Abstract
Adenosine-to-inosine (A-to-I) editing catalyzed by adenosine deaminases acting on RNA (ADARs) entails the chemical conversion of adenosine residues to inosine residues within double-stranded RNA (dsRNA) substrates. Inosine base pairs as guanosine and A-to-I editing can therefore alter the structure and base pairing properties of the RNA molecule. This has a biological significance in controlling the amount of functional RNA molecules in the cell, in expanding the functionality of a limited set of transcripts, and in defending the cell against certain RNA viruses. A-to-I editing is not limited to any specific type of RNA substrate. Instead, it can affect any RNA molecule able to attain the required double-stranded structure. This includes microRNAs, small interfering RNAs, viral RNAs, and messenger RNAs with potential for recoding events and splice site modifications.
An RNA molecule can be post-transcriptionally modified in many ways that affect its function in the cell, including splicing, polyadenylation, and RNA editing. RNA editing—the insertion, deletion, or chemical conversion of nucleotides—once seemed a quirk of trypanosome mitochondria but is now increasingly recognized as an important phenomenon affecting a diverse set of cellular pathways. By alteration of the RNA's sequence, properties of both the RNA molecule and of proteins translated from it can be modified. The cell uses this to control the amounts of biologically functional RNAs, to diversify the functionality of a limited set of unique transcripts, and to modulate viral replication of certain RNA viruses.
Chemical conversions of nucleotides commonly take the form of deaminations. Deamination of a cytidine residue replaces it with a uridine. As shown in Figure 1, deamination of adenosine (A) creates an inosine (I), a nucleotide containing hypoxanthine in place of one of the conventional nucleobases. Inosine preferentially base pairs with cytidine, effectively equating it with guanosine.
Figure 1.
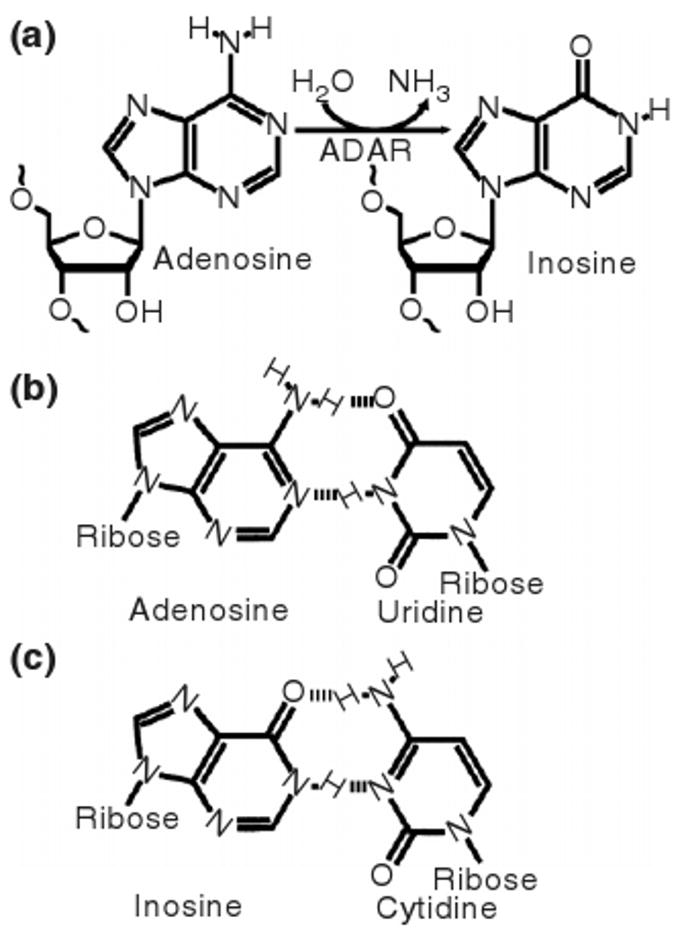
(a) The hydrolytic deamination of adenosine, as catalyzed by ADARs, converts it to inosine. (b) Whereas adenosine base pairs with uridine, (c) inosine preferentially base pairs with cytidine and the deamination therefore changes the base pairing properties of the RNA molecule.
Found in most metazoa, the adenosine deaminase acting on RNA (ADAR) family catalyzes A-to-I editing in partially or perfectly double-stranded RNA. This means that the effects of ADARs, rather than being limited to any specific functional grouping of RNAs, hold significance for a wide range of cellular functions. This review focuses on the various effects of A-to-I editing by ADARs, on the biological mechanisms underlying and controlling the editing, and on the related recent discoveries.
