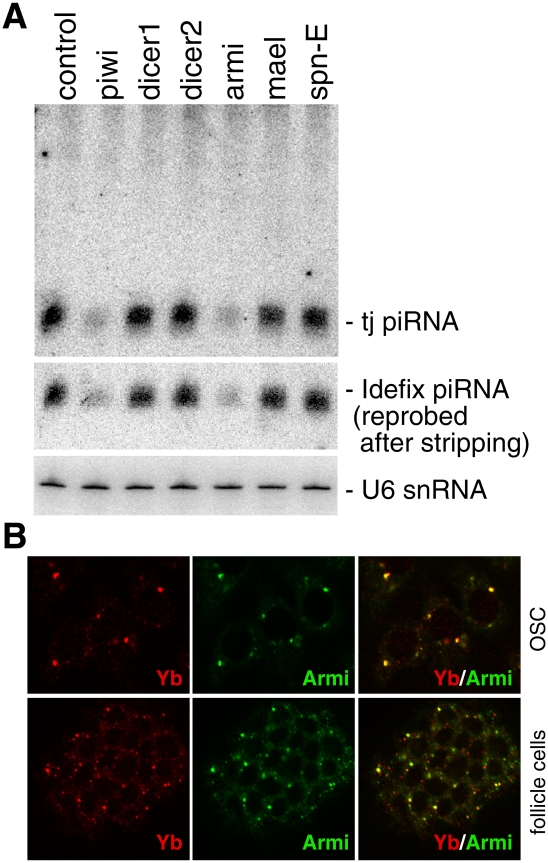
To identify the genes required for somatic primary piRNA biogenesis, we performed RNAi-based screening in OSCs. The genes screened included armitage (armi), spindle-E (spn-E), and maelstrome (mael) (Supplemental Table S1), all of which are implicated in piRNA biogenesis (Vagin et al. 2006; Lim and Kai 2007; Malone et al. 2009). However, their roles in somatic primary piRNA production remain unknown. Depletion of Armi reduced the piRNA levels to an extent very similar to that of Piwi (Fig. 1A) and Zuc depletion (Saito et al. 2009), indicating that Armi is necessary for primary piRNA biogenesis in OSCs. Depletion of Mael and Spn-E showed little or no effect on piRNA accumulation in OSCs (Fig. 1A). Mutations in both genes have been shown to significantly reduce the piRNA levels in ovaries (Vagin et al. 2006; Lim and Kai 2007; Malone et al. 2009). Thus, spn-E and mael are factors functioning in the amplification loop. Depletion of Dicer1 and Dicer2 had little or no effect on the piRNA levels (Fig. 1A), confirming that neither protein is necessary for piRNA production (Vagin et al. 2006).
Figure 1.
Armi, a novel component of Yb bodies, is necessary for primary piRNA production. (A) RNAi-based depletion of Armi, but not of Spn-E and Mael, caused a severe reduction in the expression levels of tj-piRNA and Idefix-piRNA in OSCs. (B) OSCs and ovaries immunostained with anti-Armi and anti-Yb antibodies. Merged images show that Armi colocalizes with Yb at Yb bodies.
Armi is the Drosophila ortholog of Arabidopsis Silencing-Defective 3 (SDE3) and mammalian Moloney leukemia virus 10 (MOV10) (Cook et al. 2004). These orthologs contain a conserved ATP-dependent RNA helicase domain at their C termini (Cook et al. 2004) and have been implicated in small RNA-mediated gene silencing (Dalmay et al. 2001; Cook et al. 2004; Tomari et al. 2004; Meister et al. 2005; Klattenhoff et al. 2007; Haussecker et al. 2008). However, their precise functions remain unknown. To gain further insight into the function of Armi in somatic primary piRNA processing, we produced a monoclonal antibody against Armi. Western blotting showed a discrete band in both ovary and cultured Schneider2 (S2) cell lysates (Supplemental Fig. S1A), indicating that Armi expression is not germline-specific. The ∼150-kDa protein immunopurified from S2 cells with the anti-Armi antibody was confirmed to be Armi by mass spectrometry (Supplemental Fig. S1B).
