Abstract
CCCTC-binding factor (CTCF) is a DNA-binding protein that plays important roles in chromatin organization, although the mechanism by which CTCF carries out these functions is not fully understood. Recent studies show that CTCF recruits the cohesin complex to insulator sites and that cohesin is required for insulator activity. Here we showed that the DEAD-box RNA helicase p68 (DDX5) and its associated noncoding RNA, steroid receptor RNA activator (SRA), form a complex with CTCF that is essential for insulator function. p68 was detected at CTCF sites in the IGF2/H19 imprinted control region (ICR) as well as other genomic CTCF sites. In vivo depletion of SRA or p68 reduced CTCF-mediated insulator activity at the IGF2/H19 ICR, increased levels of IGF2 expression, and increased interactions between the endodermal enhancer and IGF2 promoter. p68/SRA also interacts with members of the cohesin complex. Depletion of either p68 or SRA does not affect CTCF binding to its genomic sites, but does reduce cohesin binding. The results suggest that p68/SRA stabilizes the interaction of cohesin with CTCF by binding to both, and is required for proper insulator function.
Keywords: RNA-binding protein, noncoding RNA, CTCF, insulator function
Chromatin insulators are DNA elements that can protect a gene from outside influences, which might lead to either inappropriate activation or silencing of the gene (Kellum and Schedl 1991; Chung et al. 1993; Geyer and Corces 1992; Bushey et al. 2008). One kind of insulator action can block interaction between an enhancer and a promoter when it lies between them; a number of such DNA elements and their associated protein factors have been identified in Drosophila (Moon et al. 2005; Wallace and Felsenfeld 2007), most notably the Gypsy element and its associated DNA-binding protein, Suppressor of Hairy-wing, which recruit multiple cofactors essential to the insulator activity (Geyer and Corces 1992; Georgiev and Kozycina 1996; Pai et al. 2004). In vertebrates, the CCCTC-binding factor CTCF is the principal protein with well-established insulator function (Bell et al. 1999; Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000). Work in many laboratories has shown that CTCF-binding sites are widely distributed in vertebrate genomes (Barski et al. 2007; Kim et al. 2007; Xie et al. 2007; Cuddapah et al. 2009). Recent studies suggest that their primary function is to establish contacts between these sites, stabilizing long-range interactions (Gaszner and Felsenfeld 2006; Phillips and Corces 2009; Sandhu et al. 2009), and either separating or bringing together distant regulatory elements. In this view, insulation is a consequence of a particular configuration in which the insulator lies between the enhancer and the promoter and prevents their interaction.
CTCF is a highly conserved 11-zinc-finger, DNA-binding protein (Ohlsson et al. 2001) implicated in diverse regulatory functions, including transcriptional activation/repression and X chromosome inactivation (Filippova et al. 1996; Vostrov and Quitschke 1997; Chao et al. 2002; Phillips and Corces 2009). The role of CTCF in mediating enhancer-blocking insulation was initially identified at the 5′ DNase-hypersensitive site 4 (5′HS4) insulator of the chicken β-globin locus (Bell et al. 1999). CTCF was subsequently found to control, through its insulator activity, allele-specific expressions of IGF2 and H19 in the mouse and human IGF2/H19 loci (Bell et al. 1999; Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000). It has been shown that CTCF binds to multiple sites on the maternal allele within the imprinted control region (ICR) that lies between IGF2 and the endodermal enhancers controlling its expression, effectively blocking those enhancers and silencing IGF2 expression. In contrast, DNA methylation of the ICR on the paternal allele prevents CTCF binding and allows IGF2 expression (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000; Holmgren et al. 2001). Depletion of CTCF or mutation of its binding sites results in loss of imprinting of IGF2 and H19 (Engel et al. 2008; Wendt et al. 2008) and alters the pattern of long-range intranuclear contacts (Engel et al. 2008; Yoon et al. 2007).
CTCF insulator activity requires a number of protein cofactors that interact with CTCF. Earlier studies have identified the SNF2-like chromodomain helicase protein CHD8 and the Polycomb group subunit Suz12 as mediating CTCF insulator function, although the mechanisms of their action have not been reported (Ishihara et al. 2006; Li et al. 2008). Recent attention has focused on the cohesin complex, which interacts with CTCF and is found at a large fraction of CTCF sites in vivo (Parelho et al. 2008; Rubio et al. 2008; Wendt et al. 2008). Depletion of cohesin subunit concentration in cells strongly inhibits the insulator action of CTCF, affecting both gene expression and long-range physical contacts in the surrounding region (Hadjur et al. 2009; Nativio et al. 2009; Hou et al. 2010). Given the known properties of cohesin in bringing sister chromatids together during S phase and through G2 phase into mitosis, one attractive hypothesis is that cohesin serves an analogous function in bringing together distant CTCF-occupied sites during interphase. However, it is not known what other factors may be involved in establishing or maintaining such structures.
In this study, we report that the DEAD-box RNA helicase p68 (DDX5) and its associated noncoding RNA, steroid receptor RNA activator (SRA), are both essential in vivo for insulator function at the IGF2/H19 ICR. p68/SRA is present at the ICR in mouse and human cells. Our evidence suggests that it is important because it binds to both CTCF and cohesin and helps stabilize the cohesin–CTCF interaction.
Results
RNA helicase p68 interacts with insulator-binding protein CTCF
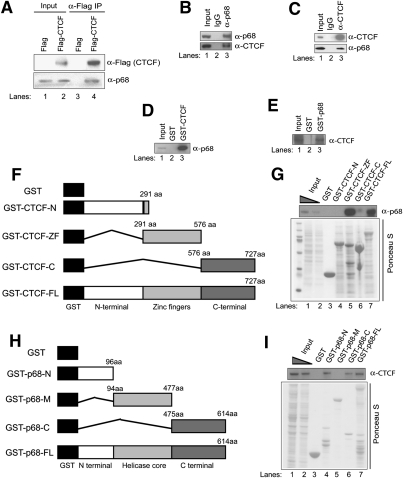
To identify previously unknown CTCF-associated cofactors, Flag-HA-tagged CTCF was purified by tandem affinity purification of nuclear extracts (NEs) from a stably transduced HeLa cell line, and copurifying factors were identified by mass spectrometry (MS). Results from MS analysis identified the DEAD-box RNA helicase protein p68, also known as DDX5, as a factor that copurified with Flag-HA-CTCF (data not shown). The MS analysis also identified p72, a protein partially homologous to p68, as well as components of the cohesin complex, which has been shown to interact with CTCF (Rubio et al. 2008) and which colocalizes with it at CTCF-binding sites in vivo (Parelho et al. 2008; Wendt et al. 2008). To confirm the MS results, we carried out Flag coimmunoprecipitation (co-IP) using NEs from the same cell line. p68 was retained in Flag-HA-CTCF-expressing cells following IP with an anti-Flag antibody, but not from cells expressing Flag-HA alone (Fig. 1A). To further verify the endogenous interaction of p68 with CTCF, co-IP analysis was carried out using NEs from wild-type HeLa cells. Both p68 and CTCF were pulled down using a rabbit anti-p68 antibody, but not with rabbit IgG (Fig. 1B). This interaction was not confined to HeLa cells; similar co-IP experiments in murine NIH3T3 NEs demonstrated that murine p68 could be copurified with CTCF using a rabbit anti-CTCF antibody (Fig. 1C). In addition, we confirmed colocalization of endogenous CTCF and p68 proteins by double immunostaining; localization was primarily nuclear (Supplemental Fig. 1).
Figure 1.
