Abstract
Flaviviruses have a single-strand, positive-polarity RNA genome that encodes a single polyprotein. The polyprotein is comprised of seven nonstructural (NS) and three structural proteins. The N- and C-terminal parts of NS3 represent the serine protease and the RNA helicase, respectively. The cleavage of the polyprotein by the protease is required to produce the individual viral proteins, which assemble a new viral progeny. Conversely, inactivation of the protease blocks viral infection. Both the protease and the helicase are conserved among flaviviruses. As a result, NS3 is a promising drug target in flaviviral infections. This article examines the West Nile virus NS3 with an emphasis on the structural and functional parameters of the protease, the helicase and their cofactors.
Keywords: dengue virus, flavivirus, helicase, NS2B, NS3, NS4A, protease, West Nile virus
Flaviviral polyprotein precursor
The Flaviviridae family currently comprises over 70 viruses, including mosquito-borne West Nile virus (WNV) and Kunjin virus (a subtype of WNV endemic to Oceania), yellow fever virus and dengue virus (DV), and tick-borne Japanese encephalitis virus (JEV). The Flavivirus genus viruses are responsible for significant human disease and mortality. The WHO estimates that there are multimillion annual cases of DV type 1–4 (DV1–4), 200,000 annual cases of yellow fever virus and 50,000 annual cases of JEV worldwide. WNV was first isolated in 1937 in the West Nile district of Uganda. Since 1999, when the virus was identified in the USA, the virus has spread rapidly throughout the country [1]. WNV has been detected in 46 states of the USA. According to the US CDC, the virus has already infected 30,000 people and has been the cause of approximately 1150 deaths (1999–2008). WNV may cause serious CNS damage unless specific treatment is administered [2,3]. There is a significant level of probability that the number of flaviviral infections will grow and that their geographical incidence will spread as the continued warming of the planet will provide a more extensive and benign environment for the flavivirus-carrying mosquito. To date, there is no specific and effective therapy available for any flavivirus infection.
Following infection of the host, the flavivirus positive strand 11-kb RNA genome is transcribed into a negative-strand RNA. The daughter genomic RNA is then synthesized using a negative-strand RNA template. Reports of sequence analysis of several flavivirus RNAs, including the yellow fever virus genome [4], DV4 [5,6], DV2 [7–10], Kunjin virus [11] and WNV [12], firmly established that flavivirus genomes share similar genomic organization (Figure 1). Naturally, the respective individual flaviviral proteins are also homologous across the family (Figure 2). As a result, the fundamental structural and regulatory parameters of the individual flaviviral proteins are also similar but not identical.
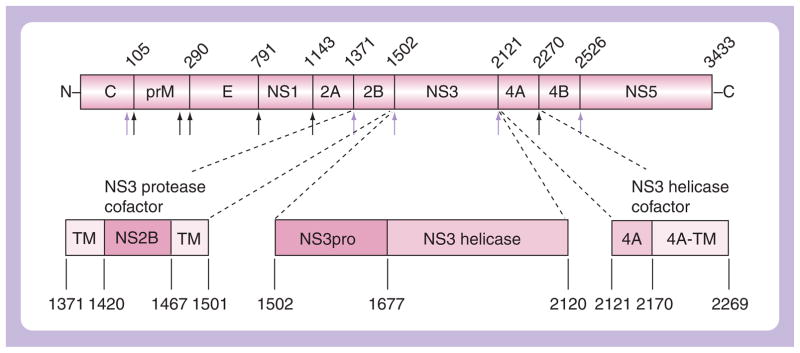
Figure 1. Organization of the capsid–membrane–envelope–NS1–NS2A–NS2B–NS3–NS4A–NS4B–NS5 polyprotein precursor, showing cleavage sites by the viral NS2B–NS3pro (gray arrows) and host cell secretase and furin (black arrows), with detail of the NS2B and NS3 sequences.
The hydrophilic central region of the NS2B cofactor is gray. The West Nile virus residue numbering is shown above the polyprotein structure.
C: Capsid; E: Envelope; M: Membrane; NS: Nonstructural; TM: Transmembrane domain.
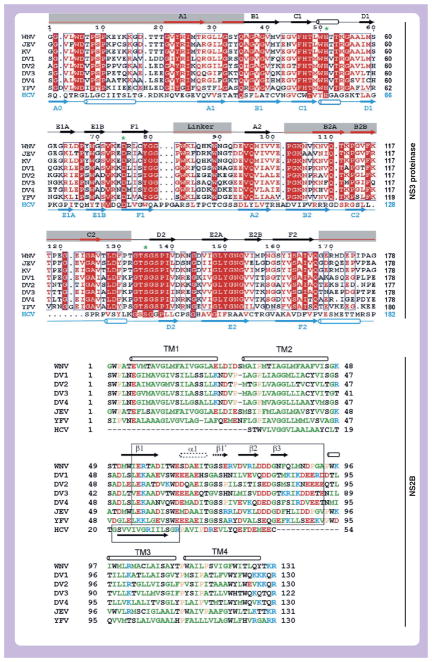
Figure 2. NS3 sequences of the flaviviruses.
(A) Sequence alignment of the NS3pro domain. Sections highlighted in red indicate identity; red letters indicate homology. Secondary structure elements above the sequences are for the WNV NS2B–NS3pro; those below are for the HCV NS3pro–NS4A (PDB entry 1JXP). Gray rectangles highlight regions where the folds of the WNV NS2B–NS3pro and the HCV NS3pro-NS4A differ. The residues of the Asp-His-Ser catalytic triad are indicated by green asterisks. (B) Sequence alignment of flaviviral NS2B and HCV NS4A. Boxes indicate minimal cofactor segments required for activation of NS3pro in vitro. TM1–TM4 are predicted transmembrane regions. Hydrophobic and aromatic residues are green, polar are black, acidic in red and basic in blue. Secondary structure elements are for the WNV NS2B–NS3pro–aprotinin complex; the alternate elements (α1 and β19) found in the inhibitor-free WNV and DV NS2B–NS3pro are drawn with dotted lines.
DV: Dengue virus; JEV: Japanese encephalitis virus; KV: Kunjin virus; TM: Transmembrane; WNV: West Nile virus; YFV: Yellow fever virus.
The genomes of flaviviruses are translated into polyproteins, which then undergo proteolytic processing. This proteolytic processing takes place both cotranslationally and post-translationally, and it involves both the host and viral proteases. As a result of this extensive proteolytic processing, the polyprotein precursor is transformed into mature viral proteins [13,14]. The genomic flaviviral RNA encodes a polyprotein precursor that consists of three structural proteins (capsid, membrane and envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) arranged in the order capsid protein–membrane protein–envelope protein–NS1–NS2A–NS2B–NS3–NS4A–NS4B–NS5 (Figure 1). Crawford et al. provided the early experimental evidence that flavivirus polyproteins are synthesized as a large precursor polyprotein, which are subsequently processed into mature polypeptides [15]. The precursor is inserted into the endoplasmic reticulum membrane and processed by the host cell and viral proteases to transform the precursor into individual, functionally potent proteins.
During evolution, viruses usurped multiple host cell components to maintain infectivity. Thus, flaviviruses employ human proteases (furin and secretase), in addition to the viral NS3 protease (NS3pro), to transform the polyprotein precursor into mature viral proteins [16–19]. It is also likely that flaviviruses efficiently use human miRNAs to regulate the genomic RNA translation and to maintain the required balance between the number of the early viral proteins, such as the capsid, membrane and envelope proteins, and the late proteins, including the NS5 RNA polymerase. If the balance between the early and the late genes is improperly maintained, the virus propagation would not be as efficient as it is.
NS3 protease–helicase protein
The full-length viral NS3 peptide sequence represents a multifunctional protein in which the N-terminal residues encode serine protease (NS3pro) and the C-terminal residues code for an RNA triphosphatase, an NTPase and an RNA helicase (NS3hel) [20]. Owing to its enzymatic activities, NS3 is implicated in both the polyprotein processing and RNA replication (Figure 1) [21,22]. The RNA triphosphatase activity likely contributes to RNA capping [23,24]. The NTP-helicase activity unwinds the viral RNA 3′-region secondary structure, separates nascent RNA strands from the template [25–28] and facilitates the initiation of viral replication [29].