Immunostaining of OSCs and ovaries with the anti-Armi antibody confirmed an earlier observation that Armi is a cytoplasmic protein (Supplemental Fig. S1C; Cook et al. 2004). The Armi signals were detected in both somatic and germ cells of ovaries (Supplemental Fig. S1C), as has been reported previously (Cook et al. 2004). The somatic signal was considered a background signal because it did not disappear even in armi homozygous mutant egg chambers (armi72.1/armi72.1) (Cook et al. 2004). In the present study, the cytoplasmic signal in OSCs mostly disappeared when Armi was depleted by RNAi (Supplemental Fig. S1D). Thus, we conclude that Armi is expressed in both somatic and germ cells in ovaries.
The subcellular localization of Armi in the armi trans-heterozygous mutants (armi72.1/armi1) appeared very similar to that in the homozygous mutants (armi72.1/armi72.1) (Supplemental Fig. S1E; Cook et al. 2004). In addition, Western blotting revealed a band corresponding to Armi in the armi ovaries (Supplemental Fig. S1F). By what mechanisms Armi is expressed in the mutant somas remains unclear. The simplest explanation is that the armi gene uses two distinct genomic elements as promoters in ovarian somas. In fact, the armi homozygous mutants armi1/armi1 and armi72.1/armi72.1 weakly express a shorter armi transcript than that expressed in the wild-type strain (Supplemental Fig. S1G; Cook et al. 2004).
The Armi signal in germ cells was rather weak, and only a small proportion of Armi accumulated at, or near, the nuage, an electron-dense structure associated with nurse cell nuclei (Supplemental Fig. S1C). Thus, Armi might not be a component of the nuage per se. This correlates well with the fact that armi mutations barely affected the ability of the ovaries to amplify endogenous piRNAs (Malone et al. 2009). In ovarian somas, Armi accumulated strongly at discrete cytoplasmic foci (Supplemental Fig. S1C), as has been shown previously (Cook et al. 2004). Each somatic cell contained one or several foci. Interestingly, the Armi-positive foci were often located near the nucleus in both ovaries and OSCs.
Armi is a novel component of Yb bodies
We noted that the immunostaining patterns of the Armi-positive foci in the ovaries looked very similar to those of Yb (Szakmary et al. 2009). Genetic studies have shown that Yb is necessary for the self-renewal of germline stem cells and somatic stem cells via the hedgehog- and piwi-mediated niche signaling pathways, respectively (King and Lin 1999; King et al. 2001). Yb accumulates strongly at cytoplasmic foci, which do not overlap with known cytoplasmic granules such as P bodies and U bodies (Supplemental Fig. S2A; Szakmary et al. 2009). Thus, the Yb-positive foci were deemed novel structures and termed Yb bodies (Szakmary et al. 2009). Yb bodies are often located close to spherical structures highly enriched in RNAs, suggesting the involvement of Yb in RNA metabolism (Szakmary et al. 2009). Immunostaining with antibodies for Yb and Armi clearly indicated that the two proteins colocalize at Yb bodies in OSCs (Fig. 1B). Such colocalization was also observed in the follicle cells in ovaries (Fig. 1B). Thus, Armi is a novel component of Yb bodies. To determine if Armi localization in Yb bodies depends on Yb, or vice versa, immunofluorescence was performed in OSCs in which Yb or Armi was depleted by RNAi. Depletion of Armi in the OSCs did not cause any notable change in Yb localization in the Yb bodies (Supplemental Fig. S2B). However, depletion of Yb drastically affected Armi localization; Armi became distributed evenly throughout the cytoplasm (Supplemental Fig. S2B). Thus, Yb and Armi have different requirements for their localization at Yb bodies.