RNA-binding protein p68 interacts with insulator-binding protein CTCF. (A) NEs from HeLa cells stably expressing Flag or Flag-tagged human CTCF were used in Flag co-IP experiments and analyzed by immunoblotting with anti-Flag and anti-p68 antibodies. (B) HeLa NEs were precipitated with rabbit IgG or rabbit anti-p68 antibody, shown on the top of the panel, and precipitates were immunoblotted with antibodies, shown on the right of the panel. (C) NIH3T3 NEs were precipitated with mouse IgG or mouse anti-CTCF antibody, shown on the top of panel, and precipitates were immunoblotted with antibodies, shown at the right of panel. (D) Western blot of input, GST-associated, and GST-CTCF-associated proteins isolated from HeLa NEs probed with anti-p68 antibody. (E) Western blot of input, GST-associated, and GST-p68-associated proteins isolated from HeLa NEs probed with anti-CTCF antibody. (F) Schematic representation of GST-CTCF and its deletions used in GST pull-down assays. (G, top panel) GST pull-down assay using immobilized GST only or recombinant GST-CTCF N terminus (GST-CTCF-N), zinc fingers (GST-CTCF-ZF), C terminus (GST-CTCF-C), and full-length (GST-CTCF-FL) proteins probed with anti-p68 antibody by Western blot. (Bottom panel) Ponceau S stain showing total protein present on the membrane before Western blot. Lanes 1 and 2 had 5% and 2% of the input protein samples, respectively. (H) Schematic representation of GST-p68 and its deletions used in GST pull-down assays. (I, top panel) GST pull-down assay using immobilized GST only or recombinant GST-p68 N terminus (GST-p68-N), RNA helicase core region (GST-p68-M), C terminus (GST-p68-C), and full-length (GST-p68-FL) proteins probed with anti-CTCF antibody by Western blot. (Bottom panel) Ponceau S stain showing total protein present on the membrane before Western blot.
Domain mapping
To investigate whether p68 also interacts with CTCF in vitro, we performed glutathione-Stransferase (GST) pull-down assays using GST-CTCF and GST-p68 fusion proteins expressed and purified from Escherichia coli BL21 cells. The results showed that GST-CTCF but not GST could pull down p68 (Fig. 1D). Similarly, GST-p68 but not GST could pull down CTCF (Fig. 1E). We undertook domain mapping of both CTCF and p68 using GST fusion proteins and HeLa NEs to identify the regions of the proteins responsible for their interaction. Full-length GST-CTCF and a GST construct containing only the zinc fingers (amino acids 291–576) pulled down endogenous p68 in HeLa NEs, whereas GST constructs of the N-terminal (amino acids 1–291) or C-terminal (amino acids 576–727) regions of CTCF did not pull down p68 (Fig. 1F,G). To determine the domains of p68 responsible for binding to CTCF, a series of GST-p68 deletion constructs was used to pull down CTCF from HeLa NEs (Fig. 1H). Both the N-terminal and C-terminal domains of p68 (amino acids 1–96 and amino acids 475–614, respectively) bound to CTCF (Fig. 1I). In contrast, the central RNA helicase core domain (amino acids 94–477) did not (Fig. 1I).
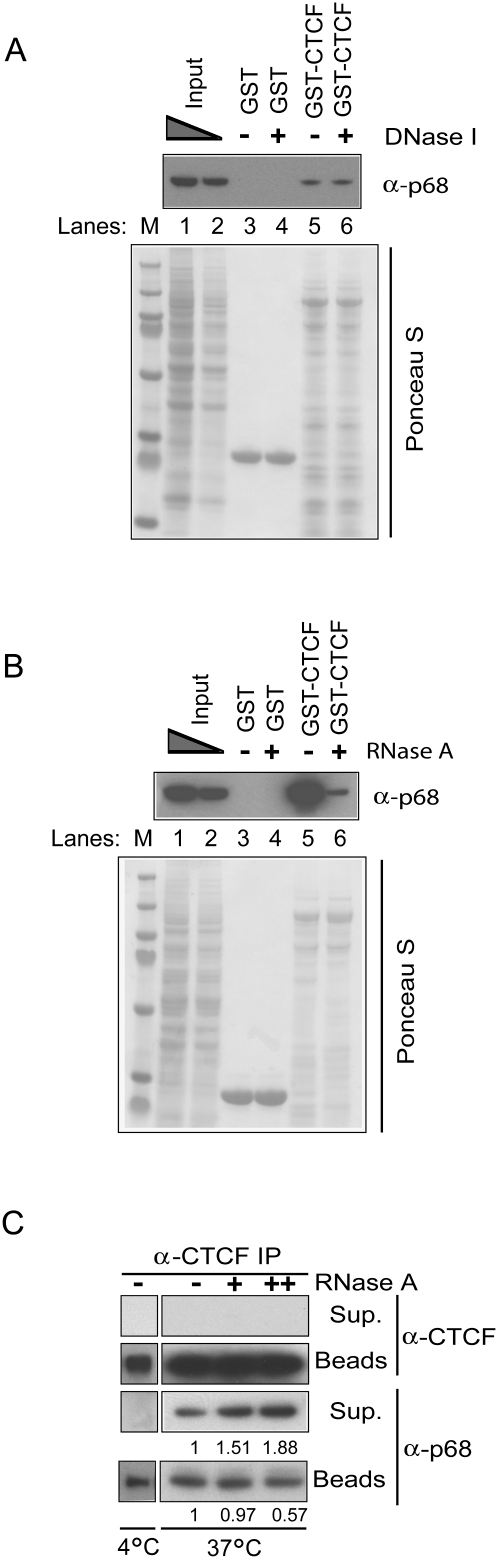
The binding of p68 to CTCF is RNA-dependent
p68 is an RNA-binding protein and is required for processing of rRNA and a subset of microRNAs (Fukuda et al. 2007). To examine whether the binding of p68 to CTCF was DNA- or RNA-dependent, we first performed GST or GST-CTCF pull-down assays with HeLa NEs in the presence or absence of DNase I or RNase A. Interactions between p68 and CTCF were not changed following DNase I digestion (Fig. 2A), indicating that this interaction was DNA-independent. In contrast, we found that the binding of p68 to CTCF decreased compared with control when RNase A was included in the reaction, indicating that RNA may play a role in this interaction (Fig. 2B). To further confirm this result, we treated magnetic bead immunoprecipitated CTCF complexes with RNase A and then isolated the tightly bound proteins of the bead fraction and the released proteins of the supernatant fraction. Although incubation at 37°C in the absence of RNase A partially released CTCF-bound p68, in the presence of RNase A, 1.51-fold to 1.88-fold more p68 was released from the CTCF-associated bead fraction with the increasing amount (50–200 μg/mL) of RNase A in the reaction (Fig. 2C). A corresponding decrease of p68 in the CTCF-associated bead fraction was seen, consistent with the results in Figure 2B and suggesting that RNA mediates the binding of p68 to CTCF.
Figure 2.
p68 binding to CTCF is RNA-dependent. HeLa NEs were incubated with purified GST or full-length GST-CTCF fusion protein immobilized on glutathione beads overnight at 4°C in the presence (+) or absence (−) of DNase I (A) or RNase A (B). Blots were then probed with anti-p68 antibody. Ponceau S stains in A and B show total protein present on the membranes before Western blotting. (C) CTCF co-IPs from HeLa NEs were digested with increasing amounts of RNase A, separated into supernatant (Sup.) and bead (Beads) fractions, and then subjected to immunoblot with antibody to either CTCF or p68.
