Abstract
Cosuppression is a silencing phenomenon triggered by the introduction of homologous DNA sequences into the genomes of organisms as diverse as plants, fungi, flies, and nematodes. Here we report sex-induced silencing (SIS), which is triggered by tandem integration of a transgene array in the human fungal pathogen Cryptococcus neoformans. A SXI2a-URA5 transgene array was found to be post-transcriptionally silenced during sexual reproduction. More than half of the progeny that inherited the SXI2a-URA5 transgene became uracil-auxotrophic due to silencing of the URA5 gene. In vegetative mitotic growth, silencing of this transgene array occurred at an ∼250-fold lower frequency, indicating that silencing is induced during the sexual cycle. Central components of the RNAi pathway—including genes encoding Argonaute, Dicer, and an RNA-dependent RNA polymerase—are all required for both meiotic and mitotic transgene silencing. URA5-derived ∼22-nucleotide (nt) small RNAs accumulated in the silenced isolates, suggesting that SIS is mediated by RNAi via sequence-specific small RNAs. Through deep sequencing of the small RNA population in C. neoformans, we also identified abundant small RNAs mapping to repetitive transposable elements, and these small RNAs were absent in rdp1 mutant strains. Furthermore, a group of retrotransposons was highly expressed during mating of rdp1 mutant strains, and an increased transposition/mutation rate was detected in their progeny, indicating that the RNAi pathway squelches transposon activity during the sexual cycle. Interestingly, Ago1, Dcr1, Dcr2, and Rdp1 are translationally induced in mating cells, and Ago1, Dcr1, and Dcr2 localize to processing bodies (P bodies), whereas Rdp1 appears to be nuclear, providing mechanistic insights into the elevated silencing efficiency during sexual reproduction. We hypothesize that the SIS RNAi pathway operates to defend the genome during sexual development.
Keywords: RNAi, meiosis, siRNA, transposon
RNAi is an evolutionarily conserved mechanism in which gene silencing is orchestrated by small RNAs (∼20 to ∼30 nucleotides [nt]) operating in a sequence-specific manner (Hannon 2002). This process was originally discovered following introduction of dsRNA into Caenorhabditis elegans and mediates both post-transcriptional gene silencing (PTGS) and transcriptional gene silencing (TGS) in myriad organisms, including plants, animals, fungi, and ciliates (Hannon 2002; Moazed 2009). RNAi serves critical roles in gene regulation, chromosomal structure, and genome stability and defense (Ghildiyal and Zamore 2009; Siomi and Siomi 2009).
Mobile genetic elements populate the genomes of virtually all eukaryotic organisms and challenge the genetic integrity of their hosts, necessitating mechanisms that limit their expansion. Early studies have pointed to links between RNAi and transposon control. For example, in C. elegans, several genes essential to RNAi are also involved in a transposon control pathway (Ketting et al. 1999). Additionally, a surprisingly large family of small RNAs mapping to specific transposable elements have been defined in Arabidopsis, Drosophila, C. elegans, and, recently, the budding yeast Saccharomyces castellii (Llave et al. 2002; Aravin et al. 2003; Drinnenberg et al. 2009). Among these, the piwi-interacting small RNAs (piRNAs) have been shown to bind the piwi proteins of the Argonaute superfamily and are required for silencing transposons in the animal germline (Brennecke et al. 2007). Thus, one conserved function of the RNAi pathway is to defend the genome from invasion by mobile elements.
Both PTGS and TGS mechanisms have been implicated in transposon control. piRNA has the capacity to silence transposons solely at a post-transcriptional level (Brennecke et al. 2007). Other species of small RNAs could similarly control transposons at the transcriptional level. For example, DNA methylation and histone modifications are abundant at transposon-related sequences in plants and animals, and these modifications are dependent on siRNAs (Wassenegger et al. 1994; Lippman et al. 2003; Zilberman et al. 2003; Pal-Bhadra et al. 2004). Remarkably, heterochromatin formation in the fission yeast Schizosaccharomyces pombe is guided by sequence-specific siRNAs present in an Argonaute complex with similarities to the RNA-induced silencing complex (RISC) in RNAi, suggesting that PTGS and TGS are coupled during this process (Volpe et al. 2002; Zofall and Grewal 2006; Moazed 2009).
The fungus Neurospora crassa has three distinct silencing pathways that operate at different stages of the life cycle: quelling, repeat-induced point mutation (RIP), and meiotic silencing of unpaired DNA (MSUD) (Selker 1997; Cogoni and Macino 1999b; Borkovich et al. 2004; Galagan and Selker 2004; Kelly and Aramayo 2007). Among these, quelling and MSUD share a similar molecular mechanism. Both processes are induced by siRNAs and require the core RNAi components, including Argonaute, Dicer-like proteins, and RNA-dependent RNA polymerase (RdRP) (Cogoni and Macino 1999a; Lee et al. 2003; Catalanotto et al. 2004; Fulci and Macino 2007). The distinction is that quelling operates in mitosis, whereas MSUD occurs during meiosis. RIP is a mechanistically distinct repeat silencing pathway that occurs premeiotically during the sexual cycle and inactivates repeated sequences by the introduction of C-to-T (G-to-A) transitions (Selker et al. 1987). Up to 30% of the GC base pairs in duplicated sequences can be mutated to AT pairs via RIP after a single passage through the sexual cycle (Cambareri et al. 1991). Therefore, RIP serves as a powerful genome defense mechanism against repetitive transposable elements, as evidenced by numerous nonfunctional transposon relics in the N. crassa genome (Galagan and Selker 2004).
Phenomena similar to quelling have also been described in Arabidopsis, Drosophila, and C. elegans (Pal-Bhadra et al. 1997; Vaucheret et al. 2001). Collectively, these are called cosuppression and can be induced by either highly expressed single-copy transgenes or moderately expressed gene arrays present in the genome in more complex configurations. Ectopic transgenes can also induce silencing of the endogenous gene in trans. In many cases, essential components in the RNAi pathway have been linked to cosuppression (Cogoni and Macino 1999a; Dalmay et al. 2000; Dernburg et al. 2000). A general PTGS model has been proposed in which a dsRNA intermediate homologous to the target gene is generated and processed into siRNAs of 21–25 nt in length. siRNAs subsequently serve as guides for the RISC, evoking degradation of the homologous transcripts (Catalanotto et al. 2002).
Orthologs of the core components in the RNAi pathway are found in most sequenced fungal genomes, implying that a wide range of fungi possess the machinery for RNA-mediated gene silencing (Nakayashiki 2005; Nakayashiki et al. 2006). The genome of the human fungal pathogen Cryptococcus neoformans encodes the core components of the RNAi pathway, including one Argonaute (Ago1), two Dicers (Dcr1 and Dcr2), and one RdRP (Rdp1) (Loftus et al. 2005). Gene targeting by dsRNA has been successfully applied to this fungal pathogen, suggesting the existence of a functional RNAi pathway (Liu et al. 2002; Cottrell and Doering 2003; Reese and Doering 2003). However, silencing phenomena such as quelling, RIP, or MSUD have not been reported.
Here, we report the discovery of sex-induced silencing (SIS), a transgene-induced RNAi-dependent gene silencing process that occurs during sexual reproduction of C. neoformans. The suite of RNAi machinery components was found to be translationally induced during mating, establishing a mechanism by which sex activates RNAi. Notably, a group of retrotransposons was highly expressed during the sexual cycle in rdp1 mutants, leading to elevated transposition and mutation. Our findings forge a link between RNAi and genome defense during sexual reproduction.
Results
Transgene silencing during sexual reproduction
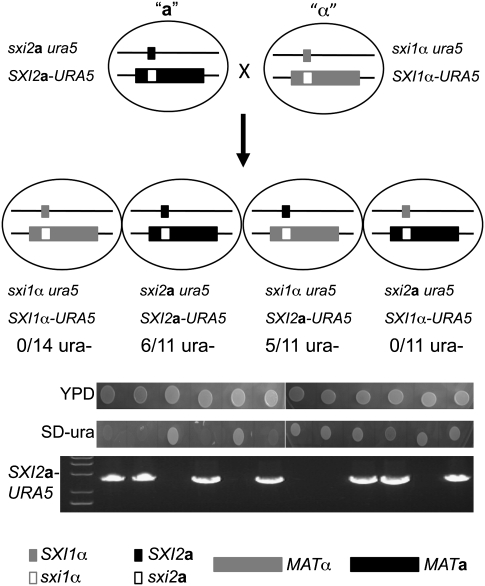
A novel example of transgene silencing was fortuitously discovered during studies on mating type determination in C. neoformans. To recapitulate the proposed ancestral tetrapolar mating system, the homeodomain genes SXI1α or SXI2a were relocated from the mating type locus to the URA5 locus on an unlinked chromosome. This generated modified “a” and “α” cells in which the pheromone/receptor genes resident at MAT were unlinked to the homeodomain genes at the URA5 locus (Hsueh et al. 2008). When f1 progeny were isolated from a cross of the a × α strains, 11 of 48 progeny were found to be uracil-auxotrophic. This was unexpected because both parents harbor the wild-type URA5 gene integrated at the ura5 locus, and thus all progeny should have been Ura+. Genotypic PCR analyses showed that the ura− phenotype is linked to the SXI2a-URA5 allele from parental strain JF289; all 11 ura− progeny carried an intact version of this allele based on PCR and Southern analysis, and in no case was the ura− phenotype attributable to loss of the integrated URA5 marker (see below). Interestingly, among the progeny that inherited the SXI2a-URA5 allele, 50% (11 of 22) were auxotrophic for uracil (Fig. 1).
Figure 1.
Silencing of the SXI2a-URA5 transgene following sexual reproduction. The homeodomain genes SXI1α and SXI2a were deleted from the MAT locus (large black and gray boxes), and the wild-type copies of the genes were integrated at the ura5 locus (small black and gray boxes), in which a G-to-A substitution results in a conserved Asp-to-Val substitution in position 225 that confers resistance to 5-FOA. The resulting strains were named JF289 a and JF306 α to reflect that the MAT configuration differs from wild-type a and α isolates. The total number of progeny isolated of each genotype is indicated, followed by the number of progeny that are uracil-auxotrophic. PCR analysis showed that uracil auxotrophy is linked to the SXI2a-URA5 transgene. Cells were grown on YPD or SD medium without uracil.