Armi and Yb are required for Piwi nuclear localization and piRNA association
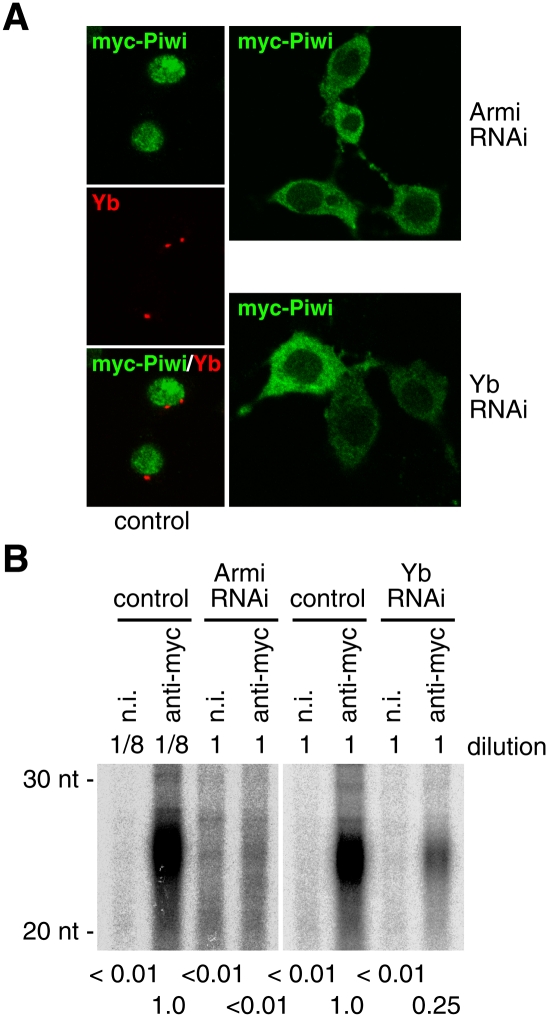
Loss of Yb function caused a severe reduction in the level of Piwi in the ovaries, implying that Yb controls the expression of Piwi in ovaries (Szakmary et al. 2009). We therefore examined how depletion of Yb affects the level of Piwi in OSCs. Yb depletion did not affect the expression level of Piwi protein (Supplemental Fig. S3A). Yb might not be necessary for maintaining Piwi stability once it is expressed. Loss of Armi affected the subcellular localization of Piwi: Although the majority of Piwi remained in the nucleus, some were distributed throughout the cytoplasm (Supplemental Fig. S3B). The nuclear localization of Traffic Jam (TJ), the only large Maf transcriptional factor in Drosophila (Li et al. 2003), which regulates Piwi expression in gonadal somas (Saito et al. 2009), was unaffected by Armi depletion (Supplemental Fig. S3B), suggesting that the effect is specific to Piwi. Piwi depletion in OSCs did not affect Armi localization (data not shown). We speculated that the nuclear signal for Piwi found in Armi-depleted cells (Supplemental Fig. S3B) might reflect Piwi that was localized to this cellular compartment before Armi depletion. Therefore, we knocked down Armi in OSCs and subsequently expressed myc-tagged Piwi (Saito et al. 2009). Under such conditions, the cytoplasmic distribution of Piwi was clearer: myc-Piwi was detected almost exclusively in the cytoplasm (Fig. 2A). myc-Piwi behaved similarly when Yb was depleted instead of Armi (Fig. 2A). We also found that Yb depletion caused a severe reduction (∼25% of the control signal) in piRNA levels in OSCs, as did Armi depletion (Supplemental Fig. S3C). When Armi or Yb was depleted, myc-Piwi was loaded with far fewer piRNAs than under normal conditions (Fig. 2B). We propose a model in which Armi and Yb act as novel primary piRNA factors, both of which have indirect effects on piRNA loading and nuclear localization of Piwi. The Piwi PAZ mutant (myc-Piwi-PAZmt), where the tyrosine residues required for piRNA loading are mutated to alanines (Y327A and Y328A), also accumulated in the cytoplasm (Supplemental Fig. S3D). Thus, in OSCs, piRNA loading is required for the nuclear localization of Piwi.
Figure 2.
Depletion of Armi and Yb causes Piwi to be mislocalized to the cytoplasm. (A) After depletion of Armi and Yb, myc-tagged Piwi was expressed in OSCs by transfection. myc-Piwi accumulated predominantly in the cytoplasm, suggesting that the nuclear signals observed in Supplemental Figure S3B (green) reflect endogenous Piwi that was localized to the nucleus prior to Armi and Yb depletion. (B) 32P-labeling of RNAs showed that, when Armi or Yb was depleted, myc-Piwi was loaded with few or no piRNAs. (Left panel) Note that the complex immunoprecipitated from control cells was diluted 1:8 prior to RNA isolation. The amount of Piwi in the immunoprecipitated complex from control cells was eightfold higher than that from Armi RNAi cells (data not shown).