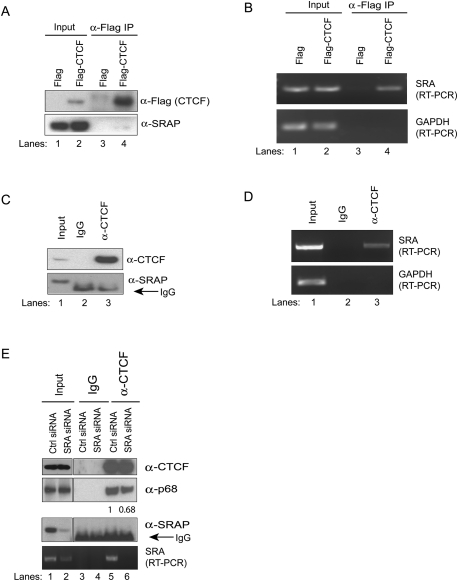
Noncoding SRA is present in a CTCF complex
Previous studies have shown that p68/p72 interacts with the noncoding RNA SRA (Watanabe et al. 2001). SRA can be present as both an RNA transcript and a stably expressed protein (SRAP), and appears to be the first example of a molecule active at both the RNA and protein level (Lanz et al. 1999; Chooniedass-Kothari et al. 2004; Colley et al. 2008; Colley and Leedman 2009). Most recently, it has been shown that SRAP is a transcriptional modulator of estrogen receptor α (ERα) and is recruited to specific promoter regions where it acts as a transcriptional repressor (Chooniedass-Kothari et al. 2010a,b); however, the function of SRAP still remains largely unknown. In order to determine if either SRA or SRAP was present in the CTCF–p68 complex, cell extracts from HeLa cells expressing Flag only or Flag-CTCF were immunoprecipitated with anti-Flag antibody and separated into two fractions for RNA and Western blot analysis. We could not detect a band using an antibody for SRAP in the Flag-CTCF co-IP, although it was present in the input (Fig. 3A), indicating that the protein encoded by SRA was not associated with CTCF. Noncoding SRA transcripts generated through alternative splicing of intron 1 have been characterized previously (Hube et al. 2006; Leygue 2007). Extracted RNAs from the co-IP material were treated with DNase I and subjected to reverse transcription, and the cDNA was amplified with primers specific for either SRA or GAPDH (as a negative control). Our data showed that the noncoding RNA for SRA coimmunoprecipitated with Flag-CTCF, but not with Flag alone (Fig. 3B). In addition, we found that SRA coimmunoprecipitated with the endogenous human CTCF following CTCF co-IP in HeLa cells, but SRAP did not (Fig. 3C,D, respectively). Since CTCF is a DNA-binding protein and SRA binds directly to p68 (Watanabe et al. 2001), the recruitment of SRA to the CTCF complex may be mediated by p68.
Figure 3.
The noncoding RNA SRA binds to CTCF. NEs from HeLa cells stably expressing Flag or Flag-tagged human CTCF were used in Flag co-IP experiments and either analyzed by immunoblotting with anti-SRAP and anti-Flag antibodies (A), or total RNA was extracted and subjected to RT–PCR with the primers specific to SRA and GAPDH (B). (C,D). HeLa NEs were precipitated with mouse IgG or mouse anti-CTCF antibody. Samples were then either immunoblotted with anti-CTCF or anti-SRAP antibodies (C), or total RNA was extracted and subjected to RT–PCR with the primers specific to SRA and GAPDH (D). (E) Control (Ctrl) siRNA- and SRA siRNA-depleted HeLa NEs were precipitated with mouse IgG or mouse anti-CTCF antibody and immunoblotted with anti-p68 antibody (top) or anti-SRAP antibody (middle), or RNA was extracted and samples were analyzed by RT–PCR with the primers specific to SRA (bottom).
Next, we investigated if SRA mediates the interaction between p68 and CTCF. We performed co-IP using an anti-CTCF antibody after SRA knockdown in HeLa cells and found that the binding of p68 to CTCF was decreased by ∼30% following SRA knockdown (Fig. 3E), suggesting that either small amounts of SRA present after knockdown are sufficient to give partial CTCF/p68 interaction, or one or more other RNA species can partially substitute for SRA.
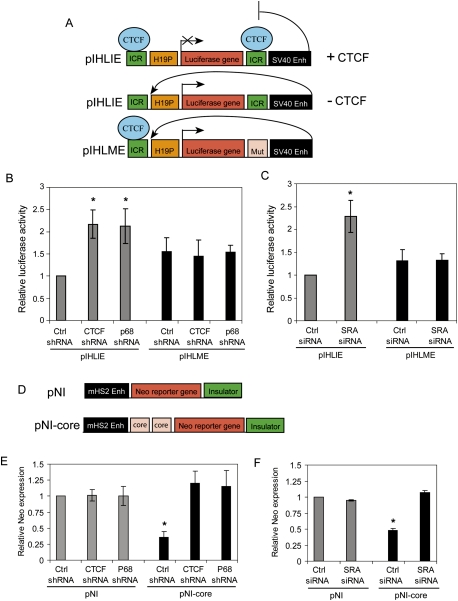
p68 and SRA are required for CTCF insulator function
Previous studies have shown that CTCF cofactors such as CHD8 and the cohesin complex are required for CTCF-mediated transcriptional insulation (Ishihara et al. 2006; Parelho et al. 2008; Wendt et al. 2008). The observation that p68, SRA transcript, and CTCF have the potential to form an RNA–protein complex prompted us to test whether either p68 or SRA was involved in CTCF-mediated insulator function. To test insulator activity, we used the previously published pIHLIE reporter construct, which carries a luciferase gene driven by the H19 promoter and is flanked by wild-type mouse ICR insulator sequences (Fig. 4A, top panel), and the pIHLME control reporter, in which insulating activity is destroyed by mutation of the CTCF-binding sites of one of the ICRs (Fig. 4A, bottom panel) (Ishihara et al. 2006). When CTCF is bound to the ICR sequences of pIHLIE, enhancer activity from a nearby SV40 enhancer is blocked. The HeLa cells were first transduced with either shRNA lentiviral particles targeted to p68 or CTCF, or siRNA targeted to SRA, then the pIHLIE or pIHLME plasmid was cotransfected into HeLa cells with a renilla luciferase control plasmid to measure relative luciferase activity. The knockdown efficiencies of CTCF, p68, and SRA RNA in pIHLIE or pIHLME transfected HeLa cells were determined by quantitative RT–PCR (qRT–PCR) (Supplemental Fig. 3). Our data indicated that the knockdown of p68, SRA, or CTCF enhanced luciferase activity by twofold in cells transfected with pIHLIE compared with that in control shRNA or siRNA (Fig. 4B,C). When the pIHLME control reporter was used (Fig. 4A, bottom panel)—in which insulating activity was destroyed by mutation of the CTCF-binding sites—neither p68, SRA, nor CTCF knockdown had any effect on luciferase activity (Fig. 4B,C).
Figure 4.
Both p68 and SRA are required for CTCF-mediated insulator function. (A) Schematic diagrams of the reporter constructs pIHLIE and pIHLME that were used in enhancer-blocking assays. In pIHLIE, CTCF binds to the H19 ICR and represses luciferase gene expression by blocking enhancer access to the promoter. In pIHLME, CTCF-binding sites have been mutated. (Enh) Enhancer; (H19P) mouse H19 promoter; (Mut) mutated ICR. (B) CTCF or p68 shRNA-depleted HeLa cells were transfected with either pIHLIE or pIHLME and a renilla luciferase control plasmid. Cells were harvested and luciferase activities were measured 48 h after transfection of the luciferase plasmids. Relative luciferase activity was determined for each cell lysate and normalized against control shRNA. (C) HeLa cells were depleted by SRA siRNA and then, 24 h later, transfected with either pIHLIE or pIHLME and a renilla luciferase control plasmid. Cells were harvested and luciferase activities were measured 24 h after transfection of luciferase plasmids. Relative luciferase activity was determined for each cell lysate and normalized against control siRNA. (D) Schematic representation of the pNI control and pNI-5′HS4 core insulator plasmids that were used in insulator assays. The plasmid backbone of pNI or pNI-core is pGEM4Z. pNI-core contains two copies of the chicken β-globin HS4 core insulator fragments, which are located between the mouse HS2 enhancer and Y-neomycin (Neo) reporter gene and which block the contact between the HS2 enhancer and Neo reporter gene. (E) K562 cells were depleted by CTCF or p68 shRNA and transfected with either pNI or pNI-core plasmid together with a GFP plasmid. Cells were harvested and RNAs were isolated for reverse transcription to examine the expression of the Neo reporter gene after 24 h of transfection. The expression of Neo was normalized against the expression of GFP. (F) SRA siRNA-depleted K562 cells were transfected with either pNI or pNI-core. Cells were harvested and RNAs were isolated for reverse transcription to examine the expression of the Neo reporter gene after 24 h of transfection. Relative neomycin expression was determined for each cell lysate and normalized against control shRNA (E) or control siRNA (F) samples. Data were analyzed by Student's t-test. For all panels, bars represent mean ± SD of n = 3. (*) P < 0.01.
