Abstract
The majority of bacterial gene regulators bind as symmetric dimers to palindromic DNA operators of 12–20 base pairs (bp). Multimeric forms of proteins, including tetramers, are able to recognize longer operator sequences in a cooperative manner, although how this is achieved is not well understood due to the lack of complete structural information. Models, instead of structures, of complete tetrameric assembly on DNA exist in literature. Here we present the crystal structures of the multidrug-binding protein TtgV, a gene repressor that controls efflux pumps, alone and in complex with a 42-bp DNA operator containing two TtgV recognition sites at 2.9 Å and 3.4 Å resolution. These structures represent the first full-length functional tetrameric protein in complex with its intact DNA operator containing two continuous recognition sites. TtgV binds to its DNA operator as a highly asymmetric tetramer and induces considerable distortions in the DNA, resulting in a 60° bend. Upon binding to its operator, TtgV undergoes large conformational changes at the monomeric, dimeric, and tetrameric levels. The structures here reveal a general model for cooperative DNA binding of tetrameric gene regulators and provide a structural basis for a large body of biochemical data and a reinterpretation of previous models for tetrameric gene regulators derived from partial structural data.
Keywords: Protein–DNA complex, tetrameric gene regulator, cooperative DNA binding, multidrug-binding protein, antibiotic resistance
Bacterial gene regulators are model systems to study protein–DNA and protein–ligand interactions as well as principles of gene regulation. The majority of bacterial gene regulators bind as symmetric dimers to palindromic DNA operators in the range of 12–20 base pairs (bp), often acting by altering the access of RNA polymerase in either a positive or negative fashion (Browning and Busby 2004). These specialized gene regulators, such as the Trp repressor and the CAP activator, bind to highly conserved DNA operator sequences with high affinity through specific interactions between the protein and DNA. However, some global regulators bind to a wider range of less-conserved DNA sequences, and more complex regulatory systems have evolved to ensure binding specificity. One strategy is to recognize a longer region of DNA using multiple less-conserved recognition sites and reduced specificity within each individual site. The larger interaction surface between the protein and DNA compensates for the relatively weak interactions at a single interaction site. A large number of bacterial gene regulators adopt this strategy and use tetramers to recognize two DNA sites. Some of the best-studied examples include the Lac repressor (LacI or LacR), the λ repressor (λcI), members of the LysR family, and a few members of the TetR and IclR regulator families (Lewis et al. 1996; Schumacher et al. 2001; Molina-Henares et al. 2006; Stayrook et al. 2008; Monferrer et al. 2010). Although this mode of recognition is widespread, there is no structural information on a tetrameric protein bound to a continuous DNA operator containing two or more binding sites, which has hindered our understanding of cooperative binding by tetrameric gene regulators and the mechanism of their activation. However, tetrameric models of protein/DNA complexes have been proposed based on partial domain structures and incomplete DNA sequences. One such example is the tetrameric LacI/DNA complex model derived from the 4.8 Å resolution structure of a dimeric LacI/DNA complex and a tetrameric structure of the C-terminal domain (CTD). This model consists of two dimers of LacI, each bound to a separate 21-bp DNA duplex (Lewis et al. 1996). A model for the tetrameric λcI/DNA complex was extrapolated from a DNA complex structure of a λcI dimer and the tetramer structure of its CTD alone (Stayrook et al. 2008). The dimer DNA complex was obtained from a mutant form of λcI deficient in tetramer formation in complex with a 17-bp DNA duplex. These structures provided insights into how a tetrameric gene regulator can bind to two sites. However, in both cases, a single DNA site was used instead of two continuous DNA sites and, consequently, the incomplete structural information required some filling in of the gaps to create plausible models (Lewis et al. 1996; Stayrook et al. 2008).
Cooperativity is observed widely in biological systems that involve protein oligomers. The general model for cooperativity in ligand binding proposes that two distinct functional states (Tense [T-state] or Relaxed [R-state]) exist in equilibrium. These two states differ in their energies and affinities for the ligand. The T-state is more stable but incompetent in ligand binding. The R-state, on the other hand, is less stable but has higher affinity for the ligand. In the absence of a ligand, the protein predominately exists in the more stable T-state. However, upon ligand binding, the favorable binding energy between the protein and the ligand can offset the higher energy cost of the R-state, switching the protein to the R-state. When the energy difference between the R-state and the T-state is sufficiently large, multiple cooperative binding events are required to overcome the energy barrier. The larger the energy difference is between the T-state and the R-state, the higher the cooperativity is.
Pseudomonas putida DOT-T1E can grow in the presence of high concentrations of a wide variety of organic solvents (Ramos et al. 1995, 1998). The most important adaptation to permit this unusual property is the extrusion of the toxic compounds to the outer medium, an energy-dependent process that is mediated by a set of efflux pumps (Ramos et al. 2002). The pumps involved in solvent extrusion are TtgGHI and TtgDEF, which exhibit a wide range of substrate specificities and belong to the RND family of efflux transporters. The expression of ttgGHI and ttgDEF is regulated by the TtgV repressor (Teran et al. 2007), which belongs to the IclR family of regulators (Krell et al. 2006; Molina-Henares et al. 2006) and recognizes a large number of effector compounds that contain one or two aromatic rings (Guazzaroni et al. 2005). Effector binding releases TtgV from its operator DNA and results in an increased expression of ttgDEF and ttgGHI.
Analytical ultracentrifugation and DNA footprinting assays revealed that TtgV is a tetramer that protects a 42-bp-long DNA sequence covering the −10 to −35 region of the ttgG promoter (Rojas et al. 2003; Guazzaroni et al. 2004; Fillet et al. 2009). In order to understand the mechanism by which TtgV binds to its long DNA operator and the mechanism of induction by effectors, we determined the crystal structures of full-length TtgV alone and in a complex with its cognate 42-bp DNA operator. The structures reveal that TtgV binds to its DNA operator as a dimer of dimers with asymmetric dimer and tetramer interfaces. The binding of TtgV induces a significant distortion of the DNA that includes an overall 60° bend in the DNA. To date, models of tetrameric regulators assembled on complete operator sequences have been constructed from partial structures using shorter DNA sequences containing a single recognition site and/or truncated protein domains. Our full-length TtgV in complex with an intact DNA operator shows that binding to two continuous DNA sites imposes significant constraints on the tetramer, resulting in very different quaternary structures in the presence and absence of DNA. These structures allow us to propose a general model for cooperative binding of many tetrameric gene regulators.