Loading of primary piRNAs onto Piwi occurs in Yb bodies
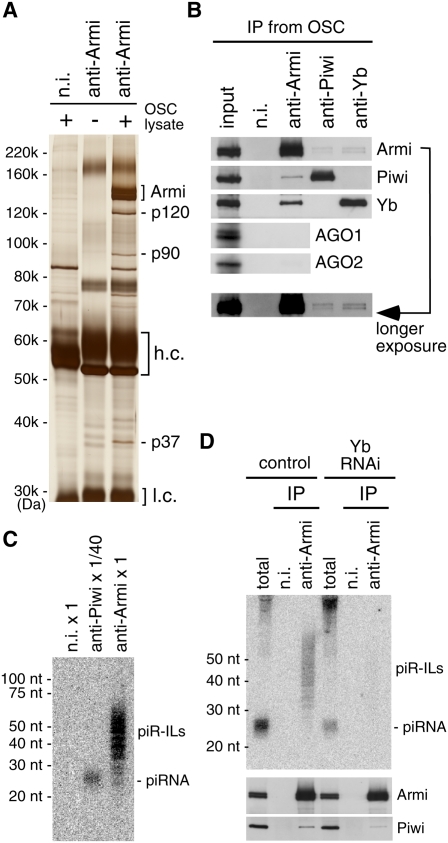
Silver staining of proteins coimmunoprecipitated with Armi from OSCs revealed three proteins (p120, p90, and p37) bound to Armi (Fig. 3A). Mass spectrometry identified p120 and p90 as Yb and Piwi, respectively, while p37 remains unidentified. Western blotting of the immunoprecipitates confirmed that Yb and Piwi exist in the Armi complex (Fig. 3B). Neither AGO1 nor AGO2 was detected in the complex (Fig. 3B), indicating that the association between Piwi and Armi is specific. Reciprocal experiments in which the complexes immunoprecipitated with anti-Piwi or anti-Yb antibody were probed for Armi, Piwi, and Yb showed that both Piwi and Yb interacted with Armi (Fig. 3B). However, Yb was not detected in anti-Piwi immunoprecipitates, and Piwi was not detected in anti-Yb immunoprecipitates (Fig. 3B). Thus, the interaction between Yb and Piwi seems to occur only through Armi. Neither AGO1 nor AGO2 was detected in the anti-Piwi and anti-Yb immunoprecipitates (Supplemental Fig. S3E).
Figure 3.
The Armi complex contains not only Piwi and Yb, but also piR-ILs. (A) Silver staining of protein components in the immunoprecipitated complex with anti-Armi antibody from OSCs. Three proteins—p120, p90, and p37—coimmunoprecipitated with Armi. (B) The complex immunoprecipitated with anti-Armi antibody contains Piwi and Yb but not AGO1 and AGO2. Yb was not detected in the complex. In addition, Piwi was not detected in the complex immunoprecipitated by anti-Yb. The input lane contains 1% of lysates used for immunoprecipitation. (C) The Armi complex immunopurified from OSCs contains piR-ILs. RNA molecules isolated from the complexes immunoprecipitated with anti-Piwi and anti-Armi antibodies were probed with a DNA oligonucleotide recognizing tj-piRNA. Note that the Piwi complex was diluted 1:40 prior to RNA isolation. (D, top panel) Depletion of Yb resulted in the disappearance of piR-ILs from the Armi complex. (Bottom panels) Western blotting showed that the association between Piwi and Armi was maintained under these conditions.
We speculated that the existence of Yb in the Armi–Piwi complexes might affect the RNA content of the complexes. To address this, RNAs isolated from both complexes were probed with a DNA oligonucleotide recognizing a piRNA derived from the tj transcript (tj-piRNA) (Fig. 3C). The anti-Piwi immunoprecipitate was diluted 1:40 before RNA isolation to equalize the amount of Piwi in both complexes (Supplemental Fig. S3F). Unlike the anti-Piwi immunoprecipitates—in which a strong, specific signal for tj-piRNA was observed—the anti-Armi immunoprecipitates showed a smear spanning 25–70 nucleotides (nt) (Fig. 3C), presumably containing piRNA intermediate-like molecules (piR-ILs) of various lengths. The smear was also detected with a probe for the idefix transposon (data not shown), suggesting that transposon–piR-ILs may also exist in the complex.