Since the activity of NS3pro is required for several internal cleavages of the viral polyprotein precursor, NS3pro is essential for viral replication [30,31]. However, NS3pro alone is inert and it has an aberrant fold and structure [32]. The presence of the upstream viral-encoded NS2B cofactor is required for NS3pro to exhibit its functional activity [33,34]. The folding and the spatial structure of the NS3 protease domain alone are significantly different from those of the two-component NS2B–NS3 protease complex [33,34].
Since both the plus and minus strands of template RNA are highly structured, the viral replicating machinery requires the unwinding of the RNA secondary structure in the template RNAs. This important function is performed by NS3hel. NS3hel is a member of the DEAH/D-box family within the helicase superfamily 2 [21,22,35]. NS3hel has seven sequence motifs, which are conserved in the helicase superfamily 2 and are involved in nucleic acid binding and ATP hydrolysis [36]. Motifs I and II, known also as Walker A and Walker B, directly bind the ATP substrate and divalent cations and are conserved in all helicase types. Flaviviruses with impaired NS3pro or NS3hel or both are unable to replicate, making the NS3pro–hel biological system a promising drug target for antiviral therapy [37–43].
In the process of replication, the NS3pro–hel interacts with the RNA-dependent RNA polymerase, the product of the viral NS5 gene [44]. The NS5 RNA polymerase is responsible for replication of the viral genome within putative complexes comprising both viral and as-yet-unidentified host proteins [45–48]. Until recently, flaviviral replication was believed to occur within viral-induced membrane-bound replication complexes in the host cytoplasm. However, in cells infected with WNV, JEV and DV a significant proportion (20%) of the total NS5 RNA polymerase activity is resident, jointly with NS3, within the nucleus [49,50]. Accordingly, we may expect a distinctive performance of the individual NS3hel in the nucleus compared with the full-length NS2B–NS3pro–hel that resides in the membrane-bound replication complexes.
NS2B & NS4A cofactor proteins
By using their pioneering, diversified and sophisticated experiments in the various expression systems convincingly, multiple authors have demonstrated that NS2B is localized upstream of NS3 in the genome and in the flaviviral polyprotein precursor interacts functionally and physically with NS3pro in vitro and in vivo [51–59]. Therefore, a deletion in NS2B blocks the functional activity of NS3pro. The activity of NS3pro can be restored by providing NS2B in trans [60].
The region that is required for a cofactor function was identified using deletions within the NS2B gene. As a result, we now know that a conserved 40-residue hydrophilic region of NS2B flanked by hydrophobic regions is necessary for the NS3 protease activity [60–62]. To the best of our knowledge, Leung et al. [63] and Nall et al. [38] were the first to report expression of the NS2B cofactor domain linked covalently to the DV and WNV NS3pro domains, respectively, with insertion of a nine-residue flexible linker (Gly–Gly–Gly–Gly–Ser–Gly–Gly–Gly–Gly). A cohort of outstanding authors then extensively studied the biochemical properties of the NS2B–NS3pro construct. Owing to their efforts, we now know that NS3pro is responsible for the cleavage of the polyprotein precursor within the capsid protein C and at the NS2A/NS2B, NS2B/NS3, NS3/NS4A and NS4B/NS5 boundaries (Figure 1) [38,64].
Similar to NS2B, which is an essential cofactor of NS3pro, the nonstructural protein NS4A appears to play a cofactor function for NS3hel. In the polyprotein precursor sequence, NS4A is localized immediately downstream of NS3hel. NS4A is one of the least characterized flaviviral proteins [59,65,66]. NS4A is highly hydrophobic, with its C-terminal region serving as a signal sequence for the translocation of the adjacent NS4B into the endoplasmic reticulum lumen. NS4A associates with membranes via four internal hydrophobic regions. NS4A resides primarily in endoplasmic reticulum-derived cytoplasmic structures, which also contain dsRNA and other DV proteins, suggesting that NS4A is a component of the membrane-bound viral replication complex. Recent results by several laboratories suggest that NS4A regulates RNA-coupled ATP hydrolysis by NS3hel, and that this effect was mediated by residues in the C-terminal acidic domain of NS4A [67,68]. It is likely that the NS4A functionality allows NS3hel to sustain the unwinding rate of the viral RNA under the conditions of ATP deficiency. The presence of this acidic domain in the NS4A sequence of the flaviviruses and HCV family, which is also a member of the Flaviviridae, suggests that this ATP-conserving regulatory mechanism is preserved across the entire family (Figure 3).
Figure 3. Sequence alignment of the flaviviral NS4A.
The acidic C-terminal motif is in a frame. The 44–53 acidic motif of HCV NS4A is shown below the alignment. Homologous residues are in gray. Dots indicate identical residues.
DV: Dengue virus; JEV: Japanese encephalitis virus; KV: Kunjin virus; WNV: West Nile virus; YFV: Yellow fever virus.
Two-component NS2B–NS3 protease
In the frequently used two-component NS2B–NS3pro recombinant constructs, the central portion of the NS2B cofactor is linked with NS3pro via a (G)4S(G)4 linker. To facilitate its isolation, the NS2B–NS3 construct is normally C-terminally tagged with a (His)×6 tag. Owing to the autolytic cleavage at the WKG↓GG site, the NS2B–NS3pro constructs are readily converted into a noncovalently bound NS2B cofactor and NS3pro. However, NS2B remains tightly associated with NS3pro and, as a result, the functional activity is not significantly affected. However, the samples, which contain noncovalently bound NS2B and NS3pro and the residual amounts of the covalently bound NS3B–NS3pro, cannot be crystallized. In a step towards solving the structure of NS2B–NS3pro, the cleavage site sequence of the NS2B–NS3pro junction region was mutated to generate the self-proteolysis resistant, single-chain NS2B–NS3pro mutant construct. Since the mutant construct exhibits high stability and functional activity, it is well suited for the follow-up structural and drug design studies [69,70]. Using the original and the mutant constructs, several inhibitors of NS2B–NS3pro have been identified, including aprotinin (Ki ~2 nM), D-Arg-based peptides, Dec-RVKR-CMK and α1-anti-trypsin variant Portland [71]. These inhibitors blocked not only the cleavage of the fluorescent peptide substrates (from which pyroglutamic acid-Arg-Thr-Lys-Arg-7-amino-4-methylcoumarin is most routinely used) but also the protein substrates (e.g., myelin basic protein, which is highly sensitive to NS3pro proteolysis) [71].
Essential role of the NS2B cofactor
The structure of the two-component NS2B–NS3pro from WNV (PDB code 2IJO, 2GGV and 2FP7) and DV (PDB code 2FOM), which has been recently solved is an excellent model for flaviviral protease activation, with important implications for a structure-based inhibitor design for this entire class of flaviviruses [33,34].
The structure of the WNV and DV NS2B–NS3pro has a chymotrypsin-like fold with the active site located at the interface of the N- and C-terminal lobes (Figure 4). The fold of the WNV NS2B–NS3pro is similar, albeit not identical, to that of the HCV NS3pro–NS4A (PDB code 1JXP). The C-terminal part of the NS2B fragment (residues 64–96 in WNV) forms a belt that wraps around NS3pro, ending in a β-hairpin (β2–β3). This hairpin augments the upper β-barrel and inserts its tip directly into the protease active site. The importance of this interaction is clearly shown by the deleterious effects of the mutation of NS2B residues Leu75 and Ile79 (conserved hydrophobic residues in flaviviruses) in DV2 [72], which lie on the inner surface of the invading cofactor β-hairpin and anchor it to a hydrophobic cleft on NS3. In agreement, multiple mutations in the β-hairpin (β2–β3) region of the NS2B cofactor fully inactivate the functional activity of the two-component NS2B–NS3pro [73,74].