Results
Crystal structure of the apo TtgV tetramer
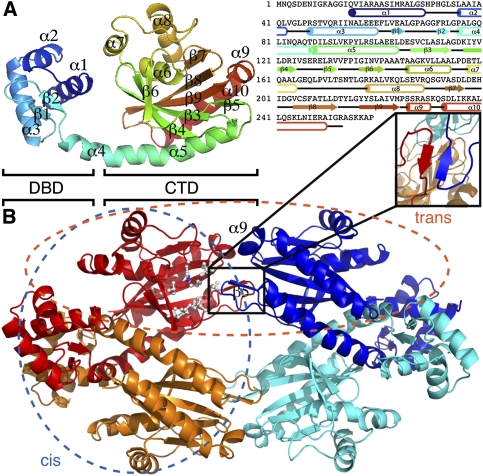
TtgV is a tetramer in solution (Guazzaroni et al. 2007b), and was crystallized in space group C2 with one dimer in an asymmetric unit. The dimer has twofold symmetry, and two dimers are related by a crystallographic twofold axis to form a compact symmetric tetramer (Fig. 1). The TtgV monomer consists of an N-terminal domain that belongs to a subgroup of the helix–turn–helix family that contains a Winged Helix (WH) motif responsible for DNA binding (hence termed DNA-binding domain [DBD]) (Gajiwala and Burley 2000). The protein also has a linker helix and a CTD that harbors the effector-binding site (Fig. 1A; Guazzaroni et al. 2005, 2007a). The WH motif contains three α helices (α1–α3) followed by two β strands (β1 and β2). The linker helix (α4) is continuous with the first helix in the CTD (α5) (Fig. 1A), forming a long, curved helix. The CTD consists of a twisted, six-stranded, anti-parallel β sheet sandwiched by two helices on one side and a three-helix bundle on the other. Based on structural and mutagenesis data, a hydrophobic pocket on the surface of the β sheet is proposed to be the effector-binding site (Fig. 1B, white ball and stick; Guazzaroni et al. 2005; Walker et al. 2006). Importantly, there are very few interactions between the CTD and the DBD within the same monomer (Fig. 1).
Figure 1.
The crystal structure of TtgV. (A) Ribbon representation of TtgV monomer colored from N termini (blue) to C termini (red). The secondary structural elements are mapped onto the amino acid sequence using the same color scheme as on the right. (B) TtgV tetramer arrangement and interactions. Each monomer is colored differently. Proposed hydrophobic residues that form the effector-binding pocket are displayed as balls and sticks. The insert shows the β5 interactions between trans-dimers. The blue ellipse indicates the cis-dimer, while the orange ellipse indicates the trans-dimer.
The CTDs between different monomers interact with one another and form a symmetric diamond shape (Fig. 1B). The CTD tetramer is in the same plane of the DBDs and is relatively flat (Fig. 1B). Within the tetramer, two distinct dimer interfaces exist (Fig. 1B). One dimer is stabilized mainly through interactions between the DBDs, which we term the cis-dimer (Fig. 1B, blue ellipse). The other dimer is formed between the β5 and α9, and we term this the trans-dimer (Fig. 1B, orange ellipse). The cis-dimer has an extensive ∼4400 Å2 buried interface, while the trans-dimer has a buried interface of ∼1100 Å2. Within the cis-dimer, the interactions between the CTDs are largely polar in nature and contribute only ∼460 Å2 to the buried interface. There are also significant polar interactions between the CTD and the DBD of the adjacent protomer within the cis-dimer. Each trans-dimer interacts by forming an anti-parallel β sheet, which is further stabilized by hydrophobic interactions between Leu 130 of one protomer (Fig. 1B, insert, red) and Val 132 from the adjacent protomer (Fig. 1B, insert, blue). The cis-dimer is more compact, with significant interactions between the DBDs, while the DBDs of a trans-dimer are located 100 Å apart. In this configuration, TtgV must undergo conformational changes in order to bind to the two sites within the operator.
Structure of the TtgV/DNA complex
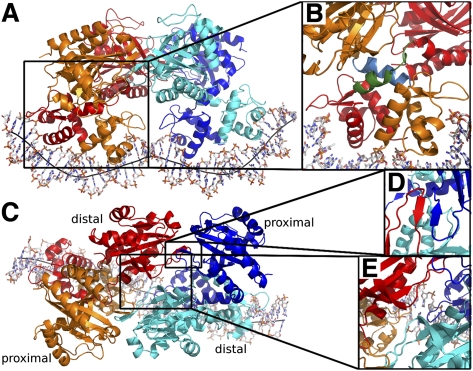
In order to understand how a TtgV binds to its operator sequence cooperatively, we determined the crystal structure of the full-length TtgV tetramer bound to its cognate 42-bp ttgG operator. The complex was crystallized in space group P65 with a complete tetramer/DNA complex in the asymmetric unit. TtgV binds to the DNA as a dimer of dimers, each bound to nonoverlapping sites on the same face of the DNA duplex (Fig. 2A). Two pairs of DBDs bind to a highly deformed DNA operator while the CTDs contact one another above, forming an asymmetric diamond shape (Fig. 2A). The tetramer contains two distinct layers of structures: one formed by the CTDs, and the other formed by the DBDs (Fig. 2A).
Figure 2.
The TtgV–DNA structure. (A) Viewed from the side. Each TtgV monomer is colored differently. The black line indicates the DNA axis. (B) Magnified view of one cis-dimer showing the different linker helix conformations. The linkers are shown in green (distal) and cyan (proximal). (C) Viewed from the top highlighting the skewed diamond shape formed by the CTDs. Proximal and distal protomers are indicated. (D) Magnified view of the trans-dimer interface. (E) Magnified view of the interaction between the diagonal distal protomers.