To determine whether the Armi–Piwi–Yb complex contains piR-ILs, we isolated small RNAs from the complex and constructed a small RNA library. Sequencing and bioinformatics analysis (using 116,680 “perfect match to the genome” clones of 184,692 total reads) showed that the library included small RNAs that are annotated as protein-coding genes, rRNAs, transposons, and others (Supplemental Fig. S4A). Small RNAs annotated to miRNAs and tRNAs were negligible (0% and 0.04%, respectively), suggesting that the Armi–Piwi–Yb complex selects small RNAs to bind. A number of small RNAs were annotated as actin 5c (act5c) mRNA (453 clones). However, Northern blotting did not detect these in the Armi–Piwi–Yb complex (Supplemental Fig. S4B). Small RNAs corresponding to the tj ORF, which does not presumably contribute to the generation of piRNAs (Saito et al. 2009), were also negative (Supplemental Fig. S4B). We assume that small RNAs annotated as act5c mRNA and tj ORF are likely background.
Clones that mapped uniquely to the piRNA locus flam were identified, and their distribution on the locus was compared with that of flam-derived piRNAs (Supplemental Fig. S5A). Interestingly, a number of the flam-derived piR-ILs overlapped with mature flam-piRNAs (497 of 1203; 41.3%). The details of the overlaps in the ∼500-nt region proximal to the dip1 gene are shown in Supplemental Figure S5B. The majority of the 5′ ends of the piR-ILs, as well as their 3′ ends, was not perfectly matched with those of mature piRNAs. piR-ILs annotated as tj are summarized in Supplemental Figure S6A. The 5′ ends of piR-ILs mapped to transposons were not U-rich (Supplemental Fig. S6B), unlike those of mature transposon–piRNAs. These results suggest that a population of RNAs residing in the Armi–Piwi–Yb complex correspond to piRNA intermediates. However, it is also possible that these RNAs in the complex may correspond to the side products of primary piRNA processing.
We then repeated the Northern blotting experiment after Yb depletion and found that the anti-Armi immunoprecipitates no longer contained piR-ILs (Fig. 3D). Yb depletion inhibited Armi accumulation at Yb bodies (Supplemental Fig. S2B), but the interaction between Armi and Piwi was maintained, as in naïve OSCs (Fig. 3D). Thus, it is likely that Yb is required for localizing the Armi–Piwi complex to Yb bodies, and that this molecular dynamic is required for the assembly of piR-ILs into the complex.
Zuc is required for piRNA maturation
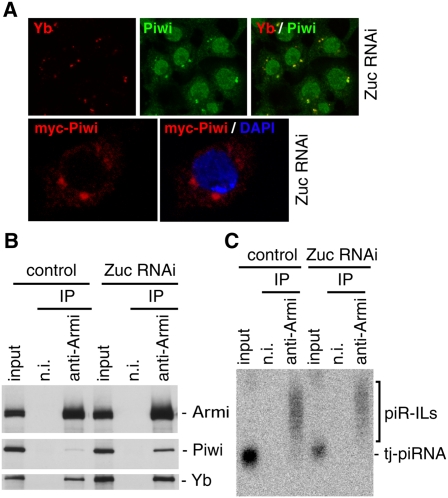
Primary piRNA accumulation in OSCs requires zuc (Saito et al. 2009). To gain further insight into the function of zuc in the processing pathway, OSCs were stained with anti-Yb and anti-Piwi antibodies under conditions in which Zuc was depleted by RNAi (Supplemental Fig. S7A). A fraction of Piwi now accumulated in the cytoplasm, particularly at Yb bodies (Fig. 4A). The cellular localization of Yb and Armi was not affected by Zuc depletion (Fig. 4A; Supplemental Fig. S7B). In OSCs where Zuc was depleted by RNAi, myc-Piwi was localized exclusively in the cytoplasm and strongly accumulated at Yb bodies (Fig. 4A). Immunoprecipitation and Western blotting showed that Zuc was not necessary for Armi to associate with Piwi and Yb (Fig. 4B); instead, more Piwi was associated with Armi when Zuc was depleted. Zuc depletion slightly decreased the amount of piR-ILs within the Armi–Piwi–Yb complex (∼70% compared with the control sample) (Fig. 4C); however, the amount of mature-like piRNAs loaded onto myc-Piwi was much lower (∼18%) than that in control cells (Supplemental Fig. S7C). We considered these as “mature-like” piRNAs because they were ∼1 nt longer than the mature piRNAs (Fig. 4C). piR-ILs in the complex also appeared to be slightly longer than those in control (Fig. 4C). Thus, the zuc gene is most likely a primary piRNA factor that plays an important role in producing piR-ILs and mature piRNAs in OSCs.