To examine further whether those progeny that escape URA5 silencing stably express the SXI2a-URA5 allele, one Ura+ f1 isolate was crossed to a strain of opposite mating type, and f2 progeny were isolated and analyzed. A high proportion of the isolated progeny (eight of 11) that inherited the SXI2a-URA5 transgene allele had again become auxotrophic for uracil. Thus, gene silencing faithfully occurs when the SXI2a-URA5 ectopic transgene is transmitted through a sexual cross.
Given that silencing occurred in only a proportion of the progeny, we addressed whether silencing is dominant or recessive and whether it could be activated in unsilenced strains in trans. To test this hypothesis, one α silenced strain (SXI2a-URA5 off) with the NEO dominant selectable marker was fused to an unsilenced a strain (SXI2a-URA5 on) with the NAT dominant selectable marker on V8 mating medium. After 24 h, the fusion products were selected on yeast extract–peptone–dextrose (YPD)/NAT + NEO medium, and doubly resistant strains were shown by FACS to be diploid. In total, 13 of 18 diploid isolates (72%) were found to be ura−; thus, silencing is dominant and trans-acting (Supplemental Fig. 1).
Tandem copies of the SXI2a-URA5 allele integrated at the ura5 locus
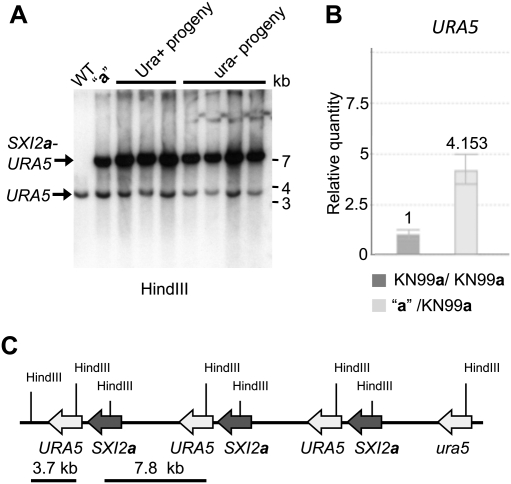
PCR analysis indicated that the SXI2a-URA5 transgene was still present in progeny with a silenced URA5 gene. To investigate whether recombination might have caused unexpected genome rearrangements, Southern analysis with a URA5 probe was conducted directly to compare genomic structures of ura− and Ura+ strains at the SXI2a-URA5 locus. As shown in Figure 2A, identical Southern patterns were observed for both, demonstrating that no rearrangement has occurred at the transgene locus. However, compared with the signal derived from the endogenous ura5 locus (3.7 kb), the signal from the introduced SXI2a-URA5 allele (7.8 kb) was more intense (Fig. 2A), indicating that multiple copies of the transgene had been inserted at ura5, resulting in a tandem repeat configuration. To determine the copy number of the SXI2a-URA5 transgene, quantitative real-time PCR was conducted with primers specific to URA5 and the actin gene ACT1 as a control. The URA5 gene is four times more abundant in the strain that carries the SXI2a-URA5 transgene when compared with wild-type cells, indicating that four copies of the URA5 gene (the endogenous ura5 and three URA5 transgene copies) are present in the genome (Fig. 2B). Hence, three copies of the SXI2a-URA5 transgene are tandemly repeated at the ura5 locus integration site (Fig. 2C).
Figure 2.
The SXI2a-URA5 transgene is tandemly integrated into the genome. (A) Southern blot analysis using URA5 as a probe detects the endogenous ura5 locus (bottom bands) and the SXI2a-URA5 transgene (top bands). (B) Quantitative real-time PCR revealed that four copies of the URA5 gene are present in the genome. (C) The deduced arrangement of the tandem integration of the SXI2a-URA5 transgene is depicted.
URA5 is post-transcriptionally silenced
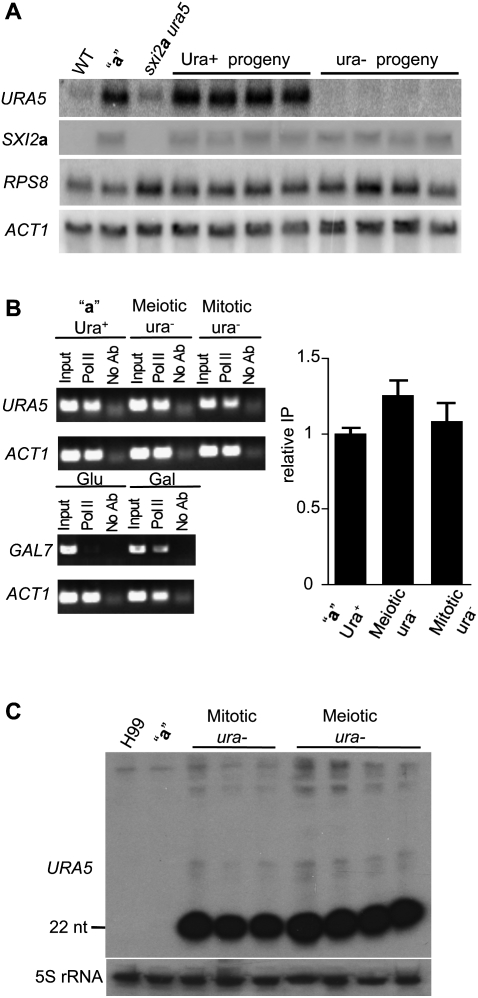
PCR and Southern analyses demonstrated that the SXI2a-URA5 transgene is present in the ura− progeny, suggesting that the transgene might be silenced. To test this hypothesis, Northern blot was performed to detect URA5 mRNA in both ura− and Ura+ isolates. As shown in Figure 3A, the steady-state levels of URA5 were higher in the original parental a strain JF289 bearing the transgene array compared with either wild-type or the sxi2a ura5 mutant. The elevated URA5 mRNA levels in the original parental strain JF289 likely reflect the presence of multiple copies of the SXI2a-URA5 transgene in the genome. Among all of the progeny that inherited the SXI2a-URA5 transgene, the Ura+ progeny showed a similar level of URA5 mRNA accumulation as in the parental strain JF289, while little or no URA5 mRNA was detected in the ura− progeny, indicating that both the endogenous and transgenic URA5 genes are silenced in the ura− isolates (Fig. 3A). In contrast, the steady-state levels of SXI2a were not decreased in the ura− progeny, indicating that SXI2a was not subject to silencing. These silenced strains were also fertile in sexual crosses (data not shown), providing additional evidence that SXI2a was not silenced, given the known roles of SXI2a in sexual reproduction (Hull et al. 2005). To examine whether the silencing effect is restricted to URA5 or spreads into other neighboring loci, we also probed for RPS8 mRNA, which flanks the URA5 gene and encodes a ribosomal protein. As shown in Figure 3A, like SXI2a, the RPS8 steady-state levels were comparable between Ura+ and ura− progeny, indicating that silencing has not propagated into this neighboring gene.
Figure 3.
URA5 is post-transcriptionally silenced in uracil-auxotrophic progeny. (A) Expression of the genes indicated on the left was examined by Northern blot analyses. The RPS8 gene flanks the URA5 locus, and the actin gene ACT1 served as the loading control. Strain JF289 (designated “a”) harbors the SXI2a-URA5 transgene, and strain JF135 (sxi2a ura5) was the recipient of the SXI2a-URA5 transgene. (B) URA5 transcription was determined by RNAPol-ChIP analysis. The panels on the left show the results of the PCR analysis of DNA extracted from input samples (input), the immunoprecipitates with anti-RNA Pol II antibody (Pol II), and samples treated without antibody (No Ab), using amplicons from the coding regions of the indicated genes. The right panel shows a real-time quantitative PCR result. (C) URA5 siRNAs are present in the ura− isolates. Northern analysis revealed that URA5 siRNAs were highly abundant. Total RNAs were isolated and resolved in a 15% denaturing polyacrylamide TBE-urea gel and probed with sense URA5 transcripts.
Silencing of the URA5 transgene array also occurred at a much lower frequency during mitotic growth. When spontaneous 5-FOA resistance was measured after the parental transgene array strain JF289 was grown in rich (YPD) medium, approximately one to three in every 1000 cells exhibited a 5-FOAR (ura−) phenotype. PCR analysis confirmed the presence of the intact transgene in these spontaneous ura− isolates. Silencing during mitotic growth occurred at a much lower frequency (1 to 3 × 10−3) than in meiotic progeny (50%). A similar low silencing rate (1.5 × 10−3) was also observed when cells were grown on V8 mating medium without a mating partner, and thus increased silencing efficiency during the sexual cycle is not solely attributable to nutrient conditions. Both meiotic and mitotic silencing of the transgene are unstable and could spontaneously revert back to Ura+ after serial passage on nonselective medium such as YPD at a low frequency (4 to 6 × 10−6) (details in the Supplemental Material).
To further address the central question of whether silencing occurs at a transcriptional or post-transcriptional level, an RNA polymerase-chromatin immunoprecipitation (RNAPol-ChIP) assay was applied. The occupancy of the URA5 coding region by RNA polymerase II (RNA Pol II) was measured by ChIP using antibodies against the phosphorylated largest subunit of RNA Pol II, which is a transcriptionally active form of RNA Pol II occurring during elongation. The binding of RNA Pol II to the coding region of URA5 was monitored by PCR, which serves as a measure of actual transcription. As shown in Figure 3B, the efficiency of product amplification was similar in the immunoprecipitates from the Ura+ and ura− strains, suggesting that URA5 is actively transcribed in the ura− strains despite the dramatic decline in mature URA5 mRNA levels. By real-time quantitative PCR, we further confirmed that the transcription is comparable between the Ura+ parental strain and its ura− progeny. To exclude the possibility that nonelongating RNA Pol II binding to the promoter region present in longer fragments of immunoprecipitated DNA that also contain the coding region was immunoselected, the GAL7 gene known to be repressed by glucose but induced by galactose (Ruff et al. 2009) was examined and demonstrated the selectivity of this RNAPol-ChIP method. As expected, GAL7 was effectively loaded with RNA Pol II in galactose medium, whereas little or no RNA Pol II occupancy was observed in glucose medium, reflecting that GAL7 was not actively transcribed in glucose (Fig. 3B). Thus, these data support a PTGS model for the SIS of URA5.