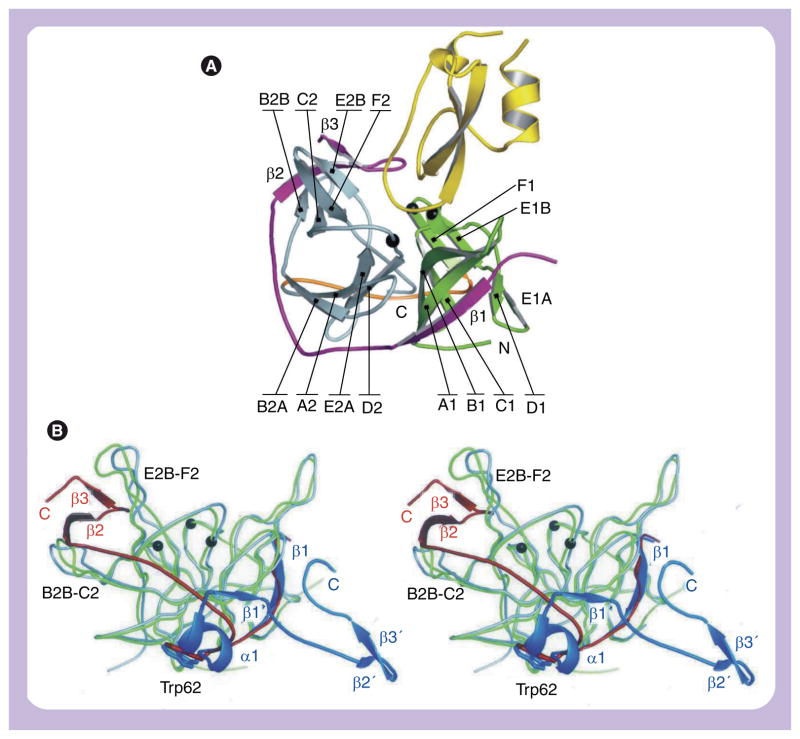
Figure 4. Structure of the West Nile virus NS2B–NS3pro.
(A) Aprotinin-bound NS2B–NS3pro with secondary structural elements and N- and C-termini indicated. The N- and C-terminal lobes of NS3 are green and gray, respectively. NS2B and aprotinin are purple and yellow, respectively. The linker between the two lobes is orange. The catalytic triad is shown as black balls. The localization of the structural elements in the peptide sequence is shown in Figure 2. (B) Two conformations of the NS2B cofactor. Superposition of WNV NS2B–NS3 proteases: substrate-free (green and blue) and aprotinin-bound (gray and red). The secondary elements of NS2B unique to the substrate-free (α1 and β1′) and the substrate-bound (β2 and β3) structures, as well as the alternative C-termini are indicated. The point of departure (Trp62) for the two NS2B elements is labeled and shown as a stick model. The elements β2′ and β3′ in the substrate-free structure are stabilized by crystal contacts.
It is clear from the structural data that the presence of the NS2B cofactor is essential for the functional activity of the flaviviral NS3 protease catalytic domain. However, alternative conformations of the NS2B cofactor relative to the NS3pro domain are also possible. While the overall fold within the protease core is similar in the NS2B–NS3pro structures of DV2 and DV4, the conformation of the NS2B cofactor, especially in its C-terminal region implicated in substrate binding, is dramatically different [75]. This alternative conformation of NS2B may be a result of a unique metal-binding site within the DV1 sequence and a first-time serotype-specific structural element in the DV subfamily of the flaviviruses.
Organization of the flaviviral NS3pro active site
The active site of chymotrypsin-like proteases includes three conserved elements [76]:
A classic His–Asp–Ser catalytic triad (His51, Asp75 and Ser135 in WNV), whose precise arrangement in space is required to enhance the nucleophilicity of the serine hydroxyl group;
The ‘oxyanion hole’, which stabilizes the developing negative charge on the scissile peptide carbonyl oxygen in the transition state;
The substrate-binding β-strands E2 and B1, which help to position the substrate in the active site.
In the WNV NS2B–NS3pro–aprotinin complex, the catalytic triad adopts a productive geometry that is virtually identical to that observed in the trypsin–aprotinin complex (PDB code 2FTL). Thus, the Ser135 OH–His51 Nε2 distance (2.8 Å) and the His51 Nδ1–Asp75 CO2− distance (2.7 Å) are indicative of strong hydrogen bonds, as is the distance (2.8 Å) between the Ser135 hydroxyl oxygen and the (nominally) scissile peptide carbonyl of aprotinin. The main chain of aprotinin residues 13–19 (PCKARII) forms antiparallel β-sheet interactions with strands E2B and B1 of NS2B–NS3, while the side chains occupy the presumed subsites S3–S1 and S19–S49, exactly as observed in the trypsin–aprotinin complex. The main-chain conformations of residues occupying the S3 and S2 sites (P3 and P2) are closely superimposable with those of the peptidic inhibitor-bound DV protease (PDB code 2FOM).
Substrate-induced fit in the oxyanion hole of NS2B–NS3pro
Intriguingly, aprotinin binding induces a catalytically competent conformation of the oxyanion hole in the WNV NS3pro. This hole, lined by main-chain nitrogens (from Gly133 to Ser135 in WNV) is not always properly formed in the substrate-free and peptidic inhibitor-bound structures; in those structures the peptide bond between Thr132 and Gly133 may be flipped [77]. The flipped bond creates a helical (310) conformation for residues 131–135, which is stabilized by two hydrogen bonds, although these are absent in the productive conformation. It is possible that the nonproductive conformation of the oxyanion hole is energetically favored in the absence of a substrate, acquiring the productive conformation only in the presence of a substrate with an appropriate P1′ residue. These structural data suggest that an ‘induced fit’ mechanism [78] contributes to substrate specificity or enzyme turnover, or both, in the NS2B–NS3pro [77].
Alternating conformations of NS2B–NS3pro
The major difference between the inhibitor-bound and inhibitor-free WNV NS2A–NS3pro structures is in the conformation of the NS2B cofactor (Figure 4). In the inhibitor-free structure, the β1 strand of NS2B is formed as in the aprotinin complex, augmenting the β-barrel of the N-terminal lobe of the NS3 protease moiety. However, beyond strand β1, the last NS2B residue that adopts the same conformation as in the aprotinin complex is Trp62, which is buried in a pocket on the C-terminal lobe of NS3. WNV Trp62 (or DV Trp61) appears to act as an ‘anchor’ for the NS2B cofactor: accordingly, the mutation of either Trp61 [72] or of the NS3 residue, Gln96 [79,80], which lines the base of the Trp62 acceptor pocket, does not permit catalytic activity. Following W62, the NS2B chain adopts a new conformation in an inhibitor-free structure: a new turn-and-a-half of helix (α1) is followed by an abrupt reversal of chain direction at Gly70 (conserved as Gly or Ala in flaviviruses), followed by a four-residue β-strand (termed β1′ in the original publication [33]), which augments the central β-sheet of the NS3 C-terminal domain by making main-chain interactions with β-strand B2A. The chain then continues in the reverse direction, toward the N-terminus of the NS2B sequence (i.e., opposite from that in the inhibitor-bound structure). Despite these major changes in the interaction with NS2B, the NS3 coordinates are very close in all of the solved flavivirus protease structures. It is now clear that both the WNV and DV proteases can adopt two distinct cofactor-bound conformations, thus raising the strong possibility that the two conformations are a conserved feature of all flaviviruses.