Figure 4.
Zuc functions in the primary piRNA processing pathway in OSCs. (A, top panels) Depletion of Zuc caused Piwi (green) to be localized to Yb bodies. Yb localization (red) was not affected by Zuc depletion. (Bottom panels) myc-Piwi (red) was expressed by transfection after Zuc was depleted in OSCs by RNAi. (B) Depletion of Zuc did not affect the association of Armi with Piwi and Yb. Under such conditions, Piwi was slightly more abundant in the Armi–Piwi–Yb complex. (C) Depletion of Zuc did not affect piRNA intermediates in the Armi complex.
We showed previously that Zuc is a cytoplasmic protein (Saito et al. 2009). In that study, the N terminus of Zuc was tagged with a myc-peptide. Zuc was reported previously to contain a mitochondrial localization signal (MLS) at the N terminus (Choi et al. 2006). Hence, the myc-peptide at the Zuc N terminus probably interfered with the mitochondrial localization and led to artefactual results. Thus, we re-examined this localization by tagging the C terminus with a myc peptide. This revealed that Zuc is localized in mitochondria (Supplemental Fig. S7D). Yb bodies have been reported to be usually associated with mitochondria (Szakmary et al. 2009). Therefore, we assessed whether Zuc signals were adjacent to Yb signals. Coimmunostaining with anti-myc and anti-Yb antibodies showed that Zuc localizes in close proximity to Yb bodies (Supplemental Fig. S7E, left). We also showed that Yb bodies were also adjacent to mitochondria (Supplemental Fig. S7E, right). These findings further support the idea that Zuc is a primary piRNA factor in processing piR-ILs and mature piRNAs in OSCs. However, the exact molecular functions of Armi, Yb, and Zuc in the somatic primary piRNA production remain to be elucidated.
piR-ILs do not contain a phosphate group at the 5′ end but do contain a 2′,3′-cyclic phosphate at the 3′ end
piR-ILs were undetectable by Northern blotting in which the RNAs were cross-linked to a nylon membrane by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) (Supplemental Fig. S8A). After phosphorylation with ATP and T4 polynucleotide kinase (PNK), piR-ILs became detectable by the EDC method (Supplemental Fig. S8A; Pall and Hamilton 2008). As expected, mature piRNAs were also detectable by this method, even without external PNK phosphorylation (Supplemental Fig. S8A). These findings suggest that piR-ILs are free from phosphate groups at both ends. piR-ILs were barely polyadenylated unless they were pretreated with PNK under acidic conditions (Supplemental Fig. S8B). Together, these results suggest that piR-ILs contain a 2′,3′-cyclic phosphate at their 3′ end. It has been shown previously that mature piRNAs loaded onto Piwi contain a phosphate group at their 5′ end. Thus, the piR-ILs that we detected in this study (Fig. 3C) are apparently not yet in a form that can associate directly with Piwi, although they did associate with the Armi–Piwi–Yb complex (Fig. 3D). Although it remains to be elucidated how piR-ILs are produced from the primary transcripts of transposons and others, including the tj gene, such characteristics of piR-ILs would help resolve this issue.
Nuclear localization of Piwi is required for transposon silencing
Piwi is required for the silencing of transposons in gonads (Ghildiyal and Zamore 2009; Malone and Hannon 2009; Siomi and Siomi 2009). In fact, Piwi depletion in OSCs caused derepression of transposons, as with Armi, Yb, and Zuc depletion (Supplemental Fig. S9A–C). Under conditions where endogenous Piwi was depleted, expression of myc-Piwi-r, which was designed to be RNAi-insensitive, rescued transposon silencing (Supplemental Fig. S9D). However, myc-Piwi-ΔN (Saito et al. 2009), which lacks 72 amino acids at the N terminus of Piwi and thus does not localize to the nucleus, did not rescue transposon silencing (Supplemental Fig. S9D), although it does associate with piRNAs to the same extent as does the wild-type Piwi (Saito et al. 2009). myc-Piwi-ΔN13, which lacks 13 amino acids at the N terminus, behaved similarly (data not shown). On the other hand, myc-Piwi-DDAA-r, a Slicer mutant of Piwi, could bind to mature piRNAs in OSCs, as does the wild-type Piwi (Saito et al. 2009), and rescued transposon silencing (Supplemental Fig. S9E). These results might suggest that Piwi must be localized in the nucleus to silence the transposable elements, and that Piwi Slicer activity is unnecessary for its function. We assume that this system has evolved to prevent nascent Piwi, not loaded with piRNAs, from being imported into the nucleus. In other words, only the functional Piwi–piRNA complex (piRISC) formed at Yb bodies could be transported to the nucleus. At present, the mechanisms of this control system remain unclear. In the nongonadal somatic S2 cell line, where the expression of piRNAs is undetectable, transfected Piwi is localized to the nucleus (data not shown), indicating that “empty” Piwi can be transported to the nucleus. It seems that the machineries necessary for the nuclear transport of Piwi might recognize different features of Piwi in different cell types.