Silencing may occur premeiotically
To elucidate whether silencing occurs premeiotically or post-meiotically during sexual reproduction, we isolated blastospores, which are yeast cells formed by budding from the dikaryotic hyphae prior to meiosis during sexual development. C. neoformans is a basidiomycete, in which nuclear fusion and meiosis are delayed and do not occur immediately after cell–cell fusion during mating. Instead, the two nuclei fuse and undergo meiosis in the basidium, the terminal structure of the dikaryotic hyphae. Therefore, blastospores represent a stage of the sexual cycle that is post-cell–cell fusion but premeiotic. The a strain JF289 bearing the SXI2a-URA5 transgene array was crossed with the congenic wild-type α strain H99, and blastospores were isolated via micromanipulation. Among the 24 MATa blastospores isolated, all inherited the transgene, supporting the conclusion that the blastospores are premeiotic and clonally derived from the parental a nucleus. Furthermore, the isolated MATα blastospores all harbored the MATa-type mitochondria, indicating that these blastospores are produced post-cell–cell fusion, based on the known uniparental inheritance of mitochondria from the MATa parent following mating or sexual reproduction in this species (Supplemental Fig. 2). Three of the 24 (12.5%) analyzed MATa blastospores were ura− when grown on synthetic dextrose (SD) medium lacking uracil, providing evidence that SIS can occur premeiotically. However, because the frequency of silencing among the blastospores was about fourfold lower compared with meiotic progeny (12.5% vs. 50%), silencing efficiency may increase during repeated cycles of hyphal cell division or during or after meiosis.
A multicopy transgene is necessary but not sufficient for SIS
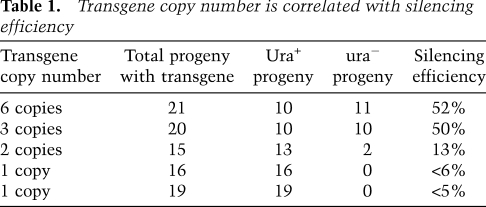
In contrast to the URA5 gene, the SXI2a gene was not silenced in the ura− progeny even though it is also present in the transgene array. Given that the tandem array includes four copies of URA5 (three URA5 transgene copies and one endogenous ura5 allele) but only three copies of SXI2a (Fig. 2C), and that transgene copy number and silencing are known to be correlated in both plants and fungi (Jones et al. 1987; Cogoni and Macino 1997), we tested if more than three tandemly repeated copies of SXI2a would be subject to SIS.
To obtain isolates harboring different copy numbers of the integrated transgene, we introduced the plasmid containing the URA5-SXI2a transgene into the same recipient strain, sxi2a ura5, by transformation. Integration of the transgene at the ura5 locus was confirmed by PCR and Southern analysis. As described above, the signals derived from the SXI2a-URA5 transgene and the endogenous ura5 allele were detected individually by Southern blot. Compared with the original JF289 transgene array strain, transformants that exhibited differing copy numbers of the URA5 transgene were chosen and analyzed by real-time PCR (data not shown). Four classes of isolates were obtained containing one, two, three, or six copies of the SXI2a-URA5 transgene integrated at the ura5 locus (Table 1).
Table 1.
Transgene copy number is correlated with silencing efficiency
One or two representative strains from each class were crossed with the wild-type α strain H99, and the resulting progeny were examined for URA5 silencing on SD-uracil and 5-FOA medium. With three transgene copies, 50% of the progeny inheriting the transgene became ura−, whereas the efficiency rate was reduced to ∼10% when only two transgenes were present. No URA5 silencing was detected in progeny derived from parental strains containing a single transgene (one URA5 transgene and the endogenous ura5 allele). This result revealed that a high transgene copy number is required for efficient SIS, and the threshold might be two copies. However, strains harboring six copies of the SXI2a-URA5 transgene remained fertile, including those in which URA5 was silenced, providing evidence that Sxi2a is still expressed and functional. Thus, the presence of multiple copies of tandemly repeated genes is not sufficient per se to elicit a silencing response, and expression levels, which differ between URA5 and SXI2a, may also contribute.
URA5 is silenced by RNAi
In the fungus N. crassa, repeat-induced gene silencing occurs via three common mechanisms: quelling, RIP, and MSUD (Selker 1997; Cogoni and Macino 1999b; Borkovich et al. 2004; Galagan and Selker 2004; Kelly and Aramayo 2007). To investigate the underlying molecular mechanism that governs SIS, we first sequenced URA5 from the silenced strains, but found no mutations indicative of RIP (GC to AT). We also excluded the possibility that this is an example of MSUD. When two strains containing the SXI2a-URA5 transgene array were crossed, URA5 was silenced efficiently in their progeny (15 of 20 isolates). Thus, unpaired DNA is not required for SIS, in contrast to MSUD (Supplemental Fig. 3).
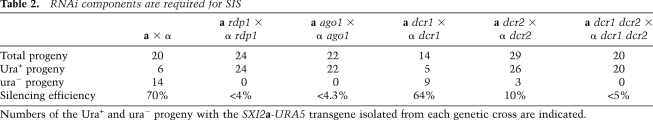
We next tested whether SIS is dependent on the RNAi machinery, similar to quelling in N. crassa that occurs during mitotic growth. The C. neoformans serotype A strain H99 genome encodes one Argonaute (Ago1), two Dicers (Dcr1 and Dcr2), and one Rdrp (Rdp1). To determine if these components are required for SIS, an rdp1 mutant was generated by deleting the RDP1 gene in the unsilenced a strain JF289 bearing the SXI2a-URA5 transgene and crossed to the wild-type α strain H99. Among 24 progeny analyzed, 12 inherited the SXI2a-URA5 allele; however, only one progeny was ura−, indicating a reduced silencing efficiency (∼8.3%) compared with the wild-type a × α cross (∼70%). When the a rdp1 strain was crossed with an α rdp1 strain, none of the 24 progeny (zero of 24, <4%) that inherited the SXI2a-URA5 allele were auxotrophic for uracil (Table 2). Thus, Rdp1 is required for silencing.
Table 2.
RNAi components are required for SIS
Numbers of the Ura+ and ura− progeny with the SXI2a-URA5 transgene isolated from each genetic cross are indicated.
That RDP1 is required for SIS prompted us to generate additional strains lacking other RNAi pathway components and test the effects of these mutations on SIS. When AGO1 was deleted in both parents, SIS was also completely abolished. Among 22 of the 40 progeny isolated from an ago1 × ago1 cross that inherited the SXI2a-URA5 transgene, none were ura− (zero of 22, <4.3%), indicating that Ago1 is required for SIS. When DCR1 was deleted in both parents, SIS efficiency was not affected. Among 14 of the 33 isolated progeny that inherited the SXI2a-URA5 transgene from a dcr1 × dcr1 cross, nine were ura−, indicating that the frequency of silencing is >50% (nine of 14, 64%). In contrast, in the dcr2 × dcr2 bilateral mutant cross, the efficiency of silencing was significantly diminished (three of 29, ∼10%). Thus, DCR2 appears to play the major role in SIS compared with DCR1. dcr1 dcr2 double mutants were generated and crossed to isolate progeny. Among 50 analyzed progeny, 20 inherited the SXI2a-URA5 transgene, all of which were uracil-prototrophic, indicating that silencing was abolished in the dcr1 dcr2 double mutants (zero of 20, <5%) and that both genes are required for SIS and are partially functionally redundant. In conclusion, the requirement of RDP1, AGO1, DCR1, and DCR2 for SIS reveals that the molecular mechanism underlying SIS involves RNAi. Similarly, we found that the RNAi components are also required for transgene-induced mitotic silencing, and, between the two Dicer genes, DCR2 again plays the major role (Table 3).
Table 3.
RNAi components are required for transgene-induced mitotic silencing
Because a distinguishing feature of RNAi is the generation of siRNA from dsRNA, we next examined the presence of siRNA in the URA5 silenced strains. RNA was extracted from both meiotically or mitotically silenced strains and analyzed by Northern hybridization. When probed with the P32-labeled sense URA5 transcript, abundant messages ∼22 nt in size were detected in both meiotically and mitotically silenced strains, but not in the wild-type strain H99 or the original parental strain JF289 bearing the SXI2a-URA5 transgene (Fig. 3C). No significant difference in the amount of accumulated small RNA was observed between samples derived from meiotically or mitotically silenced strains. The small RNAs are antisense transcripts of URA5, suggesting that they originate from a dsRNA precursor, further supporting RNAi as the underlying mechanism for both silencing processes that occur during vegetative growth or the sexual cycle in C. neoformans.
In the fission yeast S. pombe, RNAi is implicated in targeting chromatin modifications that are typical of heterochromatin, such as Histone 3 Lys 9 (H3K9) methylation, thus resulting in cotranscriptional silencing. To investigate if a similar mechanism is involved in the silencing of URA5 in C. neoformans, we examined the pattern of H3K9 methylation by using antibodies specific to dimethyl-H3K9 (H3K9me2) in the ChIP analyses. Specific primers for the URA5 gene were chosen for quantitative real-time PCR to determine the levels of URA5 found in the ChIPs. Little or no variation in the H3K9me2 pattern was detected at the URA5 locus of the Ura+ compared with the ura− strains (Supplemental Fig. 4). Because an antibody to trimethyl-H3K9 (H3K9me3) failed to detect any signal in Western blot analyses, we did not perform ChIP to test H3K9me3. Overall, these findings indicate that silencing of URA5 is unlikely to be due to heterochromatin formation, thus excluding a cotranscriptional silencing model.
siRNAs in C. neoformans
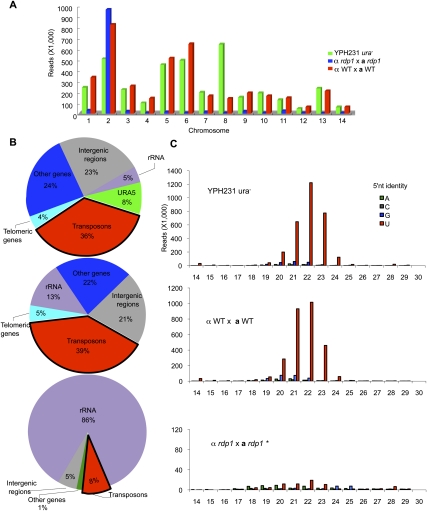
To gain more insight into the endogenous small RNA population that may function in SIS, we cloned and sequenced small RNAs with 5′ monophosphates and 3′ hydroxyls, both hallmarks of Dicer products. Three sequencing libraries for small RNAs were prepared from (1) a meiotically silenced (ura−) strain, (2) wild-type mating strains (H99α × KN99a), and (3) a rdp1 × α rdp1 mating strains, followed by high-throughput Illumina sequencing as described previously (Lau et al. 2001). The overall number of reads obtained from the ura− strain (4,229,989) and wild-type mating strains (4,683,577) was similar, and small RNA molecules were distributed throughout the genome (Fig. 4A). Fewer reads (1,979,252) were obtained from the a rdp1 × α rdp1 mating culture, and when mapped to the genome, nearly all emanate from chromosome 2, on which the rDNA genes reside (Fig. 4A).