Our laboratory previously identified and described the CTCF-dependent enhancer-blocking insulator properties at the 5′HS4 of the chicken β-globin locus (Bell et al. 1999; Recillas-Targa et al. 1999). Studies have also shown that expression of a neomycin (neo) resistance gene contained within a chicken β-globin core insulator plasmid is significantly reduced when flanked by insulators, whereas loss of the insulator activity through CTCF knockdown leads to full neo gene expression (Recillas-Targa et al. 1999; Parelho et al. 2008). To examine whether the insulator function of 5′HS4 is affected by either p68 or SRA, we transiently transfected pNI control or pNI-core insulator plasmids (Fig. 4D) into CTCF-, p68-, or SRA-depleted K562 cells (Supplemental Fig. 4A–C). As expected, in cells not depleted of these factors, neo gene expression was reduced in the pNI-core insulator plasmid when compared with the uninsulated pNI control plasmid, and depletion of CTCF returned neo gene expression to that of the uninsulated control (Fig. 4E,F). Interestingly, like CTCF depletion, depletion of either p68 or SRA also returned neo expression to levels comparable with that seen in the uninsulated control (Fig. 4E,F), suggesting that both p68 and SRA are required for 5′HS4 insulator function. In addition, we also found that knockdown of p72 increases neo gene expression in pNI-core transfected K562 cells, suggesting that p72, which typically forms a heterodimer with p68 (Caretti et al. 2006), also affects insulator activity (Supplemental Fig. 5). Taken together, our data show that knockdown of either p68/p72 or SRA results in the loss of the insulator function activities at both the H19 ICR and 5′HS4 insulators.
p68 and SRA regulate imprinted expression at the IGF2/H19 locus
p68 and SRA are required for CTCF-mediated enhancer-blocking insulator function in the plasmid assays described above. Next, we asked whether p68 and SRA affect “parent of origin”-specific transcription of the imprinted genes at the IGF2/H19 locus, which is controlled to a large extent by the IGF2/H19 ICR and mediated by CTCF binding. The ICR ensures that IGF2 is expressed from the paternal allele (DeChiara et al. 1991); H19 is transcribed from the maternal allele (Bartolomei et al. 1991). Imprinting at this locus depends mostly on the insulating activity of CTCF, which binds to ICR sites on the maternal allele, but not on the paternal allele, which is methylated (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000).
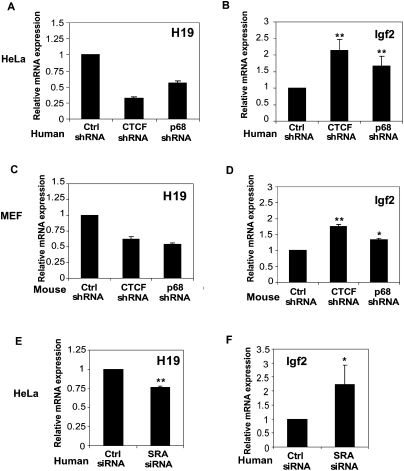
HeLa cells have been used previously to examine imprinting of the IGF2/H19 locus following cohesin depletion (Wendt et al. 2008). To determine whether p68 regulates imprinting at the IGF2/H19 locus, we measured the transcript levels of H19 (Fig. 5A) and IGF2 (Fig. 5B) in CTCF- or p68-depleted HeLa cells (Supplemental Fig. 2A,C). Although IGF2 expression is relatively low compared with H19 expression in control cells, H19 and IGF2 transcripts could be detected by RT–qPCR. H19 transcript levels were reduced by factors of ∼0.7 and 0.5 in HeLa cells after CTCF and p68 depletions, respectively (Fig. 5A), while IGF2 transcript levels were increased ∼1.5-fold to twofold in CTCF- or p68-depleted HeLa cells (Fig. 5B).
Figure 5.
p68 and SRA regulate imprinting at the IGF2/H19 locus. RT–qPCR analysis of human H19 (A) and IGF2 (B) transcripts in p68–shRNA- and CTCF–shRNA-depleted HeLa cells, mouse H19 (C) and IGF2 (D) transcripts in p68–shRNA- and CTCF–shRNA-depleted mouse primary MEF cells, and human H19 (E) and IGF2 (F) transcripts in control and SRA siRNA-depleted HeLa cells. Transcript levels were normalized to human or mouse HPRT1 levels and then the control shRNA or siRNA sample. Data were analyzed by Student's t-test. Error bars represent mean ± SD. (*) P < 0.05; (**) P < 0.01.
Primary mouse embryonic fibroblasts (MEFs) in which the maternal and paternal IGF2/H19 alleles can be distinguished have also been used to study imprinting at the IGF2/H19 locus (Engel et al. 2008). To investigate whether p68 and CTCF also regulate the imprinting of IGF2 and H19 in these MEF cells, we depleted the expression of p68 as well as CTCF (as a positive control) by using either murine CTCF–shRNA or p68–shRNA (Supplemental Fig. 2B,D), and examined the transcript levels of IGF2 and H19 in these cells. shRNA-mediated knockdowns of CTCF or p68 decreased H19 transcript levels ∼50% and 40%, respectively (Fig. 5C), while IGF2 expression was increased ∼1.76-fold and 1.35-fold, respectively, in MEF cells (Fig. 5D).
To investigate whether SRA has the same effect as p68 on H19 and IGF2 transcript levels, we depleted SRA expression in HeLa cells (Supplemental Fig. 2E). Transcript levels of H19 were decreased ∼25% (Fig. 5E) and that of IGF2 was increased about twofold (Fig. 5F). Similar effects of SRA depletion were observed in MEF cells (data not shown). Taken together, these data suggest that both p68 and SRA may have an important role in regulating imprinting at the IGF2/H19 locus, possibly through their association with CTCF.
p68 binds to the IGF2/H19 ICR at the maternal allele
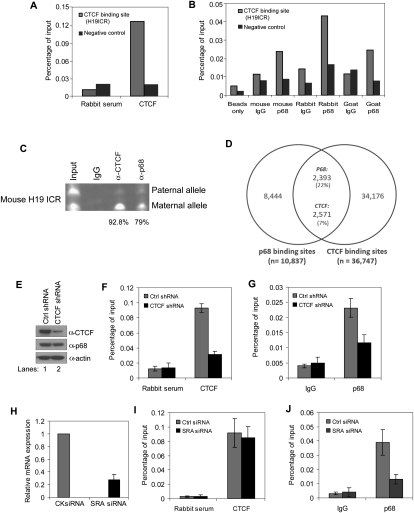
Since we found that p68 interacted with CTCF and regulated the imprinting of IGF2 and H19, we asked whether p68 could be found at CTCF-binding sites at the IGF2/H19 ICR in HeLa cells. Chromatin immunoprecipitation (ChIP) was carried out using an anti-CTCF antibody and each of three different anti-p68 antibodies, followed by qPCR. Both CTCF and p68 were bound in vivo at the IGF2/H19 ICR, but not at a CTCF-negative control site (Fig. 6A,B; Wendt et al. 2008). In order to determine if p68 binding to the IGF2/H19 ICR was allele-specific, we performed similar ChIP assays in MEF cells and found that 92.8% of the CTCF-bound fraction and 79% of p68-bound fraction were at the maternal allele (Fig. 6C). These data support the role of p68 in CTCF-mediated insulation at the IGF2/H19 locus.
