Abstract
Ca2+-sensitive NFAT (nuclear factor of activated T-cells) transcription factors are implicated in many pathophysiological processes in different cell types. The precise control of activation varies with NFAT isoform and cell type. Here we present feasibility of an in vivo assay (NFAT-RFP) that reports transcriptional activity of NFAT via expression of red fluorescent protein (RFP) in individual cells. This new tool allows continuous monitoring of transcriptional activity of NFAT in a physiological context in living cells. Furthermore, NFAT-RFP can be used simultaneously with NFAT-GFP fusion proteins to monitor transcriptional activity and subcellular localization of NFAT in the same cell.
Introduction
Transcription factors of the NFAT family are present in many cell types and regulate numerous physiological processes. In the vascular endothelium NFAT signalling has been implicated in vascular development during embryogenesis, angiogenesis, proliferation of valve endothelial cells and in inflammatory processes (for reference see Nilsson et al. 2008; Rinne et al. 2009). A rise in intracellular Ca2+ ([Ca2+]i) activates the phosphatase calcineurin, inducing dephosphorylation, nuclear translocation and transcriptional activity of NFAT (Crabtree & Olson, 2002; Wu et al. 2007). Ca2+ signals that activate NFAT have been analysed in detail in non-excitable (Macian, 2005; Rinne et al. 2009) and excitable cells (Hernandez-Ochoa et al. 2007; Rinne et al. 2010), but it needs to be substantiated whether they also induce transcriptional activity of NFAT in living cells.
The main advantage of fluorescence-based assays over the traditional luciferase (Luc) assays is their application to individual living cells, because they do not require cell lysates for quantification. By controlling expression of red fluorescent protein (RFP) with an NFAT sensitive interleukin-2 (IL2) promoter, we were able to monitor transcriptional activity of NFAT in living cells with fluorescence microscopy. We first validated this new tool (termed NFAT-RFP) in HEK 293 cells. Enhanced NFAT activity, induced by expression of wild-type NFATc1 (NFATc1-wt) or constitutively active NFATc1 (NFATc1-ca) resulted in corresponding increases in RFP positive cells and RFP intensity. Transcriptional activity was confirmed with a classical luciferase assay under the same experimental conditions. Using adenoviral expression of NFAT-RFP, we were able to measure NFAT-sensitive transcription of endogenous NFAT and of NFATc1-GFP in response to agonist stimulation in vascular endothelial cells.
Methods
Construction of plasmids and adenoviruses
NFAT-RFP-reporter plasmid pShuttle-NFAT-RFP and adenovirus Ad-NFAT-RFP
The NFAT-sensitive IL2 promoter stemming from Addgene plasmid no. 10959 (Addgene Inc., Cambridge, MA, USA) was amplified by PCR and cloned into the plasmid pShuttle using NotI and EcoRV restriction sites. A cDNA fragment encoding for monomeric red fluorescent protein (RFP) and a polyadenylation (poly A) signal was amplified from plasmid no. 13032 (Addgene Inc.) using PCR and cloned behind the IL2 promoter using EcoRV and SalI restriction sites, yielding pShuttle-NFAT-RFP. The primer sequences were 5′-GGAGACAUGCGGCCGCCCATTCAGGCTGCGCAACT-3′ and 5′-GGGAAAGUGATATCTTTACCAACAGTACCGGAATG-3′ (IL2 promoter); and 5′-GGAGACAUGATATCATGGCCTCCTCCGAGGACGTC-3′ and 5′-GGGAAAGUGTCGACTCCCCAGCATGCCTGCTATTGTC-3′ (RFP-polyA). The adenovirus Ad-NFAT-RFP was then generated using the pAdEasy system (see below).
NFAT-Luciferase reporter plasmid pShuttle-NFAT-Luc
To generate the plasmid pShuttle-NFAT-Luc (firefly luciferase), the reporter cassette NFAT-Luc, containing an interleukin IL2-promoter, cDNA encoding firefly luciferase and a translation termination signal was excised from plasmid no. 17870 (Addgene Inc.) using KpnI and SalI restriction sites and directly cloned into the plasmid pShuttle. The adenovirus Ad-NFAT-Luc was generated using the pAdEasy system (see below).