Figure 4.
siRNAs in C. neoformans. (A) Genome distribution profiles of small RNAs isolated from a ura− progeny that inherited the SXI2a-URA5 transgene array (YPH231, green), a × α wild-type (WT) (H99 × KN99) mating (red), and a rdp1 × α rdp1 mating (blue). (B) Pie chart illustrating percentages for the individual small RNA classes relative to the total amount of small RNAs sequenced from different strains: YPH231 (top), wild-type mating strains (H99α × KN99a) (middle), and a rdp1 × α rdp1 mating strains (bottom). (C) Size distribution and 5′ nucleotide of the small RNAs. Sequences homologous to rRNA are excluded. (*) The vertical axis scale is adjusted to render the value bar visible in the a rdp1 × α rdp1 mating siRNA library.
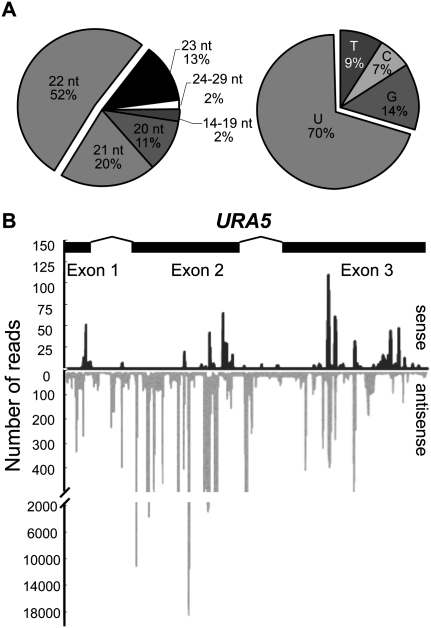
The relative abundance of different classes of small RNAs in each library is illustrated in Figure 4B. Consistent with our previous Northern result, siRNA derived from URA5 was only found in the ura− silenced strain (Fig. 4B), accounting for 8% of the total reads ranging from 14 to 29 nt in length, with 22 nt as the predominant group (Fig. 5A). Many more siRNA reads mapped to chromosome 8 in the ura− strain compared with those in wild-type mating strains (URA5 is located on this chromosome) (Fig. 4A). The vast majority of siRNA sequences matched to URA5 (99.2%) were antisense (Fig. 5B), which likely reflects that antisense small RNAs could be more stable than sense small RNAs due to loading onto the RISC. Additionally, antisense siRNAs corresponding to intron 1 and intron 2 of the silenced URA5 were observed (Fig. 5B), suggesting a cotranscriptional dsRNA synthesis. The antisense bias was also observed for small RNAs produced from most other loci (Supplemental Fig. 5). In contrast, the majority of small RNAs originating from rDNAs are sense (>95%) and are presumably products of ribosomal RNA (rRNA) degradation (Supplemental Fig. 6).
Figure 5.
URA5 siRNAs are present in the ura− isolates with a silenced transgene array. (A) Pie charts indicating the length distribution and the proportion of 5′ nucleotide of the URA5 siRNAs identified by deep sequencing. (B) Distribution and frequency of the URA5 siRNAs. Black bars represent sense siRNAs and gray bars represent antisense siRNAs. The structure of the URA5 gene is indicated above.
Abundant RNA sequences in the ura− strain (36% in total) shared homology with repetitive transposable elements, most of which are Ty or DNA transposon elements located in centromeric and telomeric regions. Only ∼5% reads in the ura− strain mapped to the rRNA. Similar distribution patterns were observed in wild-type mating strains, in which 39% mapped to transposable elements and 14% mapped to rRNA. Additionally, in both libraries, we detected siRNAs derived from all 14 putative centromeric regions, which are enriched in repetitive sequences and transposons. However, in the a rdp1 × α rdp1 bilateral mutant mating, the majority of sequences (86%) are derived from rRNAs, and small RNAs generated from transposons were dramatically reduced (Fig. 4B).
We further analyzed small RNAs obtained from the different strains after excluding all of the reads derived from rRNAs. The small RNAs from the silenced ura− and the wild-type mating strains were most enriched in 21–23 nt and had a strong preference for a 5′ uridine (75%) (Fig. 4C). Similar biases have been observed recently in other fungal small RNAs identified in N. crassa, S. castellii, and S. pombe (Buhler et al. 2008; Drinnenberg et al. 2009; Lee et al. 2009). Furthermore, Northern blot analysis showed that generation of these small RNAs was dependent on Ago1, Rdp1, Dcr1, and Dcr2 (Supplemental Fig. 7). In contrast, in rdp1 mutant strains, few small RNAs remained after those matching to rRNAs were filtered, and, more importantly, no analogous sequence bias was found, as expected for siRNAs generated from other pathways independently of Rdp1. Taken together, our results suggest that an Rdp1-dependent RNAi pathway is involved in generating siRNAs in C. neoformans and might play a role in silencing repetitive transposon elements.
Rdp1, Ago1, and Dcr1 are translationally induced during the sexual cycle
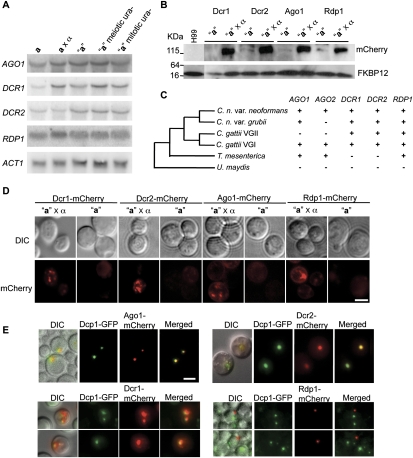
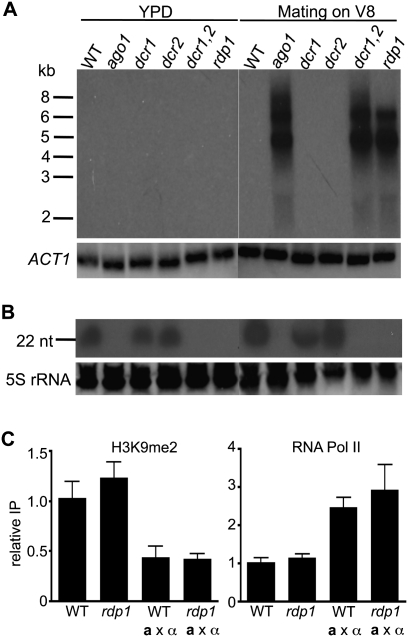
Why is transgene-induced silencing markedly induced during sexual reproduction? Because the RNAi pathway is required for SIS, we considered that the RNAi pathway components might be induced during the sexual cycle. To test this, the expression of the AGO1, DCR1, DCR2, and RDP1 genes was analyzed during vegetative growth and mating by Northern blot analysis. AGO1, DCR2, and RDP1 expression is similar during mitotic growth and mating; only DCR1 appeared to be modestly induced during the sexual cycle (∼2.5-fold) (Fig. 6A). The abundance of these transcripts was also similar in the parental Ura+ strain and the silenced ura− progeny (Fig. 6A), suggesting silencing is not regulated at the transcriptional level of the RNAi machinery.
Figure 6.
The RNAi machinery is translationally induced during the sexual cycle in C. neoformans. (A) RNA from the strains indicated (a is JF289 and α is H99) was extracted and probed in Northern blot analysis with AGO1, DCR1, DCR2, and RDP1 probes or ACT1 as a loading control. Only DCR1 transcripts were modestly increased (∼2.5-fold) under mating conditions. (B) mCherry was fused to the C terminus of Dcr1, Dcr2, Ago1, and Rdp1, and the resulting fusion proteins were expressed from their endogenous loci. Proteins were extracted from these strains under vegetative growth or mating conditions. Western blot analysis with antibodies specific to mCherry was used to determine the expression levels of the fusion proteins. Antiserum to yeast FKBP12 served as a loading control. (C) RNAi genes have been lost in related Cryptococcus species. The phylogenetic tree depicted is based on partial 18S rDNA sequences; the plus sign (+) and minus sign (−) indicate the presence or absence of the gene in the genome of JEC21 (var. neoformans), H99 (var. grubii), R265 (C. gattii VGII), WM276 (C. gattii VGI), T. mesenterica, and U. maydis. (D) Strains with C-terminal mCherry fusions were cultured in YPD alone or crossed to the wild-type α strain H99 on pH 5 V8 medium for 24 h and observed at 1000× magnification. Bar, 5 μm. (E) Colocalizaion of Ago1, Dcr1, Dcr2, and P bodies. Strains expressing Ago1-mCherry, Dcr1-mCherry, Dcr2-mCherry, or Rdp1-mCherry and Dcp1-GFP were crossed to the wild-type α strain H99 on V8 pH 5 medium for 24 h. (N.A.) 1.4 with oil. Bar, 5 μm.