Figure 6.
Binding of p68 to the maternal allele of IGF2/H19 ICR is both CTCF- and SRA-dependent. (A) Cross-linked DNA–protein complexes from HeLa cells were immunoprecipitated with anti-CTCF or normal rabbit serum, followed by qPCR amplification with primers specific to the IGF2/H19 ICR CTCF-binding region and a negative control region (chr11:1983833–1983994) (Wendt et al. 2008). Values are presented as percent of input. (B) ChIP analysis of p68 occupancy at the IGF2/H19 ICR CTCF-binding site was performed using chromatin immunoprecipitated from HeLa cells with mouse anti-p68, rabbit anti-p68, and goat anti-p68 antibodies or IgG alone. Precipitated DNA was analyzed by qPCR. (C) Allele-specific ChIP assays were carried out with antibodies against CTCF, p68, or IgG in MEF cells. Input and precipitated samples were subjected to semi-qPCR for the paternal and maternal H19 ICR alleles and then digested with a restriction enzyme, Tsp45I, a polymorphic restriction site. (D) Venn diagram showing overlap of p68- and CTCF-binding sites. (E) Western blot analysis of CTCF and p68 levels in NEs from CTCF shRNA or empty vector stably expressed HeLa cells. β-Actin was used as a loading control. (F,G) ChIP-qPCR analysis of isolated DNA associated with CTCF (F) and p68 (G) at IGF2/H19 ICR in CTCF-depleted HeLa cells (mean ± SD of n = 3). (H) RT–qPCR analysis to SRA knockdown efficiency in HeLa cells (mean ± SD of n = 3). (I,J) ChIP-qPCR analysis of isolated DNA associated with CTCF (I) and p68 (J) on the CTCF-binding site at IGF2/H19 ICR in SRA-depleted HeLa cells (mean ± SD of n = 3).
Coincidence of p68, CTCF, or cohesin binding on a genome-wide scale
We wanted to determine whether other CTCF-binding sites in the genome are also associated with p68. We identified 10,837 p68-binding sites in HeLa cells using a preliminary ChIP-seq analysis not intended to reveal all such sites. Twenty-two percent of these p68-binding sites overlap a non-tissue-specific CTCF-binding site (Fig. 6D; Schmidt et al. 2010), far more than the expected 2% (P < 10−250, one-sided binomial test) (Supplemental Material). This overrepresentation was also observed when we performed the same analysis using CTCF sites derived specifically from HeLa cells (data not shown). ChIP-qPCR validations were carried out on nine selected p68 peaks that overlapped with CTCF peaks. All of these CTCF peaks were enriched for p68, eight quite strongly (Supplemental Fig. 6A,B).
Recently, it was shown that cohesin colocalizes with ERα at DNA-binding sites independently of CTCF (Schmidt et al. 2010). Since it has been reported that p68 also interacts with ERα (Endoh et al. 1999), we asked whether p68 could also bind to sites occupied by ERα and cohesin, but not CTCF. We found that, indeed, p68 binding is significantly overrepresented at these sites (noverlap = 257; P = 10−198, one-sided binomial test).
The recruitment of p68 to CTCF-binding sites is both CTCF- and SRA-dependent
To further investigate the relationship between p68 and CTCF, ChIP analysis of the IGF2/H19 ICR was performed using anti-p68 and anti-CTCF antibodies following knockdown of p68 (Supplemental Fig. 7A) or CTCF (Fig. 6E). Knockdown of p68 or CTCF does not affect SRA expression (Supplemental Fig. 8). As expected, the recruitment of p68 to the CTCF ICR-binding sites was reduced by p68 knockdown in HeLa cells (Supplemental Fig. 7B). However, p68 depletion did not affect the binding of CTCF to the IGF2/H19 ICR sites (Supplemental Fig. 7C). ChIP assays were also performed following depletion of either CTCF (Fig. 6E) or SRA (Fig. 6H). As expected, CTCF depletion decreased the fraction of recovered CTCF at the ICR-binding sites (Fig. 6F). The recruitment of p68 to the same ICR sites was decreased not only by CTCF depletion, but also by SRA depletion (Fig. 6G,J). Conversely, the recruitment of CTCF to IGF2/H19 ICR was not affected by SRA depletion (Fig. 6I). These results indicate that p68 localization to the CTCF-binding sites in the IGF2/H19 ICR may require CTCF or SRA, but that CTCF binding does not depend on either p68 or SRA.
p68 and cohesin binding at the H19 ICR are interdependent
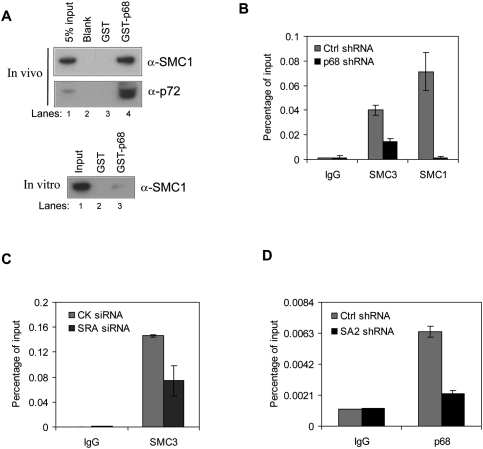
Since CTCF has also been shown to interact with the cohesin complex (Rubio et al. 2008), we next tried to determine if p68 also interacts with cohesin. To this end, we performed GST pull-down assays between p68 and components of the cohesin complex. We found that SMC1, one subunit of the cohesin complex, interacted strongly with p68 both in vivo and in vitro (Fig. 7A). Interaction with other cohesin subunits could not be detected by this method (data not shown).
Figure 7.
Functional relationships between p68 and cohesin. (A) GST pull-down assay from HeLa NEs (top panel) or in vitro translated SMC1 (bottom panel) using immobilized GST only or recombinant GST-p68. Bound proteins were analyzed by Western blot for the cohesin subunit SMC1; p72 served as a positive control for in vivo analysis. (B) ChIP-qPCR analysis of isolated DNA associated with the cohesin subunits SMC1 and SMC3 at the CTCF-binding site of IGF2/H19 ICR in p68-depleted HeLa cells. (C) ChIP-qPCR analysis of isolated DNA associated with SMC3 at the CTCF-binding site of H19 ICR in SRA-depleted HeLa cells. (CK) Control. (D) ChIP-qPCR analysis of isolated DNA associated with p68 at the CTCF-binding site of IGF2/H19 ICR in SA2-depleted HeLa cells.