Within each cis-dimer (Fig. 2, red and orange pair, blue and cyan pair), the CTDs are asymmetric in relation to their DBDs. The asymmetry in cis-dimers is due to the differences in the linker helix between the DBD and the CTD (Fig. 2B), with one of the linker helices adopting a bent conformation (Fig. 2B, blue) and the other partially unwound (Fig. 2B, green). Within each cis-dimer, there are very few polar interactions between the CTDs (only burying ∼220 Å2 interface). The two cis-dimers interact through their CTDs to form a skewed diamond shape in the tetramer (Fig. 2C). The trans-dimer interface is the same in the structure of both the apo and the TtgV/DNA complex involving the anti-parallel β sheet (β5) and helix α9 (Figs. 1B, 2C,D). However, there are additional hydrogen-bonding interactions between the diagonal CTDs (Fig. 2E, red and cyan) near the CTD tetramer center, through residues 134–136, adjacent to β5.
Comparisons between TtgV apo and TtgV/DNA structures
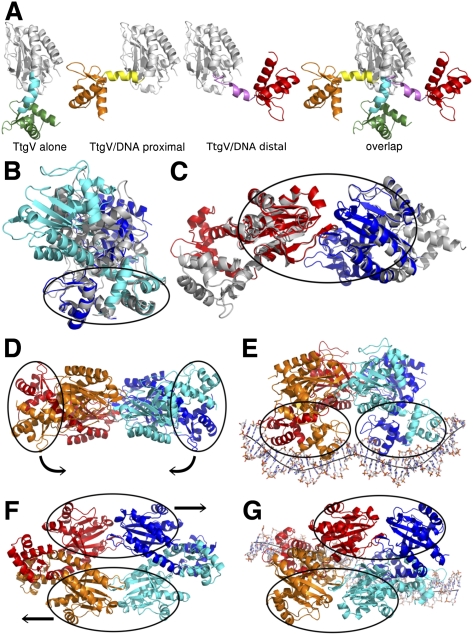
Comparison of the tetrameric structures of TtgV and TtgV/DNA reveals large conformational rearrangements at the monomeric, dimeric, and tetrameric levels. At the monomeric level, the DBD and CTD have the same structure but the protomers differ in the linker between the domains. The linker between the DBD and the CTD (α4) forms a continuous helix with α5 in the TtgV apo structure. However, in the TtgV/DNA complex, the protomers adopt two different conformations. In the protomers whose DBDs are located closer to the operator center (termed the proximal protomers), the linker helix is bent relative to α5 at residue Q86. In the protomers whose DBDs are outermost on the DNA operator (termed the distal protomers), the linker helix is partially unwound between L81 and A85 (Fig. 3A). The consequences of these changes in the linker conformation are the dramatically different orientations of the DBD in relation to the CTD. In the TtgV apo structure, the DBD is located below the CTD (Fig. 3A, left panel). In the TtgV/DNA proximal protomer, the DBD is positioned to the left of the CTD (Fig. 3A). In the TtgV/DNA distal protomer, the unwound α4 and, consequently, the DBD stretch over to the other side of the CTD compared with the proximal protomer (Fig. 3A, right).
Figure 3.
Conformational changes in TtgV. (A) Different monomeric conformations. (From left to right) Continuous linker helix (cyan), as in TtgV apo; bent linker helix (yellow), as in proximal protomer in the TtgV/DNA structure; unwound linker helix (magenta), as in distal protomer; and overlay of three conformations on their CTD (white ribbon) showing different orientations of their DBDs. (B) cis-dimer comparisons between TtgV apo (white) and TtgV/DNA (blue and cyan). Note the cis-DBDs (ellipse) have similar arrangements, while cis-CTDs differ. (C). Trans-dimer comparisons between TtgV apo (white) and TtgV/DNA (red and blue). The trans-CTDs (ellipse) have the same arrangements in both structures. (D) Side view of apo TtgV structure. Ellipses represent the cis-DBD units that rotate in opposite directions to E. (E) Side view of TtgV/DNA structure. (F) Top view of TtgV structure. Ellipses represent trans-CTD units that slide and rotate relative to each other to G. (G) Top view of TtgV/DNA structure.
The cis-dimer in the apo TtgV structure has a twofold symmetry and a larger buried interface (of ∼4400 Å2) than the cis-dimer in the TtgV/DNA structure, which is highly asymmetric and has a buried interface of ∼3400 Å2 (Figs. 1B, 2C). In both structures, the cis-DBDs have a similar arrangement (Fig. 3B, ellipse). In fact, the dimerization between the two cis-DBDs is conducted through hydrophobic residues I17, A21, M24, I53, L57, and L62 at helices α1 and α3. The interaction is further stabilized by the hydrophobic interactions between L78 and L81 at the N-terminal region of the linker helix α4. However, there are few interactions between cis-CTDs, and the relative orientations of the CTDs differ in the two structures. This suggests that the cis-DBDs move as a unit, while the cis-CTDs move independently of each other. Within the trans-dimer, the CTDs maintain a similar arrangement in both structures (Fig. 3C, ellipse; Supplemental Movie 1). We can therefore regard the trans-CTDs as a separate unit. This creates significant restrictions on the conformations within the tetramer, consisting of four rigid body units: two cis-DBD units and two trans-CTD units that are able to move relatively independently of one another (Fig. 3E,F, ellipses).
The transition from the symmetric apo TtgV structure to the asymmetric TtgV/DNA structure involves an ∼40 Å sliding and ∼60° rotation of the two trans-CTD units (Fig. 3F,G; Supplemental Movie 2). The two cis-DBD units, which are 180° and 100 Å apart from each other in the apo TtgV structure, rotate downward and adopt a configuration capable of binding to both sites in the DNA operator (Fig. 3D,E). Furthermore, although the cis-DBDs are similar in both structures, there are some small but significant differences (Fig. 3B). In the TtgV apo structure, the distance between the α2 helices and the wings in the WH motifs within each cis-DBD unit is 2–3 Å wider than that of TtgV/DNA structure, implying that TtgV undergoes local conformational changes upon binding to the DNA.
The CTD tetramer in the DNA complex structure has a larger interaction surface (∼2700 Å2) compared with that of TtgV alone (∼2200 Å2) due to the additional interactions between the diagonal protomers, implying that, within isolated CTDs, the asymmetric CTD tetramer is more stable than the symmetric CTD tetramer. However, although the symmetric CTD in the apo TtgV structure has a smaller interface than the asymmetric CTD in the TtgV/DNA structure, there is a larger interface between the DBDs, α4, and the CTDs within a cis-dimer because the DBDs are now constrained in the apo TtgV. Consequently, the full-length TtgV apo tetramer assembly has a total buried surface of ∼11,000 Å2 compared with ∼9500 Å2 in the TtgV/DNA structure. The difference in the interaction surfaces between the symmetric and asymmetric structures is almost entirely due to the loss of polar interactions between the cis-CTDs and between the CTD and the DBD within the cis-dimer. This supports the argument that the symmetric apo structure not only has a more ordered linker helix α4 and a larger interaction surface between protomers, but is indeed at a lower-energy and more stable state compared with the asymmetric structure.