Structural rationale for the substrate specificity of NS2B–NS3pro
The aprotinin-bound WNV structure [33] mimics a classic Michaelis–Menten complex, allowing for an authentic view of the enzyme–substrate precleavage complex. WNV and related flaviviral proteases have a requirement for basic residues at both the P1 and P2 positions of their substrates, and Gly, Ser or Thr at the P1′ position. In the aprotinin-bound NS2B–NS3pro structure, the P2 side chain (Arg/Lys) interacts directly with Asn84 of NS2B and indirectly with a negatively charged surface created by the invading hairpin of NS2B, including Asp80 and Asp82, as previously proposed [81]. NS2B binding also helps to define the S1 pocket in the WNV protease, by changing the conformation of NS3 residues 116–132, such that Asp129 is introduced into the base of the pocket, where it can salt-bridge with the P1 Arg/Lys. Accordingly, in WNV or DV proteases, mutation of Asp129 to Glu or Ala abrogates catalysis [64]. In trypsin, Asp189 plays an analogous role [82,83], consistent with its similar P1 specificity. The in vitro protease activity assays, in which Escherichia coli-derived Asp129Glu/Ser/Ala mutants were used, do not completely agree with the results obtained in cell-based assays [84]. These differences appear to be related to the incomplete folding of the protease mutants in E. coli [84]. The S1′ pocket in NS2B–NS3pro comprises a cavity between strand B1 and the helical turn (residues 50–53) following strand C1 and is lined on one side by the catalytic His residue and on the other by an invariant Gly residue (Gly37 in WNV). The pocket is well formed, but only large enough to accommodate small P1′ side chains, such as the consensus residues Gly, Ser or Thr. Ser and Thr have the potential to bond through hydrogen to the main-chain carbon–oxygen of the adjacent Ala36, rationalizing the P1′ preference for these residues, while glycine leaves enough space for a water molecule. The inability of the Ala side chain to interact with the hydrogen bond could explain its rare appearance in flavivirus polyprotein cleavage sequences. In trypsin, no such pocket is formed because a disulfide bridge occupies this site and, as a result, trypsin has no specificity at the P1′ position. It appears that the additional hydrogen bond in the WNV protease compared with the DV protease places stringent restraints on the substrate dihedral angles, allowing for limited ‘wiggle room’ for the P1′ side chain, which is positioned close to the catalytic His; this fact could explain the preference for glycine.
Furthermore, Trp132 in the WNV NS3pro forms a hydrogen bond with the peptide bond involving the P1′ residue. This bond is unique for WNV because, in the DV NS3 polypeptide chain, the Pro132 residue that occupies this position is incapable of making a similar hydrogen bond. Accordingly, the hydrogen bond involving Thr132 stabilizes the backbone conformations of the P1′/P2′ residues, allowing their side chains to make tight contacts with His51 and Thr132 of the WNV NS3pro. These parameters limit the mobility of the P2′ residues, thus leading to the preferred Gly at the P2′ position of the WNV NS2B–NS3pro.
Substrate cleavage preferences of WNV NS2B–NS3pro
In the viral polyprotein, the WNV NS2B–NS3 predominantly cleaves the sequence regions with positively charged amino acid residues at both the P1 and the P2 positions, and Gly at either P1′ or P2′, or both. In the P1′–P2′ positions, selectivity of DV1–4 is less evident. The presence of any residue at P2, other than positively charged Arg or Lys, makes the substrate resistant to the NS3pro cleavage (Table 1). Owing to the requirement for Arg/Lys at both the P1 and P2 positions, the WNV NS2B–NS3pro resembles the furin-like proprotein convertases from the host cells [18,85,86].
Table 1.
Known cleavage sites of NS3 protease in the flaviviral polyprotein precursor.
| Capsid C | NS2A/NS2B | NS2B–NS3 | NS3/NS4A | NS4B/NS5 | |
|---|---|---|---|---|---|
| WNV | Q101KKR↓GGTA108 | N1367RKR↓GWPA1374 | Y1498TKR↓GGVL1505 | S2117GKR↓SQIG2124 | G2522LKR↓GGAK2529 |
| JEV | Q102NKR↓GGNE109 | N1370KKR↓GWPA1377 | T1501TKR↓GGVF1508 | A2120GKR↓SAVS2127 | S2564LKR↓GRPG2571 |
| YFV | R98 KRR↓SHDV105 | F1351GRR↓SIPV1358 | G1481ARR↓SGDV1488 | E2104GRR↓GAAE2111 | T2503GRR↓GSAN2510 |
| DV1 | R97 RKR↓SVTM104 | W1341GRK↓SWPL1348 | K1471KQR↓SGVL1478 | A2090GRR↓SVSG2097 | G2489GRR↓GTGA2496 |
| DV2 | R97 RRR↓TAGV104 | S1342KKR↓SWPL1349 | K1472KQR↓AGVL1479 | A2090GRK↓SLTL2097 | N2488TRR↓GTGN2495 |
| DV3 | K97 RKK↓TSLC104 | L1340KRR↓SWPL1347 | Q1470TQR↓SGVL1477 | A2089GRK↓SIAL2096 | T2487GKR↓GTGS2494 |
| DV4 | G96 RKR↓STIT103 | A1341SRR↓SWPL1348 | K1471TQR↓SGAL1478 | S2089GRK↓SITL2096 | T2484PRR↓GTGT2491 |
The natural cleavage sites exist in the capsid protein C and at the NS2A/NS2B, NS2B/NS3, NS3/NS4A and NS4B/NS5 boundaries of the polyprotein precursor.
The P1′ and P2′ Gly residues are in bold. Arrows indicates the scissile bond.
DV: Dengue virus; JEV: Japanese encephalitis virus; WNV: West Nile virus; YFV: Yellow fever virus.
As a step towards developing a rapid and efficient inhibitor design, as well as to gain a precise understanding of the distinctive features of flaviviral proteases, our team as well as others determined the substrate cleavage pattern of NS2B–NS3pro of WNV and DV2 [64,87–94]. There is a strict requirement for the presence of either Arg or Lys at the P1 and the P2 substrate positions for NS2B–NS3pro. Consistent with the structural models of the WNV protease [33], an exclusive preference for Gly at both the P1′ and the P2′ positions is a remarkable characteristic of the WNV enzyme. By contrast, the DV NS2B–NS3pro tolerates the presence of many types of amino acid residues well, except the negatively charged Asp and Glu residues, at the P1′–P2′ positions. To support these structural and activity data, mutagenesis was then used to transform the cleavage preferences of the WNV NS2B–NS3pro into those of the DV enzyme [89]. The cleavage preferences of the mutant constructs were then assayed in a positional scanning format where the P4–P1 and the P3′–P4′ positions of the peptide substrates were fixed, and the P1′ and P2′ positions were each randomized with 18 and 15 amino acids. Guided by the available structural models of the WNV and DV NS2B–NS3pro [33,34], the authors established that WNV R1578L and P1633K-T1634P mutants acquired DV-like cleavage preferences [88,89]. These results represented the first instance of engineering a viral protease with switched substrate cleavage preferences, and a proof-of-principle that will support the redesign of other proteases.
NS3hel, but not NS2B–NS3pro–hel, exhibits the DNA unwinding activity
Little is known about the unwinding mechanism of the ATP-dependent, 3′–5′ flaviviral helicases, members of the DEXH family of helicases. Very recent structural studies captured DV4 NS3hel at several stages along the catalytic pathway, including bound to ssRNA, to an ATP analog, to a transition-state analog and to ATP hydrolysis products (PDB code 2JLQ, 2JLR, 2JLS, 2JLU, 2JLV, 2JLW, 2JLX, 2JLY and 2JLZ). These studies demonstrated, for the first time, large quaternary changes in the Flaviviridae helicase, identified the catalytic water molecule and pointed to a β-hairpin that protruded from the helicase structure as a critical element for double-stranded RNA unwinding [27,28]. RNA recognition by DV4 NS3hel appeared largely sequence independent in a way remarkably similar to eukaryotic DEAD box proteins Vasa and eIF4AIII [35,95–98]. The relative orientation of the protease and helicase domains may be drastically different in the flaviviral helicases. For example, in the structure of NS2B–NS3pro–hel from Murray valley encephalitis virus (PDB code 2WV9), the NS3pro and the NS3hel domains are spatially separated [99], while in the DV4 structure (PDB code 2VBC), these domains are packed more closely [27,28]. It is possible that this difference in acquiring alternative conformations by the recombinant NS3 constructs is dictated by a flexible interdomain linker and the small protease/helicase interface, thereby suggesting that the NS3pro and NS3hel domain are loosely tethered, although other suggestions are also plausible.