We asked whether the interaction between p68 and cohesin was important for binding of either protein to CTCF-binding sites in vivo. We measured the occupancy of cohesin components at the IGF2/H19 ICR by ChIP analysis after depleting either p68 or SRA. We found that p68 depletion in HeLa cells had only a small effect on the expression levels of the cohesin subunits Rad21, SA2, SMC1, and SMC3 (∼20% decrease for SMC3, less for the others), and SRA depletion had no effect on the expression levels of cohesin subunits (Supplemental Fig. 9A,B). In addition, pull-down of cohesin by CTCF does not depend on the presence of p68 (data not shown); however, the recruitment of the cohesin subunits SMC1 or SMC3 to the IGF2/H19 ICR was reduced by p68 knockdown (Fig. 7B). SMC3 recruitment is also impaired when SRA RNA is depleted (Fig. 7C). These data suggest that p68/SRA may play an important role in facilitating cohesin binding at the IGF2/H19 ICR. Next, we depleted SA2 by using SA2 shRNA constructs shRNA-1 through shRNA-5. We found that cells highly depleted in SA2, using SA2 shRNA-4, failed to divide (data not shown), but cells with only moderate SA2 depletion (shRNA-3 and shRNA-5) were still able to divide and grow (Supplemental Fig. 10A). In addition, cohesin depletion in HeLa cells had no effect on the expression levels of p68 (Supplemental Fig. 10B). We performed ChIP-qPCR assays on the control shRNA and SA2 shRNA-5-depleted cells using p68 antibody, and found that the occupancy of p68 at the IGF2/H19 ICR was reduced (Fig. 7D). Thus, the disruption of the cohesin complex through SA2 depletion also interferes with p68 binding to this CTCF site (Fig. 7D). These results show that, as well as interacting individually with CTCF, p68 and cohesin interact with each other in a way that appears to stabilize the binding of both cohesin and p68 to CTCF.
Depletion of p68 or CTCF increases the contact between endodermal enhancer and IGF2 promoters
Chromatin conformation capture (3C) studies have shown that long-range interactions between the downstream H19 enhancer and the IGF2 promoter region are inhibited on the maternal allele, and that the inhibition depends on the enhancer-blocking properties of CTCF bound to the ICR of that allele (Yoon et al. 2007; Engel et al. 2008). To test if p68 was involved in the CTCF-dependent organization of the IGF2/H19 locus, we depleted either mouse p68 or CTCF by shRNA in MEF cells and measured the change in contacts among sites in the locus, using the endodermal enhancer as an anchor (Supplemental Fig. 11A). Knockdown of CTCF increased the association between the H19 endodermal enhancer and IGF2 promoters compared with the control (Supplemental Fig. 11B). These results were expected based on similar measurements with mutated CTCF sites (Engel et al. 2008). While our previous experiments determined that depletion of p68 had no effect on CTCF recruitment to the IGF2/H19 site (Supplemental Fig. 7C), knockdown of p68 did result in increased contact between the H19 enhancer and the region containing the IGF2 promoters, similar to what is seen with CTCF depletion. These results are consistent with those described above showing that depletion of p68 results in overexpression of IGF2, paralleling the effects of CTCF depletion.
Discussion
CTCF is implicated in multiple roles in gene regulation, including context-dependent promoter activation/repression, enhancer-blocking and/or barrier insulation, genomic imprinting, hormone-responsive silencing, and, perhaps more fundamentally, long-range intra- and interchromosomal interactions (for review, see Gaszner and Felsenfeld 2006; Phillips and Corces 2009). A number of investigations have examined the role of factors that interact with CTCF and affect its function as an insulator protein. The chromodomain helicase protein CHD8 has been shown to be important for insulation, although its mode of action is not known (Ishihara et al. 2006). Most recently, it has been shown that CTCF recruits the cohesin complex to its binding sites, and that the presence of cohesin is essential to insulator activity, probably because it stabilizes long-range intranuclear interactions between CTCF sites (Li et al. 2008; Parelho et al. 2008; Wendt et al. 2008). In this study, we show that the DEAD-box RNA-binding protein p68 (DDX5) interacts with CTCF both in vivo and in vitro, and that the noncoding RNA SRA, a functionally important RNA known to associate with p68, immunoprecipitates with CTCF. We also showed that both p68 protein and SRA are necessary for the activity of CTCF as an insulator element in vivo.
The DEAD-box RNA helicase p68 and the partially homologous protein p72 are RNA-binding proteins that are involved in a wide variety of regulatory and biosynthetic functions. p68 is required for ribosome biogenesis, and its ATPase/helicase activities are important for pre-mRNA splicing and microRNA processing (Lin et al. 2005; Jalal et al. 2007; Salzman et al. 2007). Notably, p68 is an essential component of the Drosha complex (Fukuda et al. 2007). p68 also functions as a cofactor for a variety of transcriptional regulatory proteins, including ERα, the p53 tumor suppressor, and MyoD (Endoh et al. 1999; Métivier et al. 2003; Bates et al. 2005; Caretti et al. 2006). At least some of these do not require the helicase activity of p68, and probably involve a distinct independent mechanism. In many of these cases, the active form of p68 may involve a complex with p72 (Caretti et al. 2007; Fuller-Pace and Ali 2008). Although we focused on the role of p68, data in Supplemental Figure 5 suggest that p72 also plays a role in CTCF function. The p68/p72 protein specifically bind SRA (Watanabe et al. 2001), a functionally important RNA, and it has been shown that the coactivation of MyoD by p68 depends on the presence of SRA (Caretti et al. 2006). SRA has also been shown to bind other proteins and modulate their activity (Colley and Leedman 2009). Since some splice variants of SRA code for protein, it is necessary to distinguish the activity of the RNA from that of the protein.
We report here that the CTCF–p68 interaction is critical to CTCF function as an enhancer-blocking insulator, as demonstrated by transient expression experiments with a reporter carrying CTCF-binding sites in which p68 is depleted by shRNA. Additionally, ChIP experiments show that p68 is present at the ICR of the human IGF2/H19 locus on chromosome 11 in HeLa cells, as well as the equivalent site on mouse chromosome 7 in MEF cells. Depletion of p68 results in an increase in IGF2 expression and a decrease in H19 expression, similar to that observed in HeLa cells upon depletion of cohesin components (Wendt et al. 2008). Loss of p68 also results in an increase in genomic contacts, as measured by 3C, between the endodermal enhancer and sites upstream of IGF2, consistent with loss of insulator function. The loss of p68 is not accompanied by a decrease in CTCF binding. We also found that the binding of p68 to CTCF is RNA-dependent: Depletion of ssRNA by RNase A or down-regulation of SRA inhibited the CTCF–p68 interaction (Figs. 2, 3E). This is similar to the behavior of the interaction between p68 and p53 (Suzuki et al. 2009). It is therefore not surprising that the ability of CTCF to act as an insulator also depends on SRA. The protein SRAP did not coprecipitate with CTCF either in vitro or in vivo, suggesting that it is not involved in the CTCF–p68 interaction.
It has been reported that the Drosophila DEAD-box putative RNA helicase protein Rm62, which is homologous to p68, interacts physically with the DNA-binding insulator protein CP190 in an ssRNA-dependent manner and negatively regulates gypsy insulator function (Lei and Corces 2006). It is striking that, in the case of Drosophila, the interacting factors are different from (CP190 rather than CTCF) and the effects are the opposite of (inhibitory rather than activating) those in vertebrates. These results hint at a common mechanism of action that has diverged.
What is the role of p68 in CTCF-dependent insulator function? In Figure 7A, we showed that, in addition to its interaction with CTCF, p68 also bound to a component of the cohesin complex in vitro. As discussed above, cohesin interacts with CTCF and is essential to insulator function. Previous studies have shown that loss of cohesin does not affect CTCF binding at most sites (Parelho et al. 2008). Similarly, depletion of p68 or its associated RNA, SRA, did not affect CTCF binding to the IGF2/H19 ICR in vivo (Fig. 6; Supplemental Fig. 7); however, depletion of either did result in loss of cohesin from those sites (Fig. 7B,C), showing that the interactions observed in vitro are important in vivo. These data support a model in which cohesin binding to CTCF at the IGF2/H19 locus is further stabilized by cohesin interaction with p68/SRA. We suggest that the effects on insulator function that we observe when p68/SRA is depleted reflect, at least in part, the loss of cohesin from the site. As noted above, p68 is also found at sites occupied by ERα and cohesin. It will be interesting to determine whether, at such sites, p68 plays a role in stabilizing cohesin localization. In addition, it will be necessary to examine the findings of Schmidt et al. (2010) in light of the more recent report of cohesin recruitment to Nipbl/mediator-binding sites (Kagey et al. 2010).