Plasmids encoding for NFATc1-wt and NFATc1-ca
The cDNAs encoding for wild-type NFATc1 (NFATc1-wt) or a Ca2+/calcineurin-insensitive, constitutively active mutant of NFATc1 (NFATc1-ca) were amplified via PCR using plasmids no. 11100 and no. 11702 as templates (Addgene Inc.) and cloned into pAdTrack using KpnI and HindIII restriction sites, yielding to pAdtrack-NFATc1-wt and pAdTrack-NFATc1-ca. The primer sequences for NFATc1 were 5′-CCTGGTACCATGACGGGGCTGGAGCAGG-3′ and 5′-CCTAAGCTTTCAGTAAAAACCTCCTCTCAGCTC-3′. The plasmid pAdTrack also expresses GFP as a marker protein independent from the gene of interest.
Ad-NFATc1-GFP
The adenovirus encoding NFATc1 fused to GFP was kindly provided by Jeffery D. Molkentin, Children's Hospital Medical Center, Cincinnati, USA.
Ad-VIVIT-GFP
The cDNA encoding for the NFAT-inhibitory peptide VIVIT fused to GFP was excised from the plasmid GFP-VIVIT (Addgene Inc., plasmid no. 11106) and cloned into pShuttle-CMV using NotI and KpnI restriction sides, yielding the pShuttle-CMV-VIVIT-GFP. The adenovirus Ad-VIVIT-GFP was then generated using the pAdEasy system (see below).
Ad-GFP
The adenovirus Ad-GFP, which expresses GFP alone (infection control), was constructed by homologous recombination of the plasmids pAdTrack and pAdEasy-1.
Generation of recombinant adenoviruses
Adenoviruses were constructed using transfer plasmids as described above with the pAdEasy system. The plasmids pShuttle, pShuttle-CMV, pAdtrack, pAdEasy-1 and all other required components were purchased from the ATCC (American Type Culture Collection, Manassas, VA, USA) and adenoviruses were generated as described in Luo et al. (2007).
Culture and transfection of HEK 293 cells
HEK 293 cells were kindly provided by Dr Ruben Markosyan, Rush University, Chicago, IL, and were cultured using Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin–streptomycin (all Mediatech Inc., Manassas, VA, USA). Cells were passaged on sterile glass coverslips and transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. For NFAT-RFP experiments, cells were co-transfected with 1 μg pShuttle-NFAT-RFP and 1 μg of pAdTrack (GFP control), pAdTrack-NFATc1-wt or pAdTrack-NFATc1-ca. For luciferase assays, cells were transfected as above, but pShuttle-NFAT-RFP was replaced with pShuttle-NFAT-Luc. To analyse the time dependence of the NFAT-RFP reporter response, cells were co-transfected with pAdtrack-NFATc1-ca and pShuttle-NFAT-RFP and analysed 2 h, 4 h, 8 h and 24 h after transfections. Otherwise cells were analysed 48 h after transfections.
Cell culture and adenoviral infections of vascular endothelial cells
Calf pulmonary artery endothelial (CPAE) cells were obtained from ATCC and cultured on glass coverslips as described previously (Rinne et al. 2009). Cells were co-infected with Ad-NFAT-RFP and either Ad-GFP, Ad-NFATc1-GFP or Ad-VIVIT-GFP and imaged 2 days after infection. Only single cells from non-confluent cultures were used for experiments. To maintain a uniform endothelial phenotype, only up to six cell passages were used.
Fluorescence microscopy
For laser scanning confocal imaging experiments (Bio-Rad Bioradiance 2000/MP) the cell culture medium was replaced with a Hepes-buffered salt solution (HBSS) containing (in mm): 135 NaCl, 4 KCl, 1 MgCl2, 2 CaCl2, 10 glucose and 10 Hepes; pH 7.3 (NaOH). GFP was excited at 488 nm (argon ion laser) and GFP emission was collected between 500 nm and 520 nm. RFP was excited at 535 nm using a He–Ne laser line and emission was collected at λ≥ 570 nm. To avoid bleed-through of GFP emission into the RFP channel, GFP and RFP images were taken separately, i.e. without simultaneous excitation of GFP and RFP. The mean intensity of RFP was averaged over the surface area of the cell (determined from the GFP positive cell surface) using the image analysis software Image J (Wayne Rasband, National Institutes of Health, Bethesda, MD, USA).
Luciferase assays
HEK 293 cells were transfected as described above. Two days after transfection, cells were lysed and luciferase activity was measured using the Luciferase Assay System (Promega, Sunnyvale, CA, USA) with a single tube luminometer (type Modulus, Turner Biosystems, Sunnyvale, CA, USA) according to the manufacturer's instructions.
Application of agonists, antagonists and ionomycin treatment
For short-term application, agonists and antagonists were prepared in HBSS and applied acutely. For overnight incubations, agonists and antagonists were added to the cell culture medium. To activate endogenous NFAT in endothelial cells infected with Ad-NFAT-RFP, cells were incubated with medium containing 10 μm ionomycin for 3 h. Images were taken 24 h later to allow for RFP expression.