How is piRNA-free Piwi restrained in the cytoplasm in OSCs? One possibility is that some unknown protein binds the N-terminal end of Piwi, where its NLS (nuclear localization signal) resides, and interferes with the nuclear import machinery's ability to recognize Piwi as a cargo. The nuclear localization inhibitory factors may be retained on Piwi until a functional Piwi–piRNA complex is formed at Yb bodies. Once the complex is formed, a conformational change in Piwi would be induced, which would release the regulatory factors and reveal the Piwi NLS for recognition by the nuclear import machinery. It would be very interesting to determine the proteins that are associated with Piwi in OSCs under conditions of Armi or Zuc depletion, thus identifying the protein factors that restrain Piwi in the cytoplasm until it is loaded with mature piRNAs at Yb bodies.
Materials and methods
Production of the anti-Armi antibody and Western blotting
A 200-amino-acid fragment of the N terminus of Armi fused with glutathione-S-transferase was used as the antigen to immunize mice. An anti-Armi monoclonal antibody was produced essentially as described previously (Saito et al. 2006). Western blotting was performed as described previously (Saito et al. 2006). Anti-tubulin was obtained from the Developmental Studies Hybridoma Bank and was used at 1:1000 dilution.
Immunofluorescence; immunoprecipitation; analysis of piRNA-intermediate chemical structures, Drosophila strains, myc-Piwi expression, and RNAi in OSCs; cloning of small RNAs and processing sequence tags; genome mapping, annotation, and frequency mapping; and Northern blotting are described in the Supplemental Material.
Acknowledgments
We are grateful to H. Lin and D. Godt for antibodies, W. Theurkauf (University of Massachusetts) for armi mutant lines, and I. Sagawa for mass spectrometry. We thank Y. Ono and T. Yamada for bioinformatics analyses. We also thank T. Okada and S. Inagaki for their technical assistance, and other members of the Siomi laboratory for discussions and comments on the manuscript. This work was supported by MEXT (Ministry of Education, Culture, Sports, Science, and Technology of Japan) grants to K.S., H.S., and M.C.S. H.I, Y.K., and M.K.N. are supported by the JSPS (Japan Society for the Promotion of Science). M.C.S. is supported by CREST from the JST.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1989510.
Supplemental material is available at http://www.genesdev.org.
References
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA 2006. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol 8: 1255–1262 [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE 2004. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116: 817–829 [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H 2000. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC 2001. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J 20: 2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM 2001. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128: 2823–2832 [DOI] [PubMed] [Google Scholar]
- Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA 2008. Capped small RNAs and MOV10 in human hepatitis δ virus replication. Nat Struct Mol Biol 15: 714–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King FJ, Lin H 1999. Somatic signaling mediated by fs(1)Yb is essential for germline stem cell maintenance during Drosophila oogenesis. Development 126: 1833–1844 [DOI] [PubMed] [Google Scholar]
- King FJ, Szakmary A, Cox DN, Lin H 2001. Yb modulates the divisions of both germline and somatic stem cells through piwi- and hh-mediated mechanisms in the Drosophila ovary. Mol Cell 7: 497–508 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W 2008. Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 12: 45–55 [DOI] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D 2003. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol 5: 994–1000 [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T 2005. Identification of novel argonaute-associated proteins. Curr Biol 15: 2149–2155 [DOI] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC 2007. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ 2008. Improved Northern blot method form enhanced detection of small RNA. Nat Protoc 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2009. On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- Szakmary A, Reedy M, Qi H, Lin H 2009. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol 185: 613–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD 2004. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831–841 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]