Previous biochemical studies of flavivirus helicases predominantly concentrated on truncations that included only the helicase domain of NS3 but not the protease region of NS3 and the NS2B cofactor. However, the presence of a properly folded and functional NS2B–NS3pro domain is required for the selective unwinding activity of NS2B–NS3pro–hel. Thus, the NS2B–NS3pro–hel construct and the individual NS3hel domain exhibit a similar RNA unwinding activity. The unwinding efficiency of the DNA duplex by the NS2B–NS3pro–hel construct is exceedingly low. By contrast, the individual NS3hel was highly active in the DNA unwinding reactions. In addition, the NS3pro–hel construct that lacks the NS2B cofactor and represents the C-terminal portion of the NS3pro domain sequence and the intact NS3hel domain acquired the ability to unwind the DNA duplex as efficiently as NS3hel. It is likely that the correct, productive fold of the N-terminal NS2B–NS3pro portion is essential for the RNA selectivity of the full-length NS2B–NS3pro–hel construct [100]. It is tempting to hypothesize that this regulatory mechanism plays an important role in WNV replication, which occurs both within the virus-induced membrane-bound replication complexes in the host cytoplasm and in the nuclei of infected cells. It appears that in the course of WNV replication within virus-induced, membrane-bound replication complexes in the host cytoplasm the strict RNA unwinding activity of the WNV NS3 is required for coordinating the helicase activity with the activity of the NS5 RNA-dependent RNA polymerase and for avoiding interference by the host DNA. In turn, when a significant fraction of NS3 that is no longer associated with the membrane NS2B cofactor relocalizes jointly with NS5 to the nucleus in infected cells [49,50], the capacity of this viral helicase to unwind DNA opens a wide spectrum of possibilities beyond its role in viral RNA replication. Furthermore, in-depth studies are required to elucidate the precise mechanisms and the functional role of the NS3 protein in viral replication in vivo.
Implications for drug design
In all crystal structures of NS2B–NS3pro to date, the NS2B cofactor is located either at a distance from the substrate-binding site (open conformation) in the absence of an inhibitor and lining the substrate-binding site (closed conformation) in the presence of an inhibitor. In other words, the open conformation signifies the protease with a low level of functional activity, if any. In turn, the closed conformation is a characteristic of the functionally potent enzyme. The unfortunate consequence is that the protease can be activated by low concentrations of the active site-targeting inhibitors, a phenomenon recently reported with the structurally and functionally similar NS3 protease from HCV [101]. As a result, there is a challenging opportunity for the design of the highly selective antagonists that target the NS2B and NS4A cofactor functions, and which, through targeting the cofactor functions, would affect the activity of the viral NS3pro–hel enzymes. The recent development of the NS4A HCV antagonists (ACH-806 and ACH-1095, Achillion Pharmaceuticals, CT, USA), the activity of which is synergistic with NS3pro and NS5B RNA polymerase inhibitors, supports this suggestion [102,103].
Similarly, it is possible to identify the NS2B–NS3pro inhibitors that would affect the NS2B/NS3pro interface and would stimulate the conformation transition leading to the inactivation of the proteolytic activity of NS2B–NS3. Recent attempts by several groups suggest that this opportunity is now well recognized and appreciated [31,39,41,104–112]. Other researchers have been on a quest to identify the active site-targeting NS3pro antagonists, including the small molecule- and the peptide-based inhibitors [31,113–119] and, more recently, the highly selective human recombinant antibodies [120].
However, the focused drug optimization efforts, are still in their infancy, suggesting, optimistically, that the more potent, optimized small-molecule inhibitors of the flaviviral proteases will be available in the near future. Coordinated efforts similar to the EU Vizier Project [121], which covers topics from viral genomics to structural biology and inhibition mechanisms, are required to shed light on the design and development of antiviral drug leads.
Future perspective
The current structural studies of NS2B–NS3pro and NS2B–NS3pro–hel provided a start-up based on which structure-based drug design could be initiated. However, the realworld knowledge of how the protease and the helicase function in the virus lifecycle requires the precise biochemical and structural analysis of the multicomponent membrane-embedded complexes, which involve the full-length NS2B, NS3 and NS4A rather than their individual hydrophilic domains. Much work remains to be completed in order to unravel the structural–functional parameters of these enzyme complexes in a molecular detail. This structural analysis represents the next hurdle that virologists, enzymologists, structural biologists and medicinal chemists have to overcome jointly in order to design novel, potent and safe antivirals that will be effective in human patients.
Executive summary.
West Nile virus is an emerging pathogen
Owing to climate change, there is a significant probability for further outbreaks of flaviviral infections, including West Nile virus worldwide and, especially, dengue virus in the Americas.
There is no specific and effective therapy available for flavivirus infections, including West Nile virus.
Novel, potent and safe antivirals are urgently required.
Flaviviral NS3 protease is required for the proteolytic processing of the viral polyprotein precursor
The flaviviral NS3 is a multifunctional enzyme.
The serine protease domain represents the N-terminal portion of NS3.
The ATP-dependent RNA helicase domain represents the C-terminal portion of NS3.
NS3 protease cleaves the polyprotein at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, NS4A/NS4B (probably) and NS4B/NS5 boundaries and in the capsid protein C.
To exhibit the proteolytic activity, the NS3pro requires the NS2B cofactor
NS2B is encoded by the upstream gene in the flaviviral genome.
The cofactor activity of the hydrophilic central portion is approximately equivalent to that of the entire NS2B sequence.
There are alternating structural folds of the NS2B–NS3 protease constructs.
The NS4A protein is a likely cofactor of the NS3 helicase.
NS4A is encoded by the downstream gene in the flaviviral genome
It is likely that NS4A helps the helicase to sustain the unwinding rate of the viral RNA under the conditions of ATP deficiency.
This effect was mediated by residues in the C-terminal acidic domain of NS4A.
NS3 is a promising drug target in the Flaviviridae family
The multicomponent membrane-embedded NS2B–NS3–NS4A complexes, which involve the full-length viral proteins, should be analyzed in the near future.
The allosteric inhibitors that target the cofactors of the NS3 protease and helicase may lead to the development of selective and safe antivirals.