Data analysis and representation
Data are presented as individual observations or as means ±s.e.m. and were statistically analysed using Student's t test. Unless stated otherwise, n represents the number of individual cells and differences were considered significant at P < 0.05.
Results
NFAT-RFP reports transcriptional activity of NFAT in HEK cells
NFAT-RFP was first validated in HEK 293 cells. Control cells expressing NFAT-RFP and GFP showed only barely detectable RFP expression 2 days after transfection (Fig. 1Ab). In contrast, coexpression of wild-type NFATc1, an isoform that displays basal activity (Shen et al. 2006), resulted in 5-fold higher expression of RFP (Fig. 1Ac and Ad). This effect was further enhanced in cells coexpressing constitutively active NFATc1 (NFATc1-ca) (∼20-fold higher than control, summary data Fig. 1B). Thus, NFAT-RFP reports transcriptional activity in a concentration (NFAT levels)-dependent manner. Similar results were obtained using an NFAT-sensitive luciferase assay (NFAT-Luc). Lysates of cells expressing NFATc1 resulted in a 2-fold higher luciferase expression compared to control, which could be further increased by NFATc1-ca (Fig. 1C). To analyse the time dependence of the reporting behaviour of NFAT-RFP in response to the duration of NFAT activity, we analysed RFP expression in cells that expressed NFATc1-ca at 2, 4, 8 and 24 h after transfection. Expression of NFATc1-ca was detectable 4 h after transfections (Fig. 1Da) and its expression increased with time. In NFATc1-ca/GFP-positive cells, NFAT-dependent expression of RFP was detected after 8 h, reaching a robust signal 24 h after transfections (Fig. 1Db).
Figure 1. Transcriptional activity of NFATc1 in HEK 293 cells.
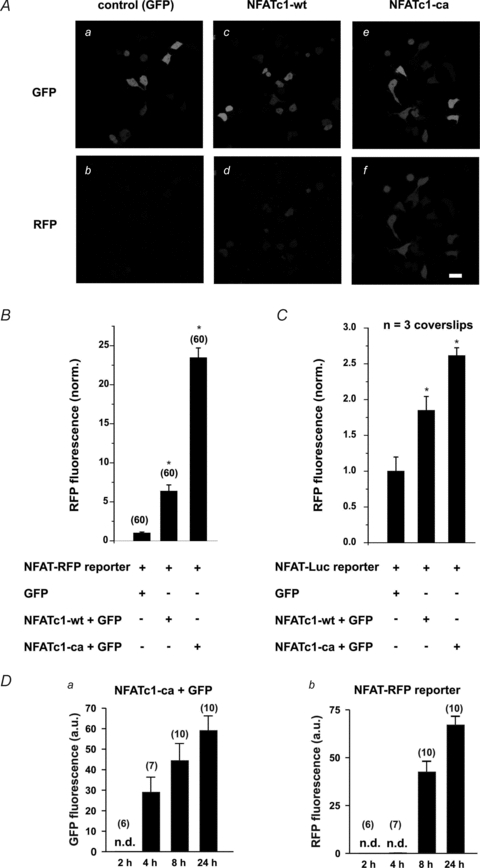
A, expression of NFATc1 wild-type (NFATc1-wt) resulted in detectable RFP expression (Ad) compared to control cells (GFP, Ab). This effect was further enhanced in the presence of constitutively active NFATc1 (NFATc1-ca) (Af). a, c and e, corresponding GFP images (transfection control). Scale bars: 30 μm. B, summary data indicating an increase in RFP fluorescence corresponding to enhanced NFATc1 activity (NFATc1-ca > NFATc1-wt > GFP-control). C, summary data of a luciferase assay (NFAT-Luc) under the same experimental conditions (3 coverslips per condition). Data were normalized to the average RFP fluorescence measured in cells expressing only GFP and NFAT-RFP (B) or NFAT-Luc (C). D, time dependence of transcriptional activity reported by NFAT-RFP in response to stimulus duration. In cells co-expressing NFATc1-ca and NFAT-RFP, GFP (a measure of NFATc1-ca expression) was detected after 4 h and further increased with time (Da). Db, in GFP-positive cells, RFP was detected after 8 h, further increasing over 24 h. Note that the expression of NFATc1-ca precedes the NFAT-RFP response by 4 h. Times indicated are hours after transfections. Numbers in parentheses indicate the number of individual cells tested. n.d.: not detectable.
These data suggest that NFAT-RFP represents a sensitive tool to monitor transcriptional activity of NFAT in intact living cells with temporal information.