We speculated whether the abundance of the RNAi components might vary at the protein level because, in many cases, transcript abundance is not directly correlated with protein abundance. The Ago1, Dcr1, Dcr2, and Rdp1 C terminus was fused to the mCherry protein, and protein abundance was examined during mating and vegetative growth by Western blot. Surprisingly, all of the RNAi components were found to be significantly more abundant under mating conditions, in which a and α cells were coincubated on V8 or Murashige and Skoog (MS) medium (Fig. 6B). Interestingly, the increased protein levels were also observed when cells were grown on V8 or MS medium without a mating partner (data not shown), suggesting that protein induction is initiated in response to nutritional cues. Accordingly, we also detected strong direct fluorescence signals in cells expressing Ago1-mCherry, Dcr1-mCherry, Dcr2-mCherry, or Rdp1-mCherry fusion proteins when incubated under mating conditions (Fig. 6D), whereas expression was barely detectable when these cells were grown on rich YPD growth medium. Thus, the core components in the RNAi machinery are translationally induced by nutrient cues and during the sexual cycle, which likely contributes, at least in part, to the higher efficiency of transgene-induced silencing observed during the sexual cycle.
Given that Argonaute localizes to cytoplasmic processing bodies (P bodies) in mammalian cells and C. elegans (Liu et al. 2005; Sen and Blau 2005), we examined if Ago1, Rdp1, Dcr1, and Dcr2 colocalize with an established P-body component in C. neoformans. Dcp1 is a decapping enzyme that removes the 5′ mRNA cap prior to exonucleolytic degradation, and Dcp1 is found in P bodies in both yeast and mammals (Sheth and Parker 2003; Cougot et al. 2004). A plasmid expressing a green fluorescent protein (GFP)-tagged DCP1 allele was introduced into cells expressing Ago1-mCherry, Dcr1-mCherry, Dcr2-mCherry, or Rdp1-mCherry. As shown in Figure 6E, by confocal microscopy, we observed colocalization of Ago1-mCherry, Dcr1-mCherry, and Dcr2-mCherry with Dcp1-GFP in discrete, brightly staining cytoplasmic foci in cells under mating conditions. No colocalization was detected between Rdp1-mCherry and Dcp1-GFP. Instead, we found that Rdp1 localized mostly to the stained nuclei (Supplemental Fig. 8). Thus, Ago1, Dcr1, and Dcr2 are associated with P bodies in C. neoformans, suggesting that RNA silencing and RNA decay pathways may occur in the same cellular compartment. However, the initial steps of generating dsRNA by RdRP may occur in the nucleus.
Retrotransposons are highly expressed during an rdp1 × rdp1 mating
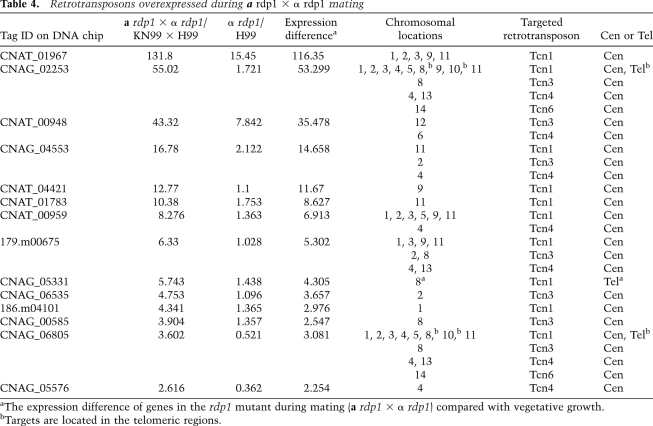
Transposon silencing is one function of RNAi in several organisms, especially in germ cells (Slotkin and Martienssen 2007). Here, through deep sequencing, we found abundant siRNA derived from transposable elements, suggesting that RNAi might also be involved in suppressing transposable elements in C. neoformans. In addition, we discovered that the sexual cycle contributes to highly efficient silencing of repetitive transgenes by RNAi. This prompted us to investigate whether the RNAi pathway plays a novel role in silencing transposable elements during sexual development. We performed a comparative transcriptome analysis between rdp1 and wild-type strains undergoing vegetative growth on YPD medium or under mating conditions involving coincubation of a and α cells on V8 medium. In each comparison, many probe tags showed much stronger hybridization when probed with rdp1 cDNAs than with those from wild-type cells, whereas very few signals became weaker, indicating that the majority of Rdp1-dependent regulation is repression.
In the first level of analysis, we considered genes that were highly expressed during an a rdp1 × α rdp1 mating, but were expressed at a lower level in the α rdp1 mutant during vegetative growth. More than 70 independent tags yielded a more than twofold stronger signal in the rdp1 mutant under mating conditions than when grown on YPD medium (Supplemental Table 1). When these enriched probes were aligned to the C. neoformans H99 genome database, the majority matched hypothetical protein genes or intergenic sequences. When the probe tags were aligned to the transposon database, 14 were found to correspond to retrotransposon sequences with a total of 62 corresponding sites in the genome due to their repetitive distribution on different chromosomes (Table 4). All of these retrotransposons are Ty elements with long terminal repeats (LTR), including Tcn1, Tcn3, Tcn4, and Tcn6. Most interestingly, the majority of the retrotransposons whose expression increased during mating of the rdp1 mutants corresponds to elements located in the presumptive centromeric regions.
Table 4.
Retrotransposons overexpressed during a rdp1 × α rdp1 mating
aThe expression difference of genes in the rdp1 mutant during mating (a rdp1 × α rdp1) compared with vegetative growth.
bTargets are located in the telomeric regions.
The expression levels of the Tcn1, Tcn3, and Tcn4 elements were elevated during mating of the rdp1 mutants based on comparative transcriptome analysis. In accord, by Northern analyses, Tcn1, Tcn3, and Tcn4 expression was barely detectable in the mutant strains grown on YPD medium, but was dramatically expressed during the a rdp1 × α rdp1 mating. A similar overexpression was detected in ago1 and dcr1/2 mutant strains during mating or growth on V8 medium without mating partners, but not in wild-type strains (Fig. 7; Supplemental Fig. 9). Abundant antisense small RNAs originating from Tcn1 were observed in wild-type and single dcr1 or dcr2 mutant strains under both vegetative and mating conditions (Figs. 4, 7), suggesting that the RNAi pathway is involved in silencing retrotransposons.
Figure 7.
Retrotransposon Tcn1 is highly expressed during mating of the RNAi mutant cells and post-transcriptionally silenced by RNAi in wild-type (WT) mating cells. (A) Total RNAs were isolated from the indicated strains shown in the top panel. Northern blot analyses were conducted to analyze the expression of Tcn1. ACT1 was used as a loading control. (B) siRNAs were enriched from individual total RNAs and resolved in a 15% denaturing polyacrylamide TBE-urea gel. A P32-labeled in vitro transcribed sense Tcn1 transcript was first hydrolyzed and then used to probe antisense Tcn1 siRNAs. (C) Wild-type and rdp1 mutant strains were collected after growth on YPD medium or mating for 24 h on V8 medium. Cell extracts were analyzed for H3K9me2 (left) and RNA Pol II (right) by ChIP assays with primers specific for the Tcn1 gene. Enrichment was quantified by RT–PCR. Results are expressed as the fold increase of the ratio to the ACT1 control relative to the results for wild type (H99), the value of which was arbitrarily set to 1.
Our results illustrate an interesting correlation between production of RNAi components and repression of retrotransposons in centromeric regions under mating conditions that is reminiscent of the role that RNAi plays in maintaining the heterochromatic state of centromeric repeats by targeting H3K9 methylation in fission yeast, plants, and animals (Wassenegger et al. 1994; Lippman et al. 2003; Zilberman et al. 2003; Pal-Bhadra et al. 2004). This led us to examine changes in the H3K9 methylation status at Tcn1 by ChIP analyses as described above. Quantitative real-time PCR revealed that the levels of H3K9me2 were comparable between wild type and the rdp1 mutant. However, an approximately threefold decrease in H3K9me2 was detected during mating at 24 h compared with growth on YPD medium. We also assessed the transcriptional activity of Tcn1 by an RNAPol-ChIP assay. Similar to our observations for H3K9me2, the occupancy of RNA Pol II at Tcn1 was not affected by the rdp1 mutation, but differed when cells started to mate. The level of the RNA Pol II signal at Tcn1 was enriched twofold to threefold in the wild-type mating and rdp1 × rdp1 strains, suggesting that Tcn1 is more actively transcribed during mating (Fig. 7). This could be due at least in part to the reduction in heterochromatin modifications. That the RNA Pol II levels are not altered at Tcn1 in the rdp1 mutant suggests a similar PTGS mechanism operates as described in the case of URA5 silencing. In addition, we did not find evidence that RNAi is involved in heterochromatin formation.
Transposons are more active in the RNAi mutant strains during mating
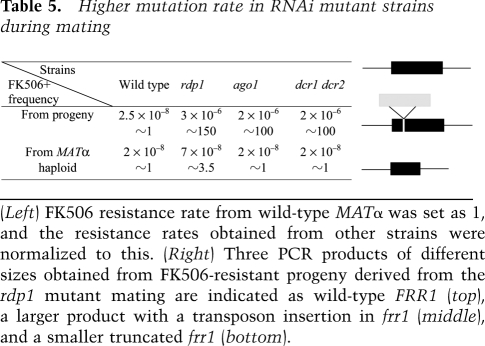
The overexpression of retrotransposons in RNAi-deficient strains during mating suggested that some transposons might be more active in these strains during sexual development. To address this, the FKBP12-encoding gene FRR1 was used as a transposon trap, as described in a previous study (Cruz et al. 1999). The rate of spontaneous FK506 resistance was examined by germinating spores obtained from the wild-type and RNAi mutant crosses on YPD medium with or without FK506. FK506 is an immunosuppressant with antifungal activity that is toxic to C. neoformans when bound to FKBP12 (Odom et al. 1997). Based on mutant fluctuation tests, the progeny obtained from the rdp1 mutant crosses yielded the highest rate of FK506 resistance (∼3 × 10−6), which is ∼120 times higher than that of the wild-type cross (∼2.5 × 10−8). The ago1 and dcr1/2 mutant crosses generated fewer FK506-resistant progeny than the rdp1 mutant but the frequency was still greater than wild type (Table 5).
Table 5.