Footnotes
Financial & competing interests disclosure
The work was supported by NIH grants AI055789 and AI061139 (to AYS). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.van der Meulen KM, Pensaert MB, Nauwynck HJ. West Nile virus in the vertebrate world. Arch Virol. 2005;150(4):637–657. doi: 10.1007/s00705-004-0463-z. [DOI] [PubMed] [Google Scholar]
- 2.Madden K. West Nile virus infection and its neurological manifestations. Clin Med Res. 2003;1(2):145–150. doi: 10.3121/cmr.1.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10(12):1366–1373. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- 4.Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- 5.Mackow E, Makino Y, Zhao BT, et al. The nucleotide sequence of dengue type 4 virus: analysis of genes coding for nonstructural proteins. Virology. 1987;159(2):217–228. doi: 10.1016/0042-6822(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 6.Zhao B, Mackow E, Buckler-White A, et al. Cloning full-length dengue type 4 viral DNA sequences: analysis of genes coding for structural proteins. Virology. 1986;155(1):77–88. doi: 10.1016/0042-6822(86)90169-8. [DOI] [PubMed] [Google Scholar]
- 7.Hahn YS, Galler R, Hunkapiller T, Dalrymple JM, Strauss JH, Strauss EG. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology. 1988;162(1):167–180. doi: 10.1016/0042-6822(88)90406-0. [DOI] [PubMed] [Google Scholar]
- 8.Yaegashi T, Vakharia VN, Page K, Sasaguri Y, Feighny R, Padmanabhan R. Partial sequence analysis of cloned dengue virus type 2 genome. Gene. 1986;46(2–3):257–267. doi: 10.1016/0378-1119(86)90410-5. [DOI] [PubMed] [Google Scholar]
- 9.Irie K, Mohan PM, Sasaguri Y, Putnak R, Padmanabhan R. Sequence analysis of cloned dengue virus type 2 genome (New Guinea-C strain) Gene. 1989;75(2):197–211. doi: 10.1016/0378-1119(89)90266-7. [DOI] [PubMed] [Google Scholar]
- 10.Deubel V, Kinney RM, Trent DW. Nucleotide sequence and deduced amino acid sequence of the nonstructural proteins of dengue type 2 virus, Jamaica genotype: comparative analysis of the full-length genome. Virology. 1988;165(1):234–244. doi: 10.1016/0042-6822(88)90677-0. [DOI] [PubMed] [Google Scholar]
- 11.Coia G, Parker MD, Speight G, Byrne ME, Westaway EG. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69(Pt 1):1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 12.Castle E, Leidner U, Nowak T, Wengler G. Primary structure of the West Nile flavivirus genome region coding for all nonstructural proteins. Virology. 1986;149(1):10–26. doi: 10.1016/0042-6822(86)90082-6. [DOI] [PubMed] [Google Scholar]
- 13.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 14■■.Chambers TJ, Weir RC, Grakoui A, et al. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990;87(22):8898–8902. doi: 10.1073/pnas.87.22.8898. The first publication that identifies the N-terminal domain of flaviviral NS3 as a serine protease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford GR, Wright PJ. Characterization of novel viral polyproteins detected in cells infected by the flavivirus Kunjin and radiolabelled in the presence of the leucine analog hydroxyleucine. J Gen Virol. 1987;68(Pt 2):365–376. doi: 10.1099/0022-1317-68-2-365. [DOI] [PubMed] [Google Scholar]
- 16.Preugschat F, Strauss JH. Processing of nonstructural proteins NS4A and NS4B of dengue 2 virus in vitro and in vivo. Virology. 1991;185(2):689–697. doi: 10.1016/0042-6822(91)90540-r. [DOI] [PubMed] [Google Scholar]
- 17.Yamshchikov VF, Trent DW, Compans RW. Upregulation of signalase processing and induction of prM-E secretion by the flavivirus NS2B–NS3 protease: roles of protease components. J Virol. 1997;71(6):4364–4371. doi: 10.1128/jvi.71.6.4364-4371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18■.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3(10):753–766. doi: 10.1038/nrm934. A comprehensive review on the furin-like proprotein convertases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3(1):13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 20.Padmanabhan R, Mueller N, Reichert E, et al. Multiple enzyme activities of flavivirus proteins. Novartis Found Symp. 2006;277:74–84. doi: 10.1002/0470058005.ch6. discussion 84–76, 251–253. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Clum S, You S, Ebner KE, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73(4):3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wengler G, Wengler G. The NS 3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology. 1993;197(1):265–273. doi: 10.1006/viro.1993.1587. [DOI] [PubMed] [Google Scholar]
- 24■.Mastrangelo E, Milani M, Bollati M, et al. Crystal structure and activity of Kunjin virus NS3 helicase; protease and helicase domain assembly in the full length NS3 protein. J Mol Biol. 2007;372(2):444–455. doi: 10.1016/j.jmb.2007.06.055. Structural analysis of the Kunjin virus full-length NS3 protein. [DOI] [PubMed] [Google Scholar]
- 25.Cui T, Sugrue RJ, Xu Q, Lee AK, Chan YC, Fu J. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology. 1998;246(2):409–417. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- 26.Borowski P, Niebuhr A, Mueller O, et al. Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme. J Virol. 2001;75(7):3220–3229. doi: 10.1128/JVI.75.7.3220-3229.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82(1):173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28■■.Luo D, Xu T, Watson RP, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27(23):3209–3219. doi: 10.1038/emboj.2008.232. Structural analysis of the dengue virus serotype-4 NS3 helicase at several stages along the catalytic pathway, including bound to single-stranded RNA, to an ATP analog, to a transition-state analog and to an ATP hydrolysis product. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frick DN. Helicases as antiviral drug targets. Drug News Perspect. 2003;16(6):355–362. doi: 10.1358/dnp.2003.16.6.829307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahl G, Sandstrom A, Akerblom E, Danielson UH. Effects on protease inhibition by modifying of helicase residues in hepatitis C virus nonstructural protein 3. FEBS J. 2007;274(22):5979–5986. doi: 10.1111/j.1742-4658.2007.06120.x. [DOI] [PubMed] [Google Scholar]
- 31■.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. West Nile Virus NS2B/NS3 protease as an antiviral target. Curr Med Chem. 2008;15(27):2771–2784. doi: 10.2174/092986708786242804. Review paper on the NS3 protease from the drug design perspective. [DOI] [PubMed] [Google Scholar]
- 32.Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275(14):9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 33■■.Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16(5):795–806. doi: 10.1110/ps.072753207. Describes in detail the structure of the two-component NS2B–NS3 West Nile virus protease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34■■.Erbel P, Schiering N, D’Arcy A, et al. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13(4):372–373. doi: 10.1038/nsmb1073. Describes the structure of the two-component NS2B–NS3 dengue virus protease. [DOI] [PubMed] [Google Scholar]
- 35.Luking A, Stahl U, Schmidt U. The protein family of RNA helicases. Crit Rev Biochem Mol Biol. 1998;33(4):259–296. doi: 10.1080/10409239891204233. [DOI] [PubMed] [Google Scholar]
- 36.Benarroch D, Selisko B, Locatelli GA, Maga G, Romette JL, Canard B. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology. 2004;328(2):208–218. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Maga G, Gemma S, Fattorusso C, et al. Specific targeting of hepatitis C virus NS3 RNA helicase Discovery of the potent and selective competitive nucleotide-mimicking inhibitor QU663. Biochemistry. 2005;44(28):9637–9644. doi: 10.1021/bi047437u. [DOI] [PubMed] [Google Scholar]
- 38.Nall TA, Chappell KJ, Stoermer MJ, et al. Enzymatic characterization and homology model of a catalytically active recombinant West Nile virus NS3 protease. J Biol Chem. 2004;279(47):48535–48542. doi: 10.1074/jbc.M406810200. [DOI] [PubMed] [Google Scholar]
- 39.Johnston PA, Phillips J, Shun TY, et al. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B–NS3 proteinase of West Nile virus. Assay Drug Dev Technol. 2007;5(6):737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- 40.Ray D, Shi PY. Recent advances in flavivirus antiviral drug discovery and vaccine development. Recent Patents Antiinfect Drug Disc. 2006;1(1):45–55. doi: 10.2174/157489106775244055. [DOI] [PubMed] [Google Scholar]
- 41.Keller TH, Chen YL, Knox JE, et al. Finding new medicines for flaviviral targets. Novartis Found Symp. 2006;277:102–114. discussion 114–109, 251–103. [PubMed] [Google Scholar]
- 42.Sampath A, Xu T, Chao A, Luo D, Lescar J, Vasudevan SG. Structure-based mutational analysis of the NS3 helicase from dengue virus. J Virol. 2006;80(13):6686–6690. doi: 10.1128/JVI.02215-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu T, Sampath A, Chao A, et al. Towards the design of flavivirus helicase/NTPase inhibitors: crystallographic and mutagenesis studies of the dengue virus NS3 helicase catalytic domain. Novartis Found Symp. 2006;277:87–97. doi: 10.1002/0470058005.ch7. discussion 97–101, 251–103. [DOI] [PubMed] [Google Scholar]