Transcriptional activity of NFATc1-GFP in vascular endothelial cells
In vascular endothelial cells, extracellular application of the vasoactive agonist ATP or treatment with the Ca2+ ionophor ionomycin generates a rise in [Ca2+]i that induces nuclear localization of NFAT (for details see Rinne et al. 2009). Short application of extracellular ATP (5 μm for 5 min, [Ca2+]o= 2 mm) or treatment of the cells with ionomycin (10 μm for 5 min, [Ca2+]o= 2 mm) induced translocation of the fusion protein NFATc1-GFP from the cytoplasm to the nucleus (Fig. 2A: before (a) and 30 min after (b) ATP application; before (c) and 30 min after (d) ionomycin treatment).
Figure 2. Transcriptional activity of NFATc1-GFP and endogenous NFAT in vascular endothelial cells.
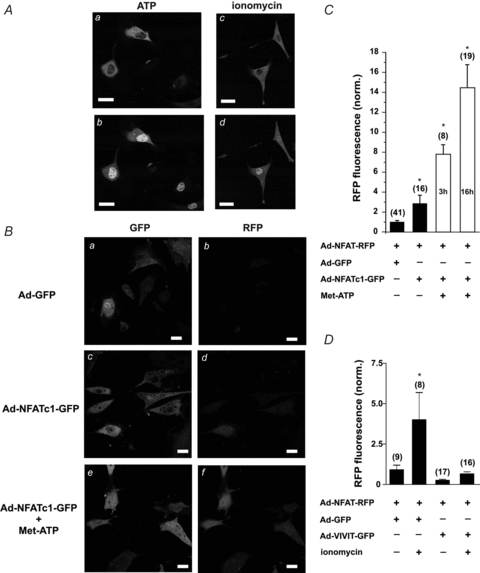
A, nuclear localization of NFATc1-GFP fusion proteins was induced by extracellular ATP or ionomycin treatment. Localization of NFATc1-GFP before (a) and 30 min after (b) application of 5 μm ATP. Treatment of cells with ionomycin (10 μm) induced nuclear localization of NFATc1-GFP. Localization of NFATc1-GFP before (c) and 30 min after (d) ionomycin treatment. B, transcriptional activity of NFATc1 in CPAE cells expressing NFAT-RFP. Expression of NFATc1-GFP induced 3-fold higher expression of RFP (Bd) compared to control (cells expressing GFP, Bb). This effect was further enhanced in a time-dependent manner, when CPAE cells were stimulated with 100 nm Met-ATP for 3 h or overnight (16 h; Bf), which also induced nuclear localization of NFATc1-GFP (Bc vs. Be). a, c and e, GFP images as infection control. Scale bars: 30 μm. C, summary data from B. D, activation of endogenous NFAT. Stimulation with ionomycin induced detectable expression of RFP in CPAE cells that express GFP plus NFAT-RFP (first two columns). The same stimulation failed to induce RFP expression in the presence of VIVIT-GFP, an inhibitor of calcineurin (last two columns). Numbers in parentheses indicate the number of individual cells tested. Data were normalized to the average RFP fluorescence measured in unstimulated cells expressing only Ad-GFP and Ad-NFAT-RFP.
To test, whether these stimuli would also induce transcriptional activity of NFATc1, CPAE cells were co-infected with Ad-NFAT-RFP and Ad-GFP or Ad-NFATc1-GFP 2 days before RFP expression was analysed. Control cells expressing GFP (Fig. 2Ba) showed only modest RFP expression (Fig. 2Bb). Expression of NFATc1-GFP (Fig. 2Bc) resulted in a 3-fold higher expression of RFP (Fig. 2Bd, summary data Fig. 2C), which was further increased to about 14-fold by overnight (16 h) stimulation with the stable ATP analogue Met-ATP (2-(methylthio)adenosine 5′-triphosphate tetrasodium salt, Sigma, St Lois, MO, USA; 100 nm; Fig. 2Bf). A shorter (3 h) exposure to Met-ATP, however, only led to an approximately 8-fold increase in RFP fluorescence, indicating that NFAT-RFP is suitable for the temporal studies of transcriptional activity. Note that stimulated cells also display GFP-positive nuclei under these conditions (Fig. 2Be, as compared to Fig. 2Bc). These data indicate that application of ATP induced both nuclear translocation (NFATc1-GFP) and transcriptional activity of NFATc1. NFAT-RFP was measured simultaneously.