Our results show that CTCF sites, the majority of which recruit cohesin, may require additional components to establish long-range interactions and maintain an active insulator complex. It remains to be determined whether p68/SRA, known to have multiple regulatory activities, contributes in other ways to CTCF function.
Materials and methods
Cell culture
HeLa-S3 and NIH3T3 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (Invitrogen). A Flag-tagged CTCF HeLa S3 stable cell line was established following methods in Yusufzai et al (2004). K562 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin–streptomycin. Primary MEF cells were C57BL/6J by C7 (CAST Chromosome 7 in a B6 background), which were kindly provided by Dr. Marisa Bartolomei (University of Pennsylvania School of Medicine) and maintained in DMEM medium supplemented with 10% FBS, 1% L-glutamine, and 1% penicillin–streptomycin. All cell cultures were maintained in a 5% CO2 humidified incubator at 37°C.
Antibodies
The following antibodies were used in this study: rabbit anti-CTCF (Abcam, ab70303) and mouse anti-CTCF (Abcam, ab37477) for Western blot and co-IP; rabbit anti-CTCF (Upstate Biotechnologies, 07-729) for ChIP; Flag M2 affinity gel-purified immunoglobulin (Sigma, A2220) for Flag-IP and Western blot; rabbit anti-SMC1 (Abcam, ab9262) for Western blot and ChIP; rabbit anti-SMC3 (Abcam, ab9263) for ChIP; anti-p68/DDX5 (Abcam, ab21696, ab10261, and ab53216) for Western blot, co-IP, and ChIP; goat anti-SA2 antibody (Abcam, ab4463) and rabbit anti-p72 (Abcam, ab66764) for Western blot.
Plasmid constructs
GST CTCF full-length, GST-CTCF N terminus (amino acids 1–291), GST-CTCF zinc finger (amino acids 291–576), and GST-CTCF C terminus (amino acids 576–727) were cloned into the pGEX5T1 vector (GE Healthcare). pcDNA3.1-p68 and pGEX4T1-p68 full-length constructs were kindly provided by Dr. Frances V. Fuller-Pace at University of Dundee, UK. GST-p68 N terminus (amino acids 1–96), GST-p68 RNA helicase core (amino acids 94–477), and GST-p68 C terminus (amino acids 475–614) were amplified by sticky-end PCR and cloned into the pGEX4T1 vector (GE Healthcare) , then confirmed by sequencing.
Immunoprecipitation and immunoblot assay
HeLa NEs were prepared as described (Yusufzai et al. 2004). For IP, cleared NEs were incubated with the indicated antibodies plus Protein A magnetic beads (Active Motif) for 4 h or overnight. For RNase A digestion, the immunoprecipitated protein-bound magnetic beads were washed four times with IP buffer and suspended in the RNA digestion buffer containing RNase A (Ambion). The samples were incubated for 1 h at 37°C and quickly separated by a magnetic bar into two fractions: beads containing the still-bound proteins, and supernatant containing the released proteins. The separated protein fractions were subjected to SDS-PAGE and transferred to PVDF membrane (Invitrogen). Immunoblotting was performed with the indicated antibodies.
GST pull-down assays
GST; GST-CTCF full-length and its deletions (N terminus [amino acids 1–291], zinc finger [amino acids 291–576], and C terminus [amino acids 576–727]); GST-p68 full-length and its deletions (N terminus [amino acids 1–96[; and RNA helicase core domain [amino acids 94–477]; and C terminus [amino acids 475–614]) were expressed in E. coli strain BL21 cells (Invitrogen). The expressed fusion proteins from bacteria were purified using glutathione sepharose 4 fast flow beads (GE Healthcare) and then incubated with HeLa NE in the PBS-T buffer supplemented with protease inhibitors (1 mM PMSF, protease inhibitor cocktail) for 4 h or overnight at 4°C. Beads were washed four times with PBS-T buffer, boiled in SDS loading buffer, resolved on SDS-PAGE, and transferred to nitrocellulose membrane (Invitrogen). The transferred proteins were monitored by Ponceau S staining. Then the membrane was washed with TBS-T buffer to remove Ponceau S. Immunoblotting was performed with the indicated antibodies.
RT–qPCR assays
Total RNA was extracted from cells using the RNeasy mini kit (Qiagen). RNA was treated with DNase I (Ambion) for 30 min at 37°C. cDNA was synthesized using AffinityScript qPCR cDNA Synthesis kit (Stratagene). Amplification of cDNA was monitored with the SYBR green PCR master mix (Applied Biosystems) on the ABI PRISM 7900 PCR Detection System (Applied Biosystems) according to the manufacturer's instructions. Transcript levels were normalized to HPRT1 levels. Primers used for qRT–PCR are shown in Supplemental Table 1.
Luciferase and insulator assays
The H19 ICR reporter plasmids (pIHLIE and pIHLME) were reported previously (Ishihara et al. 2006; Wendt et al. 2008) and were kindly provided by Dr. Mitsuyoshi Nakao at Kumamoto University, Japan. To perform luciferase reporter assays, the firefly luciferase reporter plasmid (pIHLIE or pIHLME) and a renilla luciferase control plasmid were cotransfected into p68–shRNA- or CTCF–shRNA-depleted or SRA siRNA-depleted HeLa cells using Fugene 6 (Roche). After 24 or 48 h, cells were harvested and lysed. The activities of both luciferases were detected using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions, and luciferase activity was determined with a Victor Multilabel Counter (PerkinElmer). Renilla luciferase was used as a transfection control. All assays were done in triplicate and repeated at least two times.
pNI empty and pNI-5′HS4-core insulator plasmids have been described previously for use in insulator analysis (Recillas-Targa et al. 1999; Parelho et al. 2008). p68–shRNA-, CTCF–shRNA-, or SRA siRNA-depleted K562 cells were transfected with pNI empty or pNI-core insulator plasmid, and the expressions of the neomycin gene were detected by RT–qPCR after 24 h. There were three replicates for each treatment, and the experiments were repeated at least twice.
ChIP and qPCR
ChIP was performed using the indicated antibodies. Briefly, cells (5 × 106) were cross-linked with 1% paraformaldehyde for 10 min at 37°C and lysed in buffer containing 50 mM Tris (pH 8.0), 10 mM EDTA, 1% SDS, and protease inhibitors. Chromatin was sheared by sonication to generate 200- to 1000-base-pair (bp) DNA fragments and used for IP steps. Indicated antibodies were incubated with Dynabeads G (Invitrogen) with rotation for 20 min at room temperature. Chromatin preparations were then incubated with the complex of antibody magnetic beads overnight at 4°C. Immune complexes were washed with low-salt buffer, high-salt buffer, LiCl buffer, and TE buffer two times each. Antibody-bound chromatin was reverse-cross-linked, and the ChIP DNA samples were purified and used for qPCR. The PCR primers used for ChIP are listed in Supplemental Table 2.
Allele-specific ChIP and semi-qPCR
MEF cells were used for allele-specific ChIPs. ChIPs were carried out with antibodies against CTCF, p68, or IgG. Precipitated DNA samples and input DNA were subjected to PCR. PCR products were purified and digested with a restriction enzyme, Tsp45I, that is polymorphic between maternal and paternal alleles. Then, digested PCR products were resolved on 8% TBE gels, and gels were stained using 0.5 μg/mL ethidium bromide in 1× TBE buffer for 30 min. The PCR primers used for ChIP are listed in Supplemental Table 2.