- 44.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270(32):19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 45.Kapoor M, Zhang L, Mohan PM, Padmanabhan R. Synthesis and characterization of an infectious dengue virus type-2 RNA genome (New Guinea C strain) Gene. 1995;162(2):175–180. doi: 10.1016/0378-1119(95)00332-z. [DOI] [PubMed] [Google Scholar]
- 46.Lindenbach B. Flaviviridae: the viruses and their replication. In: Bernard N, Fields DMK, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; PA, USA: 2007. p. 1101. [Google Scholar]
- 47.Lindenbach BD, Pragai BM, Montserret R, et al. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J Virol. 2007;81(17):8905–8918. doi: 10.1128/JVI.00937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu WJ, Sedlak PL, Kondratieva N, Khromykh AA. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J Virol. 2002;76(21):10766–10775. doi: 10.1128/JVI.76.21.10766-10775.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uchil PD, Kumar AV, Satchidanandam V. Nuclear localization of flavivirus RNA synthesis in infected cells. J Virol. 2006;80(11):5451–5464. doi: 10.1128/JVI.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin β 1 and importin α/β-recognized nuclear localization signals. J Biol Chem. 2002;277(39):36399–36407. doi: 10.1074/jbc.M204977200. [DOI] [PubMed] [Google Scholar]
- 51.Amberg SM, Nestorowicz A, McCourt DW, Rice CM. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J Virol. 1994;68(6):3794–3802. doi: 10.1128/jvi.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arias CF, Preugschat F, Strauss JH. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193(2):888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- 53.Chambers TJ, Grakoui A, Rice CM. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991;65(11):6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jan LR, Yang CS, Trent DW, Falgout B, Lai CJ. Processing of Japanese encephalitis virus non-structural proteins: NS2B–NS3 complex and heterologous proteases. J Gen Virol. 1995;76(Pt 3):573–580. doi: 10.1099/0022-1317-76-3-573. [DOI] [PubMed] [Google Scholar]
- 55.Lin C, Amberg SM, Chambers TJ, Rice CM. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J Virol. 1993;67(4):2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin C, Chambers TJ, Rice CM. Mutagenesis of conserved residues at the yellow fever virus 3/4A and 4B/5 dibasic cleavage sites: effects on cleavage efficiency and polyprotein processing. Virology. 1993;192(2):596–604. doi: 10.1006/viro.1993.1076. [DOI] [PubMed] [Google Scholar]
- 57.Wengler G, Czaya G, Farber PM, Hegemann JH. In vitro synthesis of West Nile virus proteins indicates that the aminoterminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991;72(Pt 4):851–858. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- 58.Wengler G, Wengler G. The carboxyterminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184(2):707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Mohan PM, Padmanabhan R. Processing and localization of dengue virus type 2 polyprotein precursor NS3–NS4A–NS4B–NS5. J Virol. 1992;66(12):7549–7554. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falgout B, Miller RH, Lai CJ. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B–NS3 protease activity. J Virol. 1993;67(4):2034–2042. doi: 10.1128/jvi.67.4.2034-2042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67(11):6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leung D, Schroder K, White H, et al. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem. 2001;276(49):45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- 64.Chappell KJ, Nall TA, Stoermer MJ, et al. Site-directed mutagenesis and kinetic studies of the West Nile Virus NS3 protease identify key enzyme-substrate interactions. J Biol Chem. 2005;280(4):2896–2903. doi: 10.1074/jbc.M409931200. [DOI] [PubMed] [Google Scholar]
- 65.Cahour A, Falgout B, Lai CJ. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B–NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992;66(3):1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem. 2007;282(12):8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 67■.Beran RK, Lindenbach BD, Pyle AM. The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase. J Virol. 2009;83(7):3268–3275. doi: 10.1128/JVI.01849-08. Determines that NS4A plays a cofactor role for the NS3 helicase from HCV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68■.Shiryaev SA, Chernov AV, Aleshin AE, Shiryaeva TN, Strongin AY. NS4A regulates the ATPase activity of the NS3 helicase: a novel cofactor role of the non-structural protein NS4A from West Nile virus. J Gen Virol. 2009;90(Pt 9):2081–2085. doi: 10.1099/vir.0.012864-0. Reports on the cofactor function of NS4A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shiryaev SA, Aleshin AE, Ratnikov BI, Smith JW, Liddington RC, Strongin AY. Expression and purification of a two-component flaviviral proteinase resistant to autocleavage at the NS2B–NS3 junction region. Protein Expr Purif. 2007;52(2):334–339. doi: 10.1016/j.pep.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Generation and characterization of proteolytically active and highly stable truncated and full-length recombinant West Nile virus NS3. Protein Expr Purif. 2007;53(1):87–96. doi: 10.1016/j.pep.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 71.Shiryaev SA, Ratnikov BI, Chekanov AV, et al. Cleavage targets and the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem J. 2006;393(Pt 2):503–511. doi: 10.1042/BJ20051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Niyomrattanakit P, Winoyanuwattikun P, Chanprapaph S, Angsuthanasombat C, Panyim S, Katzenmeier G. Identification of residues in the dengue virus type 2 NS2B cofactor that are critical for NS3 protease activation. J Virol. 2004;78(24):13708–13716. doi: 10.1128/JVI.78.24.13708-13716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Mutagenesis of the West Nile virus NS2B cofactor domain reveals two regions essential for protease activity. J Gen Virol. 2008;89(Pt 4):1010–1014. doi: 10.1099/vir.0.83447-0. [DOI] [PubMed] [Google Scholar]
- 74.Radichev I, Shiryaev SA, Aleshin AE, et al. Structure-based mutagenesis identifies important novel determinants of the NS2B cofactor of the West Nile virus two-component NS2B–NS3 proteinase. J Gen Virol. 2008;89(Pt 3):636–641. doi: 10.1099/vir.0.83359-0. [DOI] [PubMed] [Google Scholar]
- 75■.Chandramouli S, Joseph JS, Daudenarde S, Gatchalian J, Cornillez-Ty C, Kuhn P. Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J Virol. 2010;84(6):3059–3067. doi: 10.1128/JVI.02044-09. First report on serotype-specific structural element in the dengue virus NS3 protease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pasternak A, Ringe D, Hedstrom L. Comparison of anionic and cationic trypsinogens: the anionic activation domain is more flexible in solution and differs in its mode of BPTI binding in the crystal structure. Protein Sci. 1999;8(1):253–258. doi: 10.1110/ps.8.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robin G, Chappell K, Stoermer MJ, et al. Structure of West Nile virus NS3 protease: ligand stabilization of the catalytic conformation. J Mol Biol. 2009;385(5):1568–1577. doi: 10.1016/j.jmb.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 78.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44(2):98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matusan AE, Kelley PG, Pryor MJ, Whisstock JC, Davidson AD, Wright PJ. Mutagenesis of the dengue virus type 2 NS3 proteinase and the production of growth-restricted virus. J Gen Virol. 2001;82(Pt 7):1647–1656. doi: 10.1099/0022-1317-82-7-1647. [DOI] [PubMed] [Google Scholar]
- 80.Matusan AE, Pryor MJ, Davidson AD, Wright PJ. Mutagenesis of the dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J Virol. 2001;75(20):9633–9643. doi: 10.1128/JVI.75.20.9633-9643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Otlewski J, Jaskolski M, Buczek O, et al. Structure–function relationship of serine protease–protein inhibitor interaction. Acta Biochim Pol. 2001;48(2):419–428. [PubMed] [Google Scholar]
- 82.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102(12):4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 83.Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4(3):337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Valle RP, Falgout B. Mutagenesis of the NS3 protease of dengue virus type 2. J Virol. 1998;72(1):624–632. doi: 10.1128/jvi.72.1.624-632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85■.Remacle AG, Shiryaev SA, Oh ES, et al. Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J Biol Chem. 2008;283(30):20897–20906. doi: 10.1074/jbc.M803762200. In-depth analysis of the cleavage preferences of the individual proteinases from the proprotein convertase family. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seidah NG. Unexpected similarity between the cytosolic West Nile virus NS3 and the secretory furin-like serine proteinases. Biochem J. 2006;393(Pt 2):E1–E3. doi: 10.1042/BJ20051787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87■.Li J, Lim SP, Beer D, et al. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J Biol Chem. 2005;280(31):28766–28774. doi: 10.1074/jbc.M500588200. Extensive analysis of the cleavage preferences of the dengue NS3 protease. [DOI] [PubMed] [Google Scholar]