DNA interactions and distortion
The interactions between TtgV and DNA induce both conformational changes in the protein tetramer and significant distortions in the DNA. The extensive interactions between TtgV and DNA induce significant distortions in the DNA operator, with widened major grooves where the recognition helices are inserted (Fig. 4; Supplemental Figs. 2, 4). Overall, the central axis for the DNA double helix is W-shaped, with inward bends at the narrowed minor grooves (∼40°) that are A/T-rich and a convex kink at the widened minor groove with a G/C pair in the middle of the operator, reducing the concave bend over the entire operator to 60° (Figs. 2A, 4B; Supplemental Movie 3). This is consistent with observations in nucleosomes (Drew and Travers 1985), where, in A/T-rich sequences, the minor grooves face inward toward the center of the curvature, while, in G/C-rich sequences, the minor grooves of G/C pairs face outward. The widening of major and minor grooves is also observed when WH proteins such as Orc bind to DNA (Gaudier et al. 2007).
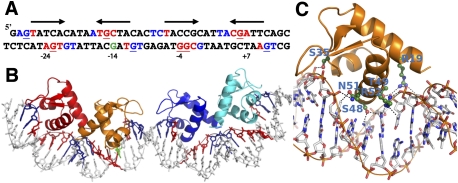
Figure 4.
TtgV–DNA interactions. (A) The 42-bp ttgG operator used in the study. The pseudosymmetry of each binding site is indicated by arrows. Red bases indicate the DNA bases that interact with TtgV in the major groove, while blue indicates the interacting bases in the minor grooves. Underlined bases are the sites protected by DMS methylation. The green G is the hypersensitive site. (B) The corresponding structure of the DNA and WH domains, with the DNA bases colored according to A. (C) Detailed interactions of one WH domain with the DNA. Residues that are involved in direct interactions are labeled.
The structure allows us to define pseudopalindromic sites within the operator (Fig. 4A; Guazzaroni et al. 2007b). The two sites span the entire DNA operator, with a 5-bp spacer in the middle. The recognition sites in the DNA major grooves are centered at positions −24, −14, −4, and +7 in the ttgGHI operator (Fig. 4A, red bases) relative to the transcription starting site of ttgG. Each major groove site spans a 5-bp region (Fig. 4A, arrows). There are further contacts with one or two flanking phosphate groups on the 5′ end of both DNA strands (Fig. 4A, blue bases). The upstream recognition site has higher palindromic symmetry (defined by the arrows) compared with the downstream recognition site. Previous biochemical studies have found that guanines at positions −27, −15, and +6 on the top strand and −23, −11, −4, −3, and +10 on the bottom strand are protected from DMS methylation upon TtgV binding (Guazzaroni et al. 2004, 2007a; Fillet et al. 2009). Strikingly, five out of the eight bases make direct hydrogen bonds with TtgV in our TtgV/DNA structure (Fig. 4A, underlined red bases), and TtgV interacts with the phosphate backbone at the other three positions (Fig. 4A, underlined blue bases). The guanine at position −14 is hypersensitive to methylation (Fig. 4A, green base; Guazzaroni et al. 2004). This site is paired with a recognized base in our structure, and is exposed by DNA distortion when TtgV binds.
TtgV interacts with the DNA major groove largely through the recognition helix (α3) of the WH motif, similar to the majority of other WH proteins (Clark et al. 1993; Gajiwala and Burley 2000). Specifically, residues S48, T49, Q51, and R52 interact with DNA via the major groove (Fig. 4C). The wings in the WH domains lie across the minor grooves and interact with the phosphate backbones at both ends and the middle of the DNA operator sequence (Fig. 4B). R19 from α1 also interacts with the phosphate backbone (Fig. 4C).
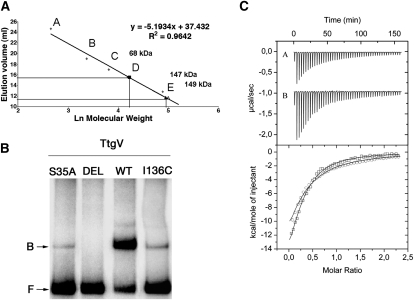
Fillet et al. (2009) identified three groups of residues within the WH domain of TtgV, based on their effects on its activity. Substitution of Group 1 residues with alanine had no effect on activity, while substitution of Group 2 residues abolished activity. Alanine substitution of Group 3 residues significantly reduced activity for the ttgG promoter, but had little effect on the ttgD promoter. The TtgV structure in complex with the ttgG promoter reveals that Group 1 residues are located largely on protein surfaces and have few interactions with the rest of the protein or DNA (Supplemental Fig. 4, white). Group 2 residues (R47, T49, and R52) are involved directly in DNA binding, and hence mutations to alanine resulted in mutant regulators that exhibit reduced affinity for the operator and a high level of expression from the target promoters. Group 3 residues are located largely at the dimer interface or interact with other parts of the protein (Supplemental Fig. 4). Mutations of these residues presumably affect dimerization or the conformation of the WH domain. Both could affect the orientation of one or both recognition helices, hence affecting DNA binding. Our structure also identifies an additional Group 2 residue (S35), which is involved in binding to the phosphate backbone. Mutating S35 to alanine reduced DNA-binding ability significantly, without affecting its quaternary structure (Fig. 5; Supplemental Table 1).
Figure 5.
Mutagenesis and in vitro studies of DNA and effector binding. (A) Oligomeric state of TtgV and its variants by gel filtration. The calibration curve was prepared using lactalbumin (14 kDa) (A), carbonic anhydrase (29 kDa) (B), chicken egg albumin (45 kDa) (C), bovine serum albumin (66 kDa) (D), and bovine serum albumin (132 kDa) (E). The molecular mass of TtgV (open triangle), TtgVS35A (closed triangle), and TtgVΔ129-131 (closed square) are indicated. (B) Electrophoretic mobility shift assay of TtgV with the ttgV/ttgG operator region. The 295-bp DNA fragment was incubated in the presence of 50 nM TtgV or the indicated TtgV mutant. (DEL) TtgVΔ129–131; (F) free DNA; (B) retarded band. (C) Isothermal titration calorimetry for the binding of 1-naphthol to TtgV (A in the top panel; circle in the bottom panel) and of TtgVΔ129–131 (B in the top panel; square in the bottom panel).