Historical Overview
Editing by ADAR was first reported1,2 as an RNA-duplex-unwinding activity in Xenopus laevis embryos in 1987. It was soon found3 that this unwinding activity was conserved through mammals. The demonstration4 that the unwinding phenomenon was in fact a deamination of adenosines to form inosines in double-stranded RNAs (dsRNAs), indirectly unwinding the RNA duplex by exchanging AU base pairs for less stable IU base pairs, alluded to a function of the activity in modifying RNA molecule information content. Indeed, the first case of such modification was soon reported5 for a glutamate receptor (GluR) subunit. Sequencing of GluR-RNA-derived cDNA showed a codon with CAG and CGG variants whereas genomic DNA sequencing could only find the CAG sequence. Editing explained this phenomenon because the reverse transcriptase machinery reads inosine as guanosine by incorporating a cytidine into the cDNA. Microsequencing of the ADAR1 protein allowed for cloning and subsequent sequencing of its gene,6 which led to the identification of ADAR27 and ADAR38 by sequence similarity. Later research has shown that ADAR editing goes far beyond recoding messenger RNAs (mRNAs), effectively acting on any substrate able to achieve the required double-strandedness, and this fuels new interest in elucidating the mysteries surrounding ADARs. As this review will show, while recent years' research has generated much new knowledge about the mechanism and significance of A-to-I editing, it has also revealed a system of which many facets still wait to be investigated.
The Adar Family Members
Although no ADAR genes have been identified in plants, yeast, or fungi, they are present in nearly all metazoa, indicating an origin early in metazoan evolution. ADAR1 and ADAR2 likely diverged not much later in evolutionary time, with distinct orthologs of both being found all the way to sea anemones.9 Many more closely related organisms, however, lack either ADAR1 or ADAR2, indicating that one of the paralogs has, on several occasions, been removed from various species. Drosophila, for an example, has a single ADAR gene most similar to ADAR2. ADAR3 is constrained to the subset of vertebrates between man and fish, and likely originated by diverging from ADAR2 to which it is most highly similar in sequence and domain structure.
All ADARs share a highly conserved catalytic deaminase domain in their C-terminal half and a number of double-stranded RNA-binding domains (dsRBDs) in their N-terminal half.6 Differences between the homologs arise mainly from the number and spacing of their dsRBDs, from the presence of various cellular localization signals, from expression patterns, and from the presence of various additional domains close to the N-terminal. The three human ADARs are illustrated in Figure 2.
Figure 2.
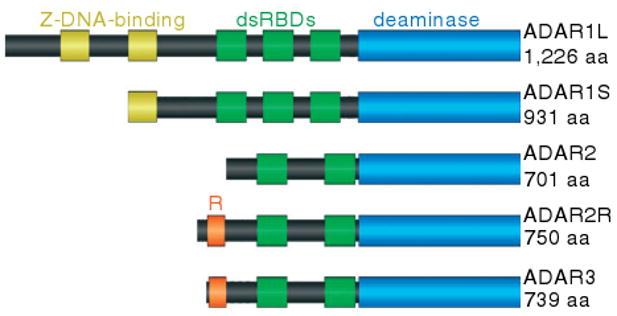
The domain structures of the human ADAR proteins show that all three proteins have a similar deaminase domain and double-stranded RNA-binding domains. The Z-DNA-binding domain is unique to ADAR1 and the R-domain is unique to ADAR3 and the ADAR2 splice variant ADAR2R.
ADAR1 has three dsRBDs and there exist two isoforms that result from alternative promoters. Compared with the shorter, 110-kDa ADAR1S which contains a single Z-DNA-binding domain, the longer, 150-kDa ADAR1L incorporates an additional Z-DNA-binding domain and nuclear export signal (NES). Although ADAR1S is transcribed from two constitutive promoters, the ADAR1L promoter is interferon-inducible.10 The consequent upregulation of ADAR1L by cellular stresses including dsRNA viruses hints at its biological function. Whereas ADAR1S localizes mainly to the nucleus, ADAR1L's additional NES causes it to localize mainly to the cytoplasm.
ADAR2 contains two dsRBDs. A novel ADAR2 splice variant, named ADAR2R, was recently reported11 to include an additional upstream exon conserved in vertebrates. This additional exon N-terminally extends the protein by 49 amino acids with high sequence similarity to the R-domain of ADAR3.
Both ADAR1 and ADAR2 are expressed in most tissues and function as homodimers.12 They have distinct yet overlapping substrate pools.13 ADAR3, in contrast, is specific to post-mitotic cells in certain parts of the central nervous system.14 Any enzymatic activity has yet to be demonstrated and all currently known editing sites are attributable to ADAR1 or ADAR2. ADAR3 does not dimerize14 and this may explain its inactivity, which cannot be explained by any obvious structural features. One possible biological function of ADAR3 lies in its ability to decrease the efficiency of ADAR1 and ADAR2, at least in vitro.14 This phenomenon relies upon both its dsRBDs and its single-stranded RNA-binding R-domain and likely results from sequestering of ADAR substrates. ADAR3 may therefore be involved in controlling editing by ADAR1 and ADAR2 in the central nervous system.