- 88■.Shiryaev SA, Kozlov IA, Ratnikov BI, Smith JW, Lebl M, Strongin AY. Cleavage preference distinguishes the two-component NS2B-NS3 serine proteinases of dengue and West Nile viruses. Biochem J. 2007;401(3):743–752. doi: 10.1042/BJ20061136. Extensive analysis of the relative cleavage preferences of the dengue and West Nile virus NS3 proteases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89■.Shiryaev SA, Ratnikov BI, Aleshin AE, et al. Switching the substrate specificity of the two-component NS2B–NS3 flavivirus proteinase by structure-based mutagenesis. J Virol. 2007;81(9):4501–4509. doi: 10.1128/JVI.02719-06. First instance of engineering a viral protease with switched substrate cleavage preferences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Insights to substrate binding and processing by West Nile virus NS3 protease through combined modeling, protease mutagenesis, and kinetic studies. J Biol Chem. 2006;281(50):38448–38458. doi: 10.1074/jbc.M607641200. [DOI] [PubMed] [Google Scholar]
- 91.Gouvea IE, Izidoro MA, Judice WA, et al. Substrate specificity of recombinant dengue 2 virus NS2B–NS3 protease: influence of natural and unnatural basic amino acids on hydrolysis of synthetic fluorescent substrates. Arch Biochem Biophys. 2007;457(2):187–196. doi: 10.1016/j.abb.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 92.Khumthong R, Angsuthanasombat C, Panyim S, Katzenmeier G. In vitro determination of dengue virus type 2 NS2B–NS3 protease activity with fluorescent peptide substrates. J Biochem Mol Biol. 2002;35(2):206–212. doi: 10.5483/bmbrep.2002.35.2.206. [DOI] [PubMed] [Google Scholar]
- 93.Khumthong R, Niyomrattanakit P, Chanprapaph S, Angsuthanasombat C, Panyim S, Katzenmeier G. Steady-state cleavage kinetics for dengue virus type 2 NS2B-NS3(pro) serine protease with synthetic peptides. Protein Pept Lett. 2003;10(1):19–26. doi: 10.2174/0929866033408228. [DOI] [PubMed] [Google Scholar]
- 94.Mueller NH, Yon C, Ganesh VK, Padmanabhan R. Characterization of the West Nile virus protease substrate specificity and inhibitors. Int J Biochem Cell Biol. 2007;39(3):606–614. doi: 10.1016/j.biocel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 95.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281(27):18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 96.Andersen CB, Ballut L, Johansen JS, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313(5795):1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 97.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126(4):713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 98.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila vasa. Cell. 2006;125(2):287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 99■.Assenberg R8, Mastrangelo E, Walter TS, et al. Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J Virol. 2009;83(24):12895–12906. doi: 10.1128/JVI.00942-09. Indicates the alternating relative folds of the protease and the helicase in the full-length NS3 protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chernov AV, Shiryaev SA, Aleshin AE, et al. The two-component NS2B–NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J Biol Chem. 2008;283(25):17270–17278. doi: 10.1074/jbc.M801719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dahl G, Arenas OG, Danielson UH. Hepatitis C virus NS3 protease is activated by low concentrations of protease inhibitors. Biochemistry. 2009;48(48):11592–11602. doi: 10.1021/bi9016928. [DOI] [PubMed] [Google Scholar]
- 102.Yang W, Zhao Y, Fabrycki J, et al. Selection of replicon variants resistant to ACH-806, a novel hepatitis C virus inhibitor with no cross-resistance to NS3 protease and NS5B polymerase inhibitors. Antimicrob Agents Chemother. 2008;52(6):2043–2052. doi: 10.1128/AAC.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wyles DL, Kaihara KA, Schooley RT. Synergy of a hepatitis C virus (HCV) NS4A antagonist in combination with HCV protease and polymerase inhibitors. Antimicrob Agents Chemother. 2008;52(5):1862–1864. doi: 10.1128/AAC.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ekonomiuk D, Su XC, Ozawa K, et al. Discovery of a non-peptidic inhibitor of West Nile virus NS3 protease by high-throughput docking. PLoS Negl Trop Dis. 2009;3(1):e356. doi: 10.1371/journal.pntd.0000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ganesh VK, Muller N, Judge K, Luan CH, Padmanabhan R, Murthy KH. Identification and characterization of nonsubstrate based inhibitors of the essential dengue and West Nile virus proteases. Bioorg Med Chem. 2005;13(1):257–264. doi: 10.1016/j.bmc.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 106.Lescar J, Luo D, Xu T, et al. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from dengue virus as a target. Antiviral Res. 2008;80(2):94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 107.Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Identification and biochemical characterization of small-molecule inhibitors of West Nile virus serine protease by a high-throughput screen. Antimicrob Agents Chemother. 2008;52(9):3385–3393. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sampath A, Padmanabhan R. Molecular targets for flavivirus drug discovery. Antiviral Res. 2009;81(1):6–15. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sidique S, Shiryaev SA, Ratnikov BI, et al. Structure-activity relationship and improved hydrolytic stability of pyrazole derivatives that are allosteric inhibitors of West Nile virus NS2B–NS3 proteinase. Bioorg Med Chem Lett. 2009;19(19):5773–5777. doi: 10.1016/j.bmcl.2009.07.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Su XC, Ozawa K, Yagi H, et al. NMR study of complexes between low molecular mass inhibitors and the West Nile virus NS2B–NS3 protease. FEBS J. 2009;276(15):4244–4255. doi: 10.1111/j.1742-4658.2009.07132.x. [DOI] [PubMed] [Google Scholar]
- 111.Tomlinson SM, Malmstrom RD, Russo A, Mueller N, Pang YP, Watowich SJ. Structure-based discovery of dengue virus protease inhibitors. Antiviral Res. 2009;82(3):110–114. doi: 10.1016/j.antiviral.2009.02.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zuo Z, Liew OW, Chen G, et al. Mechanism of NS2B-mediated activation of NS3pro in dengue virus: molecular dynamics simulations and bioassays. J Virol. 2009;83(2):1060–1070. doi: 10.1128/JVI.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chanprapaph S, Saparpakorn P, Sangma C, et al. Competitive inhibition of the dengue virus NS3 serine protease by synthetic peptides representing polyprotein cleavage sites. Biochem Biophys Res Commun. 2005;330(4):1237–1246. doi: 10.1016/j.bbrc.2005.03.107. [DOI] [PubMed] [Google Scholar]
- 114.Ekonomiuk D, Su XC, Ozawa K, et al. Flaviviral protease inhibitors identified by fragment-based library docking into a structure generated by molecular dynamics. J Med Chem. 2009;52(15):4860–4868. doi: 10.1021/jm900448m. [DOI] [PubMed] [Google Scholar]
- 115.Kiat TS, Pippen R, Yusof R, Ibrahim H, Khalid N, Rahman NA. Inhibitory activity of cyclohexenyl chalcone derivatives and flavonoids of fingerroot, Boesenbergia rotunda (L.), towards dengue-2 virus NS3 protease. Bioorg Med Chem Lett. 2006;16(12):3337–3340. doi: 10.1016/j.bmcl.2005.12.075. [DOI] [PubMed] [Google Scholar]
- 116.Knox JE, Ma NL, Yin Z, et al. Peptide inhibitors of West Nile NS3 protease: SAR study of tetrapeptide aldehyde inhibitors. J Med Chem. 2006;49(22):6585–6590. doi: 10.1021/jm0607606. [DOI] [PubMed] [Google Scholar]
- 117.Stoermer MJ, Chappell KJ, Liebscher S, et al. Potent cationic inhibitors of West Nile virus NS2B/NS3 protease with serum stability, cell permeability and antiviral activity. J Med Chem. 2008;51(18):5714–5721. doi: 10.1021/jm800503y. [DOI] [PubMed] [Google Scholar]
- 118.Yin Z, Patel SJ, Wang WL, et al. Peptide inhibitors of dengue virus NS3 protease Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg Med Chem Lett. 2006;16(1):40–43. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 119.Yin Z, Patel SJ, Wang WL, et al. Peptide inhibitors of dengue virus NS3 protease Part 1: warhead. Bioorg Med Chem Lett. 2006;16(1):36–39. doi: 10.1016/j.bmcl.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 120.Shiryaev SA, Radichev IA, Ratnikov BI, et al. Isolation and characterization of selective and potent human Fab inhibitors directed to the active-site region of the two-component NS2B–NS3 proteinase of West Nile virus. Biochem J. 2010;427(3):369–376. doi: 10.1042/BJ20100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bollati M, Alvarez K, Assenberg R, et al. Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antiviral Res. 2010;87(2):125–148. doi: 10.1016/j.antiviral.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]