Our structures suggest that tetramer formation is crucial for the cooperative binding to the operator, and β5 is a key component in the tetramer interface (Figs. 1B, 2D). In order to test the importance of this interface, we deleted β5 (residues 129–131). The mutant protein (TtgVΔ129–131) has an affinity for effectors similar to that of wild type, confirming that the mutations do not affect the overall protein structure and folding (Fig. 5C). However, the deletion mutant indeed forms dimers rather than tetramers (Fig. 5A), and consequently has lost cooperativity and is no longer able to bind to DNA (Fig. 5B).
Discussion
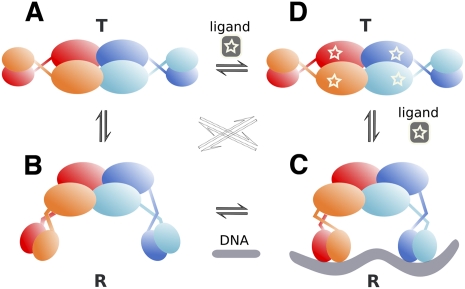
Our structures of apo TtgV at 2.9 Å and TtgV/DNA at 3.4 Å explain how a TtgV tetramer binds cooperatively to two DNA sites within an operator. A TtgV tetramer consists of a pair of cis-dimers: Each cis-dimer unit can bind to one DNA site (ligand). There are two distinct functional states for a TtgV tetramer: a stable symmetric state, represented by the symmetric TtgV apo structure, where both cis-dimers exist in the T-state, which is more stable but is unable to bind to its DNA operator; and a less stable asymmetric form, represented by the asymmetric configuration and released DBDs, where both cis-dimers exist in the R-state, which is less stable but has the correct conformation to permit simultaneous binding to two DNA sites. For simplicity, we define the symmetric apo tetramer, which consists of two T-state cis-dimers, the stable T-state for the tetramer, and the asymmetric form, which consists of two R-state cis-dimers, the less stable R-state for the tetramer (Fig. 6).
Figure 6.
A proposed cooperative binding and induction mechanism for TtgV. TtgV exists in equilibrium of a straight linker helix (symmetric T-state) (A) or bent/flexible linker helix (asymmetric R-state) (B). The T-state is more stable, while the R-state favors DNA binding. (C) Upon DNA binding, the favorable interaction energy between the protein and the DNA stabilizes the unstable R-state. (D) Upon effector binding, the CTDs slide back to the symmetric configuration and the DBDs rotate in opposite directions, releasing from DNA and returning to the stable symmetric configuration (T-state).
Without any constraint, the CTD tetramer is more stable in the asymmetric R-state. However, there are increased interactions between the CTDs and the DBDs in the apo symmetric state. The net energy balance favors the symmetric apo configuration for the TtgV tetramer. In this stable T-state, the DBDs are confined in a configuration that is not competent to bind DNA (Fig. 6A). When DBDs are removed, the CTDs can relax to a more stable asymmetric configuration (Fig. 6B). Indeed, this is supported by structures of other IclR family proteins that are truncated to contain just the CTD (Zhang et al. 2002; Lorca et al. 2007) and display asymmetric arrangements.
In solution, apo TtgV is likely to exist in equilibrium between the T-state and R-state (Fig. 6, left). In order to bind DNA, the symmetric tetramer (T-state) has to release the two pairs of DBDs from the CTDs (Fig. 6A,B). This has two conflicting effects: It costs favorable interaction energy between the CTDs and the DBDs, and it gains energy through the favorable asymmetric CTD arrangement. The energy values are poised so that binding to a single DNA site does not provide sufficient energy to compensate for the loss of two pairs of CTD/DBD interactions, but binding to two sites does. This explains the extremely weak binding affinity of TtgV to a single site, as mutating one of the two operator sites reduces the ability of TtgV to bind to its operator significantly (Guazzaroni et al. 2007b), while the TtgV dimer (as in the form of TtgVΔ129–131) is unable to bind to DNA. However, binding to two sites provides sufficient favorable interactions and allows the tight binding of the TtgV tetramer to its operator.
Unlike DNA binding, the effectors bind to each TtgV monomer independently (Guazzaroni et al. 2005) and the effector binding stabilizes the T-state (Fig. 6D), although it is possible that, upon effector binding to TtgV, further conformational changes occur that stabilize an even lower-energy T-state.
There are two types of effector-binding sites in the TtgV/DNA complex: One is located near the CTD center, and the other is adjacent to the linker helices (Supplemental Fig. 3). Effector binding to the sites near the CTD tetramer center will weaken the favorable interactions in the asymmetric CTD tetramer involving residues 134–136. Mutating F134, which is also in the effector-binding site, reduced the ability of the protein to bind to DNA (Guazzaroni et al. 2007a). Mutating I136 also significantly reduced the ability in DNA binding, although it maintained a similar affinity in effector binding compared with wild type (Fig. 5; Supplemental Table 1). Presumably, both mutations affect the asymmetric tetramer arrangement, which is key for DNA binding. Since residue F134 is involved in both effector binding and the asymmetric tetramer interface in the R-state, it is possible that effector binding induces conformational changes in F134, which destabilizes the asymmetric R-state. The energy cost in the disruption of the favorable asymmetric CTDs will need to be compensated for by the increased interactions between the CTD and the DBD, therefore promoting the return to the stable symmetric T-state. Effector binding to the other sites that are located close to the linker helices would disturb the interactions of the linker helices, helping the return to the continuous helix conformation that is observed in the T-state.