Higher mutation rate in RNAi mutant strains during mating
(Left) FK506 resistance rate from wild-type MATα was set as 1, and the resistance rates obtained from other strains were normalized to this. (Right) Three PCR products of different sizes obtained from FK506-resistant progeny derived from the rdp1 mutant mating are indicated as wild-type FRR1 (top), a larger product with a transposon insertion in frr1 (middle), and a smaller truncated frr1 (bottom).
To test if the FK506 resistance phenotype is attributable to frr1 mutations or transposon insertion into this gene, the FRR1 gene was PCR-amplified from genomic DNA isolated from the FK506-resistant progeny derived from the RNAi mutant cross and wild-type crosses. A PCR product of the expected size was obtained from the two mutant progeny obtained from the wild-type cross. However, among the FK506-resistant progeny derived from the RNAi mutants mating, three different size PCR products were observed: a wild-type size PCR product, a larger PCR product, or a smaller PCR product. We cloned and sequenced the FRR1 PCR products of larger or shorter sizes. The results showed that a 3652-base-pair (bp) sequence had inserted into the FRR1 ORF in two different FK506-resistant isolates, one from the rdp1 mutant mating and the other from the ago1 mating, resulting in a larger PCR product. Based on sequence analysis, this DNA fragment contains a transposase gene that belongs to the Harbinger transposon superfamily (Supplemental Fig. 10) and was integrated in FRR1 in two different locations in opposite orientations. Northern blot analyses showed that this transposon was highly expressed during mating of RNAi mutant strains (Supplemental Fig. 9), similar to Tcn1, Tcn3, and Tcn4 as shown above. In the case of FK506-resistant isolates yielding a shorter PCR product, a portion of the FRR1 ORF sequence is deleted, accounting for the FK506 resistance phenotype. This may reflect a higher frequency of gene rearrangements/deletions or a higher transposition rate (transposons being integrated into and excised from the genome) occurring during mating of the RNAi mutants.
We also examined the rate of FK506 resistance during mitotic growth of the MATα wild-type and RNAi mutant strains (Table 5). The rate for each strain is dramatically lower (∼100-fold) than the corresponding ones obtained from f1 progeny, and the wild-type and RNAi mutant strains exhibited comparable rates. Only the rdp1 mutant exhibited a slightly higher resistance rate (∼3.5-fold). Taken together, we conclude that a higher rate of mutation and transposition occurs during mating of the RNAi pathway mutant.
The RNAi machinery is important for spore formation but not for virulence
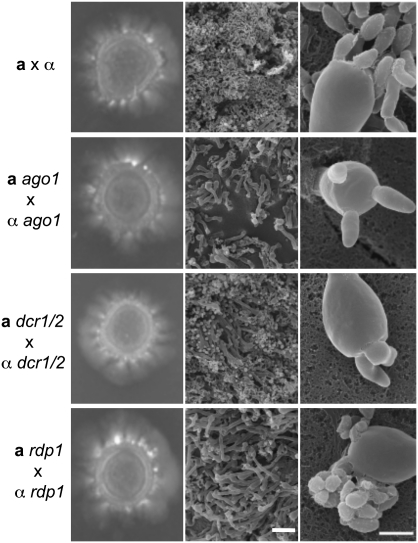
Given the involvement of Ago1, Rdp1, Dcr1, and Dcr2 in transposon silencing during the sexual cycle, we investigated the role of the RNAi components during C. neoformans sexual reproduction. In mating crosses between the a and α ago1, rdp1, dcr1/2 mutants, no deficiencies in mating-specific hyphal formation were observed (Fig. 8). However, based on light and scanning electron microscopy, all of the bilateral RNAi mutant crosses were found to produce ∼20-fold fewer spores than the wild-type mating. While basidia in wild-type crosses were decorated with four long spore chains, the basidia produced by dcr1/2 × dcr1/2 or ago1 × ago1 mutant crosses often yielded only three spores. In the rdp1 × rdp1 mutant mating, the spores were disorganized on the basidia surface rather than forming four distinct individual spore chains (Fig. 8). In summary, the RNAi machinery regulates both the efficiency of sporulation and the sites of spore emergence. Spores micromanipulated from both wild-type and RNAi mutant crosses have comparable germination rates, and no obvious growth defects were observed (data not shown).
Figure 8.
The RNAi machinery is important for sexual spore formation. (Left) Matings were performed on MS medium with incubation in the dark at 25°C, and hyphae were photographed following incubation for 2 wk. The edges of mating cultures from both wild type (H99α × KN99a) and RNAi mutant (a ago1 × α ago1, a rdp1 × α rdp1, and a dcr1/2 × α dcr1/2) cocultures were fixed and viewed by scanning electron microscope at two different magnifications: 1000× (middle) and 10,000× (right). Bars: middle, 20 μm; right, 5 μm.
Well-characterized virulence factors in C. neoformans include melanin and the polysaccharide capsule (Janbon et al. 2001). No defects in melanin or capsule production were observed in rdp1, ago1, and dcr1/2 strains compared with wild type (data not shown). A murine inhalation model of systemic C. neoformans infection was employed to test if the RNAi components contribute to virulence. Only one rdp1 mutant exhibited an enhanced virulence that met statistical significance (Supplemental Fig. 11). However, this was not observed with two other rdp1 mutants, both of which were of virulence equivalent to wild type. Hence, the RNAi machinery is not necessary for virulence.
Discussion
In this study, we report the discovery of SIS in C. neoformans, a novel repeat silencing mechanism triggered by a tandem multicopy insertion of a SXI2a-URA5 transgene. SIS requires the central components of the RNAi pathway and occurs post-transcriptionally. A group of LTR retrotransposons located in the candidate centromeric regions was identified through comparative transcriptome analysis and found to be highly induced in the RNAi-deficient strains under nutrient limitation/mating conditions but not expressed on rich growth medium or when RNAi is functional. The discovery of SIS forges a link between the RNAi pathway and genome defense during the sexual cycle of C. neoformans.
RNAi in closely related Cryptococcus species
RNAi is largely conserved among eukaryotes, yet, in the fungal kingdom, it has been lost independently in several species. One well-known example is the budding yeast Saccharomyces cerevisiae, which lacks all of the central RNAi pathway components. Interestingly, recent studies also revealed that the genomes of S. castellii and Candida albicans, two hemiascomycete species that are closely related to S. cerevisiae, encode an Argonaute gene and a novel Dicer-like gene with an RNaseIII domain, and the two species were shown to have a functional RNAi pathway (Drinnenberg et al. 2009). In the basidiomycete lineage, RNAi has been lost in the corn smut fungus Ustilago maydis but retained in the sibling species Ustilago hordei (Laurie et al. 2008). The apparently independent RNAi pathway losses may provide a selective benefit under certain conditions.
In this study, we established a link between the RNAi pathway and SIS in the C. neoformans serotype A var. grubii lineage; however, it was unclear whether similar and/or distinct silencing pathways exist or have been lost in the sibling species, including the serotype D var. neoformans lineage and Cryptococcus gattii. Thus, we examined whether the key RNAi genes (AGO1, DCR1, DCR2, and RDP1) are present in the genomes of the serotype D strain JEC21; two C. gattii strains, R265 and WM276 (which are likely two cryptic species); and the jelly fungus Tremella mesenterica as an aligned representative ancestral outgroup to the Cryptococcus species. In contrast to the single Argonaute gene found in C. neoformans var. grubii strain H99, two were identified in the serotype D strain JEC21, the C. gattii strain WM276, and the ancestral species T. mesenterica, and, more strikingly, both of the Argonaute genes AGO1 and AGO2 were not found in the C. gattii VGII strain R265 (Fig. 6C). The absence of the Argonaute genes indicates that R265 might not have a functional RNAi pathway. The success of R265 as a pathogen that is causing the C. gattii outbreak on Vancouver Island and in the Pacific Northwest may also imply that loss of the RNAi machinery could be beneficial under certain circumstances during evolution. Interestingly, in T. mesenterica, two Dicer genes have been lost, yet the two Argonaute genes are still present in the genome; this suggests that either RNAi has been lost in this species or other RNaseIII enzymes may function as noncanonical Dicers. Notably, a novel Dicer gene was recently identified and shown to process dsRNA to ∼22-nt small RNA in S. castellii (Drinnenberg et al. 2009).
On the other hand, it will be of interest to examine whether the serotype D strains have additional silencing pathways, as two Argonaute genes are encoded by the genome in contrast to the single Argonaute gene in the strains analyzed in our studies. In multicellular eukaryotes, Argonaute genes are often expressed in different tissues and loaded with distinct classes of small RNAs (Kim et al. 2009). In addition, if SIS operates in serotype D strains, the efficiency of silencing might be more robust due to the second AGO2 gene.
Transgene silencing during the sexual cycle
Our study found a PTGS phenomenon occurring in C. neoformans; when the tandemly repeated SXI2a-URA5 transgene was introduced, transcription of the URA5 gene was unaffected, but its transcripts did not accumulate as a consequence of degradation via an RNAi pathway. Similar to the findings in N. crassa (Cogoni and Macino 1997), a correlation between transgene copy number and silencing efficiency was observed; however, the silencing effect may become saturated at ∼50%, as there was very little difference in the level of silencing for transgenes present in three or six tandem repeats (Table 1). Interestingly, in our studies, silencing did not occur in the SXI2a gene, even though it lies adjacent to the silenced URA5 gene and at a copy number higher than the predicted threshold. Accordingly, only three unique small RNA reads longer than 14 nt corresponding to the sense or antisense strand of the SXI2a gene were found, but siRNAs specific to the URA5 gene were abundant, indicating that SXI2a escaped from the RNAi pathway. Given the fact that SXI2a encodes a homeodomain transcription factor whose expression is limited prior to sexual reproduction, it is possible that the low levels of transgenic mRNA are insufficient to activate silencing. This hypothesis is supported by previous studies in plants (Vaucheret et al. 2001) and the fungus Mucor circinelloides (Nicolás et al. 2009), in which higher transgene expression is a stronger inducer of PTGS, but this is not universal. For example, a high expression level was insufficient to trigger gene silencing in N. crassa (Romano and Macino 1992). Moreover, SIS is more complicated than just gene expression levels, as SXI2a is highly expressed late during mating but is still not silenced. Thus, we consider an alternative model in which a qualitatively aberrant feature of transgenic RNA or DNA–RNA hybrids, possibly via endogenous transgene DNA pairing, combined with a high level of transgenic RNA accumulation, may trigger silencing.