Adar Domain Structure
A variable number of dsRBDs of approximately 70 amino acids each are found in all ADAR family members. These have an α-β-β-β-α topology, the α-helices packing against a three-stranded anti-parallel β-sheet. Spanning 16 base pairs of dsRNA, the dsRBD interacts with two successive minor grooves and the intervening major groove on one face of A-form RNA. The N-terminal α-helix packs into one minor grove, the loop between the second and third β-sheets packs into the major grove, and the C-terminal α-helix packs into the next minor grove.15 Only a small minority of the interactions made with the dsRNA are with the nucleobases, the rest being with the phosphate and ribose backbone. This explains why dsRBD interactions depend more on the RNA's secondary structure and duplex stability than the presence of any primary structure motif. Multiple dsRBD copies per protein can be expected to increase the affinity for the dsRNA in a cooperative manner and to affect substrate specificity.
A highly conserved deaminase domain, catalyzing the hydrolytic deamination of adenosine, is present in all ADARs. This domain has a catalytic zinc center, presumably coordinating the attacking water molecule, lowering its pKa to facilitate nucleophilic attack on the adenine C6 carbon as a hydroxide.16 The domain's core also contains a tightly bound inositol hexakisphosphate approximately 8 Å removed from the catalytic site. This highly negatively charged molecule appears to be necessary for proper folding of the deaminase domain by bringing together the many positively charged side chains surrounding it in the native structure. Why such a mechanism is used is unclear. In one proposed scenario, it allows for control of enzymatic activity, or did so in an evolutionary ancestor of ADAR, through availability of inositol hexakisphosphate.16 However, the affinity of present-day ADAR for this cofactor is so strong that removal of inositol hexakisphosphate from an ADAR's surroundings does not appreciably remove already-bound cofactors. The domain's surface contains a positively charged cleft, which probably functions to bind the negatively charged dsRNA.16 Within this cleft, an even deeper pocket contains the catalytic zinc ion. It is believed that a nucleotide-flipping mechanism will point the adenine into this pocket where it would be in close contact with the zinc and its coordinated, nucleophilic water molecule.
The Z-DNA-binding domain is unique to ADAR1. This domain binds the left-handed form of DNA, Z-DNA.17 Z-DNA is temporarily found in the trail of underwound DNA left behind the transcription machinery before it is acted upon by topoisomerases. This suggests the first known biological function for Z-DNA: localizing ADAR1 to the site of transcription via the Z-DNA-binding domain, allowing it to efficiently act upon the RNA prior to splicing. It was later demonstrated18 that this domain also has the ability to bind left-handed Z-RNA. This suggests a second biological function of the Z-DNA-binding domain in localizing cytoplasmic ADAR1L to the underwound RNA in the wake of transcription on RNA viruses, allowing ADAR1L to more efficiently edit these viruses as a form of antiviral defense.
The R-domain is abundant in arginine and lysine residues and is found in ADAR3 and likely in an ADAR2 splice variant.11 It has been shown14 to be responsible for the binding of ADAR3 to single-stranded RNA, indicating it might function cooperatively with the dsRBDs to bind RNAs containing both double-stranded and single-stranded segments. However, the biological significance of this remains largely unknown.
Extended Editor Family
A variety of proteins deaminate nucleobases, either catalyzing A-to-I deaminations or cytidine to uridine deaminations. For example, A-to-I deaminations have long been known to take place on transfer RNAs. However, it was only after the sequencing of ADAR that the adenosine deaminases acting on transfer RNAs (ADATs) family was identified19 due to deaminase domain sequence similarity. ADATs, conserved through prokaryotes, are much more ancient than ADARs. Homology indicates that ADARs originated when a metazoa ancestor's ADAT acquired dsRBDs, an event which would conserve the enzyme's chemistry but alter its specificity. Today, the specificity of ADATs for transfer RNAs contrasts with ADAR's ability to edit various functional groupings of RNAs. Regardless of the difference in specificity, any future insight into the enzymatic mechanism of one is predicted to apply to the other.
Conservation of critical residues at the active sites of ADARs and ADATs implicate cytidine deaminase (CDA) as their closest relative. CDA catalyzes the formation of uridine monomers from cytidine monomers and shares a set of active site conserved residues with ADARs and ADATs, the conservation of which hints at the chemical mechanism underlying editing as described below. It is surprising to find this relationship with CDA rather than the purine equivalent adenosine deaminase (ADA), as this indicates two separate origins of adenosine deaminases. The evolutionary cause of this is not clear.
Subcellular Localization
ADAR1 contains a nuclear localization signal (NLS) in its third dsRBD which interacts with transportin-1 in a manner which does not require, and might even be slightly prevented by, dsRNA binding.20,21 All of its dsRBDs interact with exportin-5 in a manner which is not dependent on, but can be enhanced by, dsRNA