Cooperativity in ligand binding can be described by either a concerted model or a sequential model (Fersht 1999). The concerted model has been generally applied to oligomeric proteins that have identical ligand-binding sites. It argues that the protein exists in equilibrium of either of two states (T-state or R-state), and all subunits within the oligomer exist in the same states, all in the T-state or R-state. The sequential model, on the other hand, assumes that, in the absence of a ligand, the protein exists in the T-state and ligand binding induces changes in one subunit (induced fit from T-state to R-state), which could then influence ligand binding in other subunits. In most cases, the system is somewhere in between. We proposed a model for cooperative DNA binding of tetrameric proteins that more closely resembles the concerted model, with each cis-dimer acting as a single unit. This is because the structural evidence so far suggests that TtgV and other tetrameric gene regulators have two identical cis-dimer units within a tetramer and they have identical DNA-binding properties. The apo TtgV structure contains two identical cis-dimers that we termed the T-state due to its lower energy status and inability to bind to the DNA (ligand). Likewise, the TtgV structure in the DNA complex has both cis-dimers in the R-state, which is characterized by its higher-energy state and capability to bind to the DNA. However, it is important to note that, in the absence of DNA and effectors, the protein exists in an equilibrium of the T-state and R-state, so that the T-state, represented by the apo TtgV structure here, does not necessarily define the unique conformation that apo TtgV adapts in solution. This conformation, or a conformation with an even lower-energy state, is stabilized by effector binding in solution, while the R-state is stabilized by DNA binding. In the apo TtgV crystal presented here, this conformation is stabilized by the interactions between DBDs within the crystal, allowing us to capture this T-state.
Many tetrameric gene regulators contain tetrameric effector-binding domains and a linker that connects the effector-binding domains with the DBDs. We propose that these tetrameric gene regulators use a similar cooperative binding model irrespective of their exact domain structures. The T-state is represented by a more stable full-length tetramer configuration, as observed in the TtgV apo structure. In this state, both pairs of DBDs are confined and unable to bind to the DNA. In the R-state, although overall the full-length tetramer is less stable, the tetramer arrangement of the effector-binding domains is more stable, and the DBDs are released to permit binding to the DNA operator. DNA binding causes distortions of DNA, which are compensated for by the favorable interaction energy between the protein and two DNA sites. The degree of the cooperativity depends on the energy cost from the T-to-R-state transition and the degree of distortions induced in the DNA. The larger the energy cost is to switch from the T-state to the R-state, and/or the larger the distortion is in the DNA, the stronger the requirement for a cooperative binding between the protein tetramer and DNA operator is, such that favorable interactions between the protein and two DNA sites can compensate for the large energy cost. This is indeed the case for TtgV, where both DNA sites and a full tetramer are required for efficient binding. However, in some systems, where a weaker cooperativity exists, a dimeric form of the protein can bind to a single site, albeit with a weaker affinity. These protein/DNA configurations that contain just one DNA site, however, do not represent the final R-state in the tetramer, but a decoupled intermediate state.
Recent structural studies using a mutant form of λ repressor deficient in tetramer formation revealed an asymmetric arrangement of the cis-dimer in complex with its 17-bp DNA target (Stayrook et al. 2008), and asymmetry was proposed to play important roles in forming the tetramer assembly (Hochschild and Lewis 2009). Our results agree with that hypothesis, and suggest that the asymmetry in the cis-dimer is a prerequisite for tetrameric gene regulators upon binding to their DNA operator containing two adjacent sites. A tetrameric arrangement of λ repressor, when bound to two adjacent DNA sites, was proposed based on this asymmetric dimer/DNA complex and the tetrameric arrangement of CTDs (Stayrook et al. 2008). The model provided mechanistic insights into cooperative binding of λ repressor to adjacent operator sites based on the available data. However, the model used two separate, discontinuous 17-bp DNA duplexes. Consequently, in that model, although the two DNA duplexes roughly align into a continuous helix, the individual strands do not join smoothly. Furthermore, the angular alignment of the two cis-DBD units does not match that of the natural operators (Stayrook et al. 2008). Additional distortion in DNA will be required for a full assembly of protein/DNA. The energy cost in DNA distortions will have to be compensated for by additional interactions between the protein and DNA, and between protein subunits.
In summary, previous structures of tetrameric gene repressors have shed light on how they might bind to DNA cooperatively. However, due to incomplete structural information, models of the repressor/operator complex do not necessarily represent the actual stable protein/operator assembly, or the final R-state. Our structures here provide details of a complete assembly of a tetrameric repressor with a full operator sequence and reveal a general cooperative model that can be applied to explain many other tetrameric regulators. We propose that the energy difference between the T-state and the R-state and the energy cost in inducing DNA distortions are key determinants for its cooperativity. To achieve the full assembly of regulator/DNA in a cooperative fashion, conformational changes must occur to allow simultaneous binding to both sites and compensate for the energy difference between the two states.
Materials and methods
Site-directed mutagenesis, overexpression, and purification of TtgV and mutants
TtgV mutants were generated by amplification of ttgV from plasmid pANA126 using pfu turbo DNA polymerase (Stratagene) and 39 mer primers that incorporated the appropriate mismaches to introduce the desired mutation. The PCR product was digested with DpnI, ligated to pET28b(+), and transformed in Escherichia coli BL21 (DE3). The nature of each mutant allele was confirmed by DNA sequencing. TtgV and mutants were expressed and purified as described previously (Guazzaroni et al. 2005). Briefly, BL21 (DE3) cells were grown in 2-L conical flasks containing 1 L of 2× YT culture medium with 50 μg/mL kanamycin, incubated at 37°C with shaking, and induced with 1 mM IPTG when the culture reached a turbidity of ∼0.7. The cultures were grown for 3 h at 18°C and then harvested by centrifugation. Cells were lysed by French press or sonication. After centrifugation, the supernatant was filtered and loaded onto a 5-mL HisTrap HP column (GE Healthcare) and eluted with a imidazol gradient (45–500 mM). Peak fractions were then loaded onto a Superdex-200 10/300GL column (Amersham Biosciences) equilibrated in buffer A (25 mM sodium phosphate at pH 7.0, 0.5 M NaCl, 5% [v/v] glycerol). The protein sample was eluted at a constant flow rate of 0.7 mL/min. Peaks containing TtgV were collected and concentrated. The molecular weight of the protein was estimated from a plot of the elution volume against the logarithm (Ln) of the molecular weight of standard calibration proteins; namely, α-lactalbumin from bovine milk (14.2 kDa), carbonic anhydrase from bovine erythrocytes (29 kDa), albumin from chicken egg white (45 kDa), and albumin from bovine serum (66 kDa) (Sigma).