Silencing of the SXI2a-URA5 transgene occurs at a relatively low frequency (∼0.3%) during mitotic growth compared with meiotic progeny (>50%). What factors contribute to sex-specific induction of silencing? Transcription of key RNAi components is comparable between vegetative growth and mating. However, regulation was found to occur at the translational level; the proteins were detected at higher abundance during mating, suggesting that either translation of the RNAi components is enhanced or protein stability increases during mating. Interestingly, more abundant RNAi components were also observed when cells were grown on V8 mating medium without sexual partners, but the silencing rate remained low. Thus, the overexpressed RNAi machinery seems necessary but not sufficient for efficient SIS, which requires the sexual process. The model we propose here is that sexual reproduction enhances the generation or detection of aberrant RNAs derived from the multicopy transgenic locus, a prerequisite of efficient PTGS, by a genome surveillance mechanism, resulting in increased silencing efficiency. Interestingly, there are other examples of transgene silencing in C. neoformans that are more readily detectable during mitotic growth (P Wang and J Heitman, unpubl.). It will be of interest to investigate in these cases if silencing occurs at an even higher frequency during sexual reproduction, and what factors affect the efficiency of transgene-induced silencing during mitotic versus meiotic reproduction.
Given the elevated protein, but not mRNA, levels of RNAi components during sexual reproduction that contribute to SIS, it will be of interest to identify potential RNA-binding proteins involved in translational regulation. Characterizing the 5′ and 3′ untranslated region (UTR) regions of these genes may reveal common regulatory motifs. In a broader context, this finding may lead to the discovery of a specific set of genes that are translationally regulated during the sexual cycle or under stress. Functionally related transcripts can be translationally coregulated via specific RNA–protein interactions. For example, in myeloid cells, a group of inflammatory genes are translationally repressed by the IFN-γ-activated inhibitor of translation (GAIT) complex, which binds to their 3′ UTRs and inhibits translation (Mukhopadhyay et al. 2009). The fact that all of the RNAi components examined here are translationally induced supports the hypothesis that a coregulation mechanism may operate in C. neoformans as an RNA operon (Keene 2007).
The RNAi components are localized to discrete foci in cells in structures reminiscent of P bodies that are centers for RNA processing and degradation (Eulalio et al. 2007). Similar localization patterns were observed in our studies. We documented that Ago1, Dcr1, and Dcr2 are associated with P bodies, while Rdp1 is nuclear, suggesting that RNA silencing and RNA decay pathways may take place in the same cellular compartment. However, the initial steps of generating dsRNA by RdRP during the RNAi pathway likely occur in the nucleus.
SIS may be triggered prior to meiosis and does not require an unpaired DNA structure
When does SIS occur during the sexual cycle? By demonstrating that URA5 is silenced in some proportion of blastospores, cells that are generated post-cell–cell fusion but prior to nuclear fusion and meiosis, we conclude that SIS can operate prior to meiosis. Furthermore, in a unilateral a rdp1 × wild-type α cross, the efficiency of SIS was significantly reduced from ∼50% to 8.3%. The requirement of Rdp1 for efficient SIS in a unilateral cross suggests that being RNAi-deficient in the a nucleus has a significant impact on the efficiency of silencing that is not complemented by the wild-type copy of RDP1 from the opposite mating partner. This may represent a mechanism by which two nuclei silence each other's resident transposons prior to nuclear fusion.
When the SXI2a-URA5 transgene is present at the same locus in both parents, a high frequency of silencing was observed (82.5%), indicating that SIS is not triggered by an unpaired DNA structure and is therefore mechanistically distinct from MSUD. Nonetheless, we cannot exclude that C. neoformans may have another silencing pathway similar to MSUD.
siRNA and transposon silencing
Profiling small RNAs from C. neoformans revealed interesting features shared with siRNAs of other species. First, endogenous small RNAs from the C. neoformans genome were enriched mostly for those 21–23 nt in length (Fig. 4). This length bias is common to siRNAs and microRNAs (miRNAs) found in plants, animals, and fungi that are typically 21–25 nt (Moazed 2009). Second, most of the C. neoformans siRNAs have a 5′ uridine. This feature is shared with miRNAs and piRNAs identified from many different species and also observed in the endogenous siRNAs recently characterized in S. castellii and N. crassa (Grimson et al. 2008; Drinnenberg et al. 2009; Ghildiyal and Zamore 2009; Lee et al. 2009; Malone and Hannon 2009). Third, the vast majority of siRNAs derive from the antisense strands of the originating genes that was also observed in S. castellii siRNAs targeting transposons and coding sequences (Drinnenberg et al. 2009), reflecting that dsRNA is generated as an intermediate in RNAi pathways and antisense small RNAs might be more stable.
The role of small RNAs as a mechanism to defend genomes against mobile elements is broadly conserved among RNAi pathways (Brennecke et al. 2007; Malone and Hannon 2009). Here, we identified abundant small RNAs that map to repetitive transposon clusters in the C. neoformans genome, indicative of an analogous role in transposon control. This subset of siRNAs is similar to piRNAs in their function and genomic origin. piRNA has only been identified in animal germ cells and is generated through a pathway distinct from siRNAs and miRNAs. The piwi-clade Argonaute is not present in fungi, so it is possible that endogenous siRNA is an even more ancestral mechanism to defend against mobile elements. However, it is known that plants, which lack Piwi proteins, have evolved a specialized RNAi system that generates a distinct class of small RNAs for transposon control, and this is accomplished through specialized Dicer and Argonaute proteins (Slotkin and Martienssen 2007). H99 has only one Argonaute gene, but, in some other species of C. neoformans (serotype D strain JEC21), two Argonaute genes were identified. It will be of interest to examine if the other Ago functions in a distinct siRNA pathway, which may similarly control transposons.
A major finding of this study is that a group of LTR retrotransposons located in the presumptive centromeric regions were highly induced in the RNAi-deficient strains under nutrient limitation/mating conditions but were not expressed on rich growth medium or when RNAi is functional (Table 4). By ChIP analyses, we found that the levels of H3K9me2 at Tcn1 decreased in mating strains, whereas the abundance of RNA Pol II increased and was comparable between wild-type and rdp1 mating strains (Fig. 7), suggesting that Tcn1 is actively transcribed during mating but is silenced post-transcriptionally by an RNAi pathway. We did not find a relationship between RNAi and heterochromatin formation for the genomic regions examined, although this is well established for S. pombe centromeric regions (Volpe et al. 2002). These findings reveal a salient feature: Retrotransposons in centromeric regions are potentially derepressed under mating/nutritional limitation. Combined with the fact that a higher transposition rate was observed in the rdp1 mutant progeny, this leads to the proposal that an efficient silencing mechanism is necessary to defend the genome and reduce a potential mutational burden during sex. Thus, the robust nature of highly efficient SIS could be one strategy that cells deploy to silence retrotransposons. Notably, by microarray analysis, the MFα pheromone gene and RPL22α gene were overexpressed in rdp1 × rdp1 mating compared with wild type, which may reflect a role for SIS in dampening expression of genes in MAT-linked repetitive sequences (Supplemental Table 1).
In summary, we discovered and characterized RNAi-mediated SIS as a genome defense mechanism that silences transgene arrays in C. neoformans. In Cryptococcus, introduced DNA often forms long linear multimers that can integrate into the genome, and the formation of tandem arrays may facilitate silencing of foreign DNA as a genome defense mechanism. Moreover, the highly expressed retrotransposons and increased transposition rate identified in rdp1 mutant strains during the sexual cycle suggest that SIS may serve a role broader than merely silencing introduced transgenes. The difference between transgene and transposon silencing is that a complete sexual cycle is required for elevated transgene silencing but may not be necessary for transposon silencing (Supplemental Table 4). This difference could be linked to the major question that needs to be addressed in future studies: how SIS is initiated. In transgene arrays, overexpression of the RNAi machinery that occurs early during mating (or on V8 mating medium) is insufficient for efficient SIS. Accordingly, stages of the sexual cycle involving the sequence homology search that mediates meiotic recombination may accelerate the formation of aberrant DNA–RNA hybrids in tandem repeat sequences, possibly producing templates for Rdp1, thus activating silencing. At transposons, the transcription rate increases when mating begins, and the resulting overexpression may trigger recruitment of Rdp1 and initiate silencing. Thus, in this scenario, the abundant RNAi machinery may suffice to silence overexpressed transposons. Our findings illustrate a novel role for RNAi during the sexual cycle, functioning to silence transposons and repetitive sequences, and thereby promoting genome integrity in C. neoformans.
Materials and methods
Strains and media
The strains and plasmids used in this study are listed in Supplemental Table 2. Yeast cells were grown and maintained on YPD media. SD medium lacking uracil and synthetic medium containing 5-FOA (1 g/L) were used to test whether isolates of interest are auxotrophic for uracil. Mating of C. neoformans was conducted on 5% V8 juice agar medium (pH 5) or MS medium minus sucrose (Sigma-Aldrich), as reported previously (Xue et al. 2007).
Genetic cross and spore dissection
Strains of interest were cocultured on MS medium for 2 wk at 25°C in the dark. Basidiospores produced at the edges of mating colonies were randomly isolated with a micromanipulator as described (Hsueh et al. 2006). After the spores germinated, the colonies were tested for growth on SD-ura and YPD medium. DNA was extracted and the presence or absence of the SXI2a-URA5 transgene was determined by PCR with primer pairs JOHE16835/JOHE16836. Mating assays were conducted to determine the mating type of each progeny. To isolate blastospores, mating colonies were observed under a microscope, and regions of mating hyphae associated with abundant blastospores were isolated and transferred to YPD medium. The blastospores were then randomly isolated with a micromanipulator as described above.