Transcriptional activity of endogenous NFAT in CPAE cells
NFAT-RFP was also sensitive to endogenous NFAT activity. CPAE cells were co-infected with Ad-NFAT-RFP and Ad-GFP or Ad-VIVIT-GFP, a peptide that prevents NFAT–calcineurin interaction (Aramburu et al. 1999). Twenty-four hours after infections, endogenous NFAT was activated by ionomycin treatment (10 μm for 3 h) and expression of RFP was analysed 24 h later. In cells expressing GFP, ionomycin treatment induced a 4-fold higher RFP expression compared to the corresponding group of untreated cells (Fig. 2D, first two columns). In contrast, ionomycin failed to induce RFP expression in cells that expressed VIVIT-GFP (Fig. 2D, last two columns). These data suggest that NFAT-RFP also represents a sensitive tool to analyse in vivo transcriptional activity of endogenous NFAT in individual cells.
Discussion
Here we demonstrate feasibility of a novel assay to measure time- and stimulus strength-dependent transcriptional activity of NFAT in living cells (NFAT-RFP). Based on expression of RFP under control of an NFAT-sensitive promoter, this assay displayed dose-dependent sensitivity to different levels of NFAT activation in HEK 293 cells that were induced by expression of wild-type NFATc1 or constitutively active NFATc1. NFAT-RFP responsiveness was similar to that of a classical luciferase assay (NFAT-Luc). Furthermore, by incorporation of NFAT-RFP into a recombinant adenovirus, we were able to measure activity of endogenous NFAT or NFATc1-GFP fusion proteins in vascular endothelial cells. The latter was used to monitor agonist-induced transcriptional activity of NFAT and subcellular localization of NFAT-GFP simultaneously. This approach might help to identify mechanisms or stimuli that activate NFAT in cell types, where Ca2+ signals are well defined but details about NFAT activation are still elusive, such as adult cardiac myocytes (Molkentin, 2006).
Acknowledgments
This work was supported by grants from the National Institutes of Health (HL62231 and HL80101 to L.A.B.) and the American Heart Association (0820080Z to A.R.).
Glossary
Abbreviations
- CPAE cells
calf pulmonary artery endothelial cells
- GFP
green fluorescent protein
- HBSS
Hepes-buffered salt solution
- IL2
interleukin-2
- Luc
luciferase
- Met-ATP
2-(methylthio)adenosine 5′-triphosphate
- NFAT
nuclear factor of activated T-cells
- RFP
red fluorescent protein
Author contributions
Both authors contributed to the design of the study, analysis and interpretation of the data, and writing of the manuscript. A.R. conducted the experiments.
References
- Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(Suppl):S67–79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- Hernandez-Ochoa EO, Contreras M, Cseresnyes Z, Schneider MF. Ca2+ signal summation and NFATc1 nuclear translocation in sympathetic ganglion neurons during repetitive action potentials. Cell Calcium. 2007;41:559–571. doi: 10.1016/j.ceca.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, Vogelstein B, He TC. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- Molkentin JD. Dichotomy of Ca2+ in the heart: contraction versus intracellular signaling. J Clin Invest. 2006;116:623–626. doi: 10.1172/JCI27824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson LM, Nilsson-Ohman J, Zetterqvist AV, Gomez MF. Nuclear factor of activated T-cells transcription factors in the vasculature: the good guys or the bad guys? Curr Opin Lipidol. 2008;19:483–490. doi: 10.1097/MOL.0b013e32830dd545. [DOI] [PubMed] [Google Scholar]
- Rinne A, Banach K, Blatter LA. Regulation of nuclear factor of activated T cells (NFAT) in vascular endothelial cells. J Mol Cell Cardiol. 2009;47:400–410. doi: 10.1016/j.yjmcc.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne A, Kapur N, Molkentin JD, Pogwizd SM, Bers DM, Banach K, Blatter LA. Isoform- and tissue-specific regulation of the Ca2+-sensitive transcription factor NFAT in cardiac myocytes and in heart failure. Am J Physiol Heart Circ Physiol. 2010;298:H2001–2009. doi: 10.1152/ajpheart.01072.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T, Liu Y, Cseresnyes Z, Hawkins A, Randall WR, Schneider MF. Activity- and calcineurin-independent nuclear shuttling of NFATc1, but Not NFATc3, in adult skeletal muscle fibers. Mol Biol Cell. 2006;17:1570–1582. doi: 10.1091/mbc.E05-08-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Peisley A, Graef IA, Crabtree GR. NFAT signaling and the invention of vertebrates. Trends Cell Biol. 2007;17:251–260. doi: 10.1016/j.tcb.2007.04.006. [DOI] [PubMed] [Google Scholar]