Solexa sequencing and validation of ChIP-seq
Thirty-six-base-pair single-read high-throughput sequencing was performed using an Illumina GAII Genome Analyzer. Tags were aligned to the human genome (hg18) using the GAII analysis pipeline (version 1.5.0), yielding 23.3 million quality-filtered, aligned tags from the p68 ChIP sample and 35.5 million from the input DNA sample. Data have been deposited in the Gene Expression Omnibus with accession number GSE24126 .
To validate our ChIP-seq results, we used real-time PCR with SYBR green PCR master mix (Applied Biosystems) to amplify ChIP samples and input DNA for nine specific sites that were strongly positive in the ChIP-seq analysis (Supplemental Fig. 5), and calculated percentage of input for each region. It should be noted that the IGF2/H19 locus did not give a significant peak in the ChIP-seq analysis, consistent with the observation that, in the direct ChIP experiments, it gave a rather low signal compared with other sites (cf. Fig. 6B and Supplemental Fig. S5B). Primer sequences are listed in Supplemental Table 3.
Identification of ChIP-enriched regions
Input DNA ChIP-seq tags were subsetted randomly to match the number of tags in the p68 ChIP-seq set. MACS version 1.3.7 (Zhang et al. 2008) was used to identify p68-enriched sites by comparing the p68 tags with the tag-matched input tags. As the signal:noise ratio in this sample was low, a conservative false discovery rate of 1% was used to filter called peaks. Adjacent peaks separated by less than half the width of either peak were merged.
Acknowledgments
We thank Drs. Frances Fuller-Pace and Zhi-Ren Liu for p68 constructs, Dr. Mitsuyoshi Nakao for luciferase constructs, Drs. Chunhui Hou and Ann Dean for the mouse CTCF shRNA construct, Dr. Marisa Bartolomei for MEF cells, and Drs. Chunhui Hou and Zhixiong Xu for suggestions in 3C experiments. We also acknowledge Drs. Keith Giles and Zhixiong Xu for their critical reading of the manuscript, and all other members of the Felsenfeld laboratory for helpful discussions. This work was supported by the intramural research program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1967810.
Supplemental material is available at http://www.genesdev.org.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman SM 1991. Parental imprinting of the mouse H19 gene. Nature 351: 153–155 [DOI] [PubMed] [Google Scholar]
- Bates GJ, Nicol SM, Wilson BJ, Jacobs A-MF, Bourdon J-C, Wardrop J, Gregory DJ, Lane DP, Perkins ND, Fuller-Pace FV 2005. The DEAD box protein p68: A novel transcriptional coactivator of the p53 tumour suppressor. EMBO J 24: 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG 2008. Chromatin insulators: Regulatory mechanisms and epigenetic inheritance. Mol Cell 32: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V 2006. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell 11: 547–560 [DOI] [PubMed] [Google Scholar]
- Caretti G, Lei EP, Sartorelli V 2007. The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: Eclectic regulators of disparate biological functions. Cell Cycle 6: 1172–1176 [DOI] [PubMed] [Google Scholar]
- Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT 2002. CTCF, a candidate trans-acting factor for X-inactivation choice. Science 295: 345–347 [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, Hube F, Mutawe M, Watson PH, Leygue E 2004. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett 566: 43–47 [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S, Hamedani MK, Wang X, Carascossa S, Yan Y, Cooper C, Vincett D, Myal Y, Jalaguier S, Cavailles V, et al. 2010a. The steroid receptor RNA activator protein is recruited to promoter regions and acts as a transcriptional repressor. FEBS Lett 584: 2218–2224 [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S, Vincett D, Yan Y, Cooper C, Hamedani MK, Myal Y, Leygue E 2010b. The protein encoded by the functional steroid receptor RNA activator is a new modulator of ERα transcriptional activity. FEBS Lett 584: 1174–1180 [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G 1993. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Colley SM, Leedman PJ 2009. SRA and its binding partners: An expanding role for RNA-binding coregulators in nuclear receptor-mediated gene regulation. Crit Rev Biochem Mol Biol 44: 25–33 [DOI] [PubMed] [Google Scholar]
- Colley SM, Iyer KR, Leedman PJ 2008. The RNA coregulator SRA, its binding proteins and nuclear receptor signaling activity. IUBMB Life 60: 159–164 [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64: 849–859 [DOI] [PubMed] [Google Scholar]
- Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, Yanagisawa J, Metzger D, Hashimoto S, Kato S 1999. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol Cell Biol 19: 5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Engel N, Raval AK, Thorvaldsen JL, Bartolomei SM 2008. Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking. Hum Mol Genet 17: 3021–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 16: 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. 2007. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol 9: 604–611 [DOI] [PubMed] [Google Scholar]
- Fuller-Pace FV, Ali S 2008. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): Novel transcriptional co-regulators. Biochem Soc Trans 36: 609–612 [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G 2006. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Georgiev P, Kozycina M 1996. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 142: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Corces VG 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865–1873 [DOI] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Holmgren C, Kanduri C, Dell G, Ward A, Mukhopadhya R, Kanduri M, Lobanenkov V, Ohlsson R 2001. CpG methylation regulates the Igf2/H19 insulator. Curr Biol 11: 1128–1130 [DOI] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A 2010. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci 107: 3651–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube F, Guo JM, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA, Blanchard AAA, Wang XM, Deng G, Myal Y, et al. 2006. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol 25: 418–428 [DOI] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M 2006. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell 23: 733–742 [DOI] [PubMed] [Google Scholar]
- Jalal C, Uhlmann-Schiffler H, Stahl H 2007. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res 35: 3590–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10: 853–856 [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950 [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, MeKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97: 17–27 [DOI] [PubMed] [Google Scholar]
- Lei EP, Corces VG 2006. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet 38: 936–941 [DOI] [PubMed] [Google Scholar]
- Leygue E 2007. Steroid receptor RNA activator (SRA1): Unusual bi-faceted gene products with suspected relevance to breast cancer. Nuclear Recept Signal 5: e006 doi: 10.1621/nrs.05006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Hu JF, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu TH, Hoffman AR 2008. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol 28: 6473–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Yang JJ, Huang Y, Liu ZR 2005. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol Cell Biol 25: 7484–7493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F 2003. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115: 751–763 [DOI] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 5: e1000739 doi: 10.1371/journal.pgen.1000739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F, Bell AC, Felsenfeld G 1999. Positional enhancer-blocking activity of the chicken β-globin insulator in transiently transfected cells. Proc Natl Acad Sci 96: 14354–14359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A 2008. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman DW, Shubert-Coleman J, Furneaux H 2007. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J Biol Chem 282: 32773–32779 [DOI] [PubMed] [Google Scholar]
- Sandhu KS, Shi C, Sjölinder M, Zhao Z, Göndör A, Liu L, Tiwari VK, Guibert S, Emilsson L, Imreh MP, et al. 2009. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev 23: 2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K 2009. Modulation of microRNA processing by p53. Nature 460: 529–533 [DOI] [PubMed] [Google Scholar]
- Vostrov AA, Quitschke WW 1997. The zinc finger protein CTCF binds to the APBβ domain of the amyloid β-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem 272: 33353–33359 [DOI] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G 2007. We gather together: Insulators and genome organization. Curr Opin Genet Dev 17: 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, Suzawa M, Kobayashi Y, Yano T, Yoshikawa H, et al. 2001. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor α coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J 20: 1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci 104: 7145–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K 2007. Analysis of the H19ICR insulator. Mol Cell Biol 27: 3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13: 291–298 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]