RNAPol-ChIP assay
Exponentially growing cultures of strains JF289 (Ura+), YPH789 (meiotic ura−), and YPH785 (mitotic ura−) were adjusted to 1% formaldehyde and incubated for 20 min at room temperature with gentle shaking. Cells were harvested in prechilled bottles and washed twice with ice-cold phosphate-buffered saline. Chromatin was prepared as described previously, and the entire procedure was performed at 4°C as described in the Supplemental Material (Sandoval et al. 2004). Phospho-RNA Pol II (S2) antibody (Bethyl Laboratories) was used for immunoprecipitation. Immunocomplexes were recovered by adding 30 μL (bead volume) of protein G Dynabeads (Invitrogen). Input, immunoprecipitation, and NoAb fractions were analyzed by PCR and quantitative PCR with the primers pairs listed in Supplemental Table 3.
Microarray and data analysis
To obtain total RNA for microarray analysis, wild-type and rdp1 mutant cells from mating and nonmating conditions were harvested from V8 medium (24 h of incubation) and 5 mL of YPD overnight cultures, respectively. Three independent cultures for each strain or mating were prepared for total RNA isolation as biological replicates. Total RNAs were extracted with Trizol as described in the Supplemental Material. cDNA was synthesized by using AffinityScript reverse transcriptase (Stratagene), Cy3/Cy5 labeled, and hybridized to C. neoformans 70-mer microarray slides (version 2; Washington University, St. Louis, MO). The microarray used in this study was developed by the Cryptococcus Community Microarray Consortium with financial support from individual researchers and the Burroughs Wellcome Fund (http://genome.wustl.edu/services/microarray/cryptococcus_neoformans). Hybridized arrays were washed as described before (Ko et al. 2009) and then scanned with a GenePix 4000B scanner (Axon Instruments). The signals were processed with GenePix Pro (version 4.0). Three independent DNA microarrays with three independent biological replicates were performed. For statistical analysis, data transported from GenePix software were analyzed with GeneSpring software (Agilent) by employing Lowess normalization, reliable gene filtering, ANOVA analysis (P < 0.05), and Microsoft Excel software.
Small RNA library preparation and deep sequencing
Total RNA was extracted from the silenced strain YPH231, wild-type mating strains (H99 α × KN99 a), and a rdp1 × α rdp1 mating strains. Approximately 18-nt to ∼30-nt RNAs were cloned by methods described previously (Lau et al. 2001; Umbach and Cullen 2010). Briefly, total RNA was size-fractionated on a 15% TBE-urea gel. Eighteen-nucleotide to 30-nt small RNAs were excised and ligated to 3′ and 5′ linkers. cDNA libraries were submitted to the Duke IGSP DNA Sequencing Facility for Illumina deep sequencing. Customized Perl scripts were used to remove the linker sequence from the resulting sequence, and small RNA reads were mapped to the H99 genome database.
Acknowledgments
We thank Bryan Cullen and Jennifer Umbach for guidance and reagents for constructing the small RNA cDNA libraries and suggestions on deep sequencing analysis. We are indebted to Maria Cardenas for key advice about the ChIP protocol, and thank Arno Greenleaf and Beth Sullivan for antibodies to RNA Pol II and antibodies to H3K9me2. We thank Soo Chan Lee for fluorescence microscopy assistance, Kasey Carroll for technical assistance, Sun Bahn and Guilhem Janbon for strains and discussion, and Valerie Knowlton at the North Carolina State University EM facility for SEM assistance. We thank Soo Chan Lee, Edmond Byrnes, Cecelia Shertz, and Sun Sheng for their critical reading and comments on the manuscript. This work is supported by NIH RO1 grant AI39115 from NIAID to J.H. Y-.P.H. and X.W. contributed equally to this work. Y-.P.H. contributed the initial discovery, observations, and analysis of SIS. X.W. further investigated the mechanisms of SIS. Y-.P.H. constructed the small RNA libraries with assistance and advice from R.S. in B. Cullen's laboratory, and W.L. performed the bioinformatics analysis. X.W., W.L., and Y-.P.H. all participated in demonstrating a role for SIS in transposon control. A.F. contributed technical assistance. X.W., Y-.P.H., and J.H. wrote the paper. Y-.P.H., X.W., and J.H. designed the experiments. All authors discussed the results and implications and commented on the manuscript at all stages.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1970910.
Supplemental material is available at http://www.genesdev.org.
References
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T 2003. The small RNA profile during Drosophila melanogaster development. Cell 5: 337–350 [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, et al. 2004. Lessons from the genome sequence of Neurospora crassa: Tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68: 1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master ,regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D 2008. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 15: 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri EB, Singer MJ, Selker EU 1991. Recurrence of repeat-induced point mutation (RIP) in Neurospora crassa. Genetics 127: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C, Azzalin G, Macino G, Cogoni C 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev 16: 790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C, Pallotta M, ReFalo P, Sachs MS, Vayssie L, Macino G, Cogoni C 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol 24: 2536–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci 94: 10233–10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G 1999a. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169 [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G 1999b. Homology-dependent gene silencing in plants and fungi: A number of variations on the same theme. Curr Opin Microbiol 2: 657–662 [DOI] [PubMed] [Google Scholar]
- Cottrell TR, Doering TL 2003. Silence of the strands: RNA interference in eukaryotic pathogens. Trends Microbiol 11: 37–43 [DOI] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Séraphin B 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biochem 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz MC, Cavallo LM, Gorlach JM, Cox G, Perfect JR, Cardenas ME, Heitman J 1999. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol 19: 4101–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Zalevsky J, Colaiacovo MP, Villeneuve AM 2000. Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev 14: 1578–1583 [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP 2009. RNAi in budding yeast. Science 326: 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci V, Macino G 2007. Quelling: Post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr Opin Microbiol 10: 199–203 [DOI] [PubMed] [Google Scholar]
- Galagan JE, Selker EU 2004. RIP: The evolutionary cost of genome defense. Trends Genet 20: 417–423 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ 2002. RNA interference. Nature 418: 244–251 [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Idnurm A, Heitman J 2006. Recombination hotspots flank the Cryptococcus mating-type locus: Implications for the evolution of a fungal sex chromosome. PLoS Genet 2: e184 doi: 10.1371/journal.pgen.0020184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Fraser JA, Heitman J 2008. Transitions in sexuality: Recapitulation of an ancestral tri- and tetrapolar mating system in Cryptococcus neoformans. Eukaryot Cell 7: 1847–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Boily MJ, Heitman J 2005. Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot Cell 4: 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Himmelreich U, Moyrand F, Improvisi L, Dromer F 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol Microbiol 42: 453–467 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Gilbert DE, Grady KL, Jorgensen RA 1987. T-DNA structure and gene expression in petunia plants transformed by Agrobacterium tumefaciens C58 derivatives. Mol Gen Genet 207: 478–485 [Google Scholar]
- Keene JD 2007. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Kelly WG, Aramayo R 2007. Meiotic silencing and the epigenetics of sex. Chromosome Res 15: 633–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH 1999. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner Syndrome helicase and RNaseD. Cell 99: 133–141 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Ko Y-J, Yu YM, Kim G-B, Lee G-W, Maeng PJ, Kim S, Floyd A, Heitman J, Bahn Y-S 2009. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell 8: 1197–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Laurie JD, Linning R, Bakkeren G 2008. Hallmarks of RNA silencing are found in the smut fungus Ustilago hordei but not in its close relative Ustilago maydis. Curr Genet 53: 49–58 [DOI] [PubMed] [Google Scholar]
- Lee DW, Pratt RJ, McLaughlin M, Aramayo R 2003. An argonaute-like protein is required for meiotic silencing. Genetics 164: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y 2009. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459: 274–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R 2003. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1: e67 doi: 10.1371/journal.pbio.0000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Cottrell TR, Pierini LM, Goldman WE, Doering TL 2002. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC 2002. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, et al. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307: 1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL 2009. The GAIT system: A gatekeeper of inflammatory gene expression. Trends Biochem Sci 34: 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki H 2005. RNA silencing in fungi: Mechanisms and applications. FEBS Lett 579: 5950–5957 [DOI] [PubMed] [Google Scholar]
- Nakayashiki H, Kadotani N, Mayama S 2006. Evolution and diversification of RNA silencing proteins in fungi. J Mol Evol 63: 127–135 [DOI] [PubMed] [Google Scholar]
- Nicolás FE, Torres-MartÌnez S, Ruiz-Vázquez RM 2009. Transcriptional activation increases RNA silencing efficiency and stability in the fungus Mucor circinelloides. J Biotechnol 142: 123–126 [DOI] [PubMed] [Google Scholar]
- Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J 16: 2576–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA 1997. Cosuppression in Drosophila: Gene silencing of alcohol dehydrogenase by white-Adh transgenes is polycomb dependent. Cell 90: 479–490 [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672 [DOI] [PubMed] [Google Scholar]
- Reese AJ, Doering TL 2003. Cell wall α-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol 50: 1401–1409 [DOI] [PubMed] [Google Scholar]
- Romano N, Macino G 1992. Quelling: Transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol 6: 3343–3353 [DOI] [PubMed] [Google Scholar]
- Ruff JA, Lodge JK, Baker LG 2009. Three galatose inducible promoters for use in C. neoformans var. grubii. Fungal Genet Biol 46: 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J, Rodriguez JL, Tur G, Serviddio G, Pereda J, Boukaba A, Sastre J, Torres L, Franco L, Lopez-Rodas G 2004. RNAPol-ChIP: A novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res 32: e88 doi: 10.1093/nar/gnh091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU 1997. Epigenetic phenomena in filamentous fungi: Useful paradigms or repeat-induced confusion? Trends Genet 13: 296–301 [DOI] [PubMed] [Google Scholar]
- Selker EU, Cambareri EB, Jensen BC, Haack KR 1987. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell 51: 741–752 [DOI] [PubMed] [Google Scholar]
- Sen GL, Blau HM 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol 7: 633–636 [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2009. On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR 2010. In-depth analysis of Kaposi's Sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J Virol 84: 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Beclin C, Fagard M 2001. Post-transcriptional gene silencing in plants. J Cell Sci 114: 3083–3091 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL 1994. RNA-directed de novo methylation of genomic sequences in plants. Cell 76: 567–576 [DOI] [PubMed] [Google Scholar]
- Xue C, Tada Y, Dong X, Heitman J 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1: 263–273 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]
- Zofall M, Grewal SI 2006. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol 71: 487–496 [DOI] [PubMed] [Google Scholar]