Electrophoresis mobility shift assay
The DNA probes were 295-bp fragments containing the ttgV–ttgGHI intergenic region obtained from plasmid pGG1 by PCR with primers G5′E (5′-NNNNNNGAATTCGTTCATATCTTTCCTCTGCG-3′) and G3′P (5′-NNNNNNCTGCAGGGGGATTACCCGTAATGCAC-3′). Cycling parameters were 2 min at 95°C followed by 30 cycles of 1 min at 95°C, 30 sec at 50°C, and 30 sec at 72°C, ending with 10 min at 72°C. PCR products were isolated from agarose gel by use of a QiaQuick gel extraction kit (Qiagen) and radiolabeled at the 5′ end with [γ-32P] ATP and T4 polynucleotide kinase. A 1 nM concentration (∼104 counts per minute) of the labeled probe was then incubated with the indicated concentrations of purified proteins and analyzed using nondenaturing 4.5% (w/v) polyacrylamide gels as described (Guazzaroni et al. 2007b).
Isothermal titration calorimetry
Measurements were performed on a VP-Microcalorimeter (MicroCal) at 25°C. Proteins were dialyzed thoroughly against 25 mM Tris acetate (pH 8.0), 8 mM magnesium acetate, 100 mM NaCl, 10% (v/v) glycerol, and 1 mM dithiothreitol. The protein concentration was determined using the Bradford assay. Stock solutions of 1-naphthol at a concentration of 500 mM were prepared in dimethylsulfoxide and subsequently diluted with dialysis buffer to a final concentration of 0.5 mM (1-naphthol and 4-nitrotoluene). Each titration involved a single 1.6-μL injection and a series of 4.8-μL injections of effector into the protein solution. Titration curves were fitted by a nonlinear least-squares method to a function for the binding of a ligand to a macromolecule as incorporated in Origin software (MicroCal).
Crystallization
DNA oligos were purchased from MWG and dissolved to a final concentration of 0.2 mM in a buffer containing 50 mM HEPES (pH 8.0) and 40 mM NaCl. To anneal into dsDNA, complementary ssDNA fragments were mixed in equal molar ratio, heated for 10 min at 95°C, and cooled slowly to room temperature. Annealed 0.1 mM dsDNA was mixed with 50 mg/mL purified protein at a molar ratio of 1:4. The protein used in crystallization experiments contains C109S and C205S mutations in order to reduce possible protein aggregation. The mutant proteins are fully functional, as assayed by effector and DNA binding. The mixture was left for 2 h at 4°C to allow the binding between the protein and DNA. The complex was then loaded onto a Superdex-200 column, which was washed with the same buffer that the DNA samples were dissolved in. The fractions of a single peak were collected containing both TtgV and DNA, and the sample was concentrated to a final concentration of 5 mg/mL. Crystals were obtained by sitting drop vapor diffusion. TtgV alone crystals were achieved accidentally from an attempt to crystallize TtgV in complex with a 43-bp DNA fragment covering ttgGHI operator region −30 to +13. The crystallization buffer contained 15% PEG2000MME, 100 mM BisTris propane (pH 6.4), and 200 mM KNO3. The crystals appeared in 3 d at 4°C. The crystals of TtgV/42bp DNA (−29 to +13) with a 3′ extruding base at the bottom strand were grown under the condition of 7% PEG4000, 50 mM NaCl, and 100 mM Tris (pH 8.0). Crystals grew for 1 mo at 4°C.
Data collection and structure determination
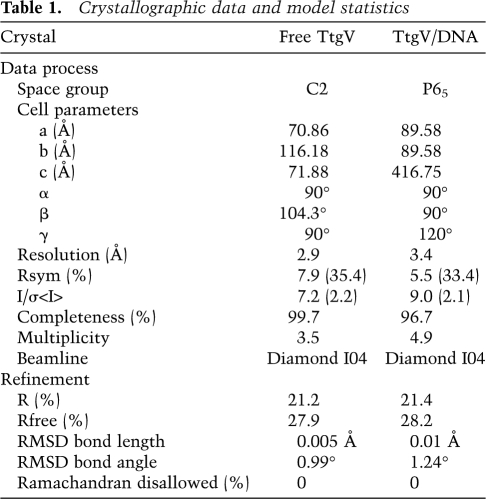
Crystals of TtgV alone and in complex with its DNA operator were transferred into cryo-buffers containing 10% or 20% ethylene glycol in addition to their crystallization buffers, respectively, before placing in liquid nitrogen. Data sets were collected at beamline I04, Diamond Synchrotron Radiation Source. Diffraction data were processed in Mosflm (Leslie 1992), scaled, and truncated in Scala in the CCP4 suite (Collaborative Computational Project, Number 4 1994). Both TtgV alone and TtgV/DNA complex structures were determined by molecular replacement method. The initial searching model for the TtgV alone structure was made in Chainsaw (Stein 2008) based on the CTD of an IclR family member protein structure (Protein Data Bank code 1MKM) (Zhang et al. 2002), and the molecular replacement solution was obtained in Phaser (McCoy et al. 2007). The remainder of the structure was built manually in Coot (Emsley et al. 2010). The refined TtgV structure was then used as the initial searching model for the complex. A solution of the CTD was obtained in Phaser. Subsequently, solutions of DBDs and small B-DNA fragments were added to the existing CTDs by using Molrep (Vagin and Teplyakov 1997). The remainder of the structure was built manually in Coot. All refinements were carried out in Phenix (Adams et al. 2010). The final models for both structures consist of residues 15–253. For apo TtgV structure, all but four residues from each protomer have their side chains in the final model. For the DNA complex structure, a total of 16 residues within the tetramer have their side chains missing.
Structural analysis
The DNA structure was analyzed using the program Curves+ (http://www.ibpc.fr/UPR9080/Curonline.html). The buried surface was calculated in PISA and the protein structure was analyzed using PROCHECK, both incorporated in CCP4 (Collaborative Computational Project, Number 4 1994). The coordinates have an estimated error of 0.5 Å for the complex structure and 0.4 Å for the apo structure. All structural alignment and measurements, as well as structural figures, were carried out in PyMol (http://www.pymol.org). Supplemental Figure 5a was created in Adobe Illustrator. Sequence alignment was carried out using MAFFT (Katoh et al. 2002).
Table 1.
Crystallographic data and model statistics
Acknowledgments
We are grateful for many stimulating discussions with Dale Wigley, Paul Freemont, and colleagues in Paul Freemont's and X.Z.'s groups. We thank Dale Wigley, Maruf Ali, and Paul Freemont for critically reading the manuscript. This project was funded by the UK Medical Research Council to X.Z. (76791), with early funding from the Human Frontier Science Program to X.Z. and M.T.G. Work in Granada was funded by EDFR from the Ministry of Science and Education (BIO2006-05668 and BIO2010), and an EDFR grant (CV344) from Junta de Andalucia.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.603510.
Supplemental material is available at http://www.genesdev.org.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Busby SJ 2004. The regulation of bacterial transcription initiation. Nat Rev Microbiol 2: 57–65 [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364: 412–420 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Drew HR, Travers AA 1985. DNA bending and its relation to nucleosome positioning. J Mol Biol 186: 773–790 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A 1999. Structure and mechanism in protein science. W.H. Freeman and Company, New York [Google Scholar]
- Fillet S, Velez M, Lu D, Zhang X, Gallegos MT, Ramos JL 2009. TtgV represses two different promoters by recognizing different sequences. J Bacteriol 191: 1901–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajiwala KS, Burley SK 2000. Winged helix proteins. Curr Opin Struct Biol 10: 110–116 [DOI] [PubMed] [Google Scholar]
- Gaudier M, Schuwirth BS, Westcott SL, Wigley DB 2007. Structural basis of DNA replication origin recognition by an ORC protein. Science 317: 1213–1216 [DOI] [PubMed] [Google Scholar]
- Guazzaroni ME, Teran W, Zhang X, Gallegos MT, Ramos JL 2004. TtgV bound to a complex operator site represses transcription of the promoter for the multidrug and solvent extrusion TtgGHI pump. J Bacteriol 186: 2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guazzaroni ME, Krell T, Felipe A, Ruiz R, Meng C, Zhang X, Gallegos MT, Ramos JL 2005. The multidrug efflux regulator TtgV recognizes a wide range of structurally different effectors in solution and complexed with target DNA: Evidence from isothermal titration calorimetry. J Biol Chem 280: 20887–20893 [DOI] [PubMed] [Google Scholar]
- Guazzaroni ME, Gallegos MT, Ramos JL, Krell T 2007a. Different modes of binding of mono- and biaromatic effectors to the transcriptional regulator TTGV: Role in differential derepression from its cognate operator. J Biol Chem 282: 16308–16316 [DOI] [PubMed] [Google Scholar]
- Guazzaroni ME, Krell T, Gutierrez del Arroyo P, Velez M, Jimenez M, Rivas G, Ramos JL 2007b. The transcriptional repressor TtgV recognizes a complex operator as a tetramer and induces convex DNA bending. J Mol Biol 369: 927–939 [DOI] [PubMed] [Google Scholar]
- Hochschild A, Lewis M 2009. The bacteriophage λ CI protein finds an asymmetric solution. Curr Opin Struct Biol 19: 79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T 2002. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell T, Molina-Henares AJ, Ramos JL 2006. The IclR family of transcriptional activators and repressors can be defined by a single profile. Protein Sci 15: 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW 1992. Recent changes to the MOSFLM package for processing film and image plate data. In Joint CCP4 and ESF-EAMCB newsletter on protein crystallography, No. 26, pp. 27–33 [Google Scholar]
- Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P 1996. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science 271: 1247–1254 [DOI] [PubMed] [Google Scholar]
- Lorca GL, Ezersky A, Lunin VV, Walker JR, Altamentova S, Evdokimova E, Vedadi M, Bochkarev A, Savchenko A 2007. Glyoxylate and pyruvate are antagonistic effectors of the Escherichia coli IclR transcriptional regulator. J Biol Chem 282: 16476–16491 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Henares AJ, Krell T, Eugenia Guazzaroni M, Segura A, Ramos JL 2006. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol Rev 30: 157–186 [DOI] [PubMed] [Google Scholar]
- Monferrer D, Tralau T, Kertesz MA, Dix I, Sola M, Uson I 2010. Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Mol Microbiol 75: 1199–1214 [DOI] [PubMed] [Google Scholar]
- Ramos JL, Duque E, Huertas MJ, Haidour A 1995. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J Bacteriol 177: 3911–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Duque E, Godoy P, Segura A 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J Bacteriol 180: 3323–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol 56: 743–768 [DOI] [PubMed] [Google Scholar]
- Rojas A, Segura A, Guazzaroni ME, Teran W, Hurtado A, Gallegos MT, Ramos JL 2003. In vivo and in vitro evidence that TtgV is the specific regulator of the TtgGHI multidrug and solvent efflux pump of Pseudomonas putida. J Bacteriol 185: 4755–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294: 2158–2163 [DOI] [PubMed] [Google Scholar]
- Stayrook S, Jaru-Ampornpan P, Ni J, Hochschild A, Lewis M 2008. Crystal structure of the λ repressor and a model for pairwise cooperative operator binding. Nature 452: 1022–1025 [DOI] [PubMed] [Google Scholar]
- Stein N 2008. CHAINSAW: A program for mutating pdb files used as templates in molecular replacement. J Appl Crystallogr 41: 641–643 [Google Scholar]
- Teran W, Felipe A, Fillet S, Guazzaroni ME, Krell T, Ruiz R, Ramos JL, Gallegos MT 2007. Complexity in efflux pump control: Cross-regulation by the paralogues TtgV and TtgT. Mol Microbiol 66: 1416–1428 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A 1997. MOLREP: An automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- Walker JR, Altamentova S, Ezersky A, Lorca G, Skarina T, Kudritska M, Ball LJ, Bochkarev A, Savchenko A 2006. Structural and biochemical study of effector molecule recognition by the E. coli glyoxylate and allantoin utilization regulatory protein AllR. J Mol Biol 358: 810–828 [DOI] [PubMed] [Google Scholar]
- Zhang RG, Kim Y, Skarina T, Beasley S, Laskowski R, Arrowsmith C, Edwards A, Joachimiak A, Savchenko A 2002. Crystal structure of Thermotoga maritima 0065, a member of the IclR transcriptional factor family. J Biol Chem 277: 19183–19190 [DOI] [PMC free article] [PubMed] [Google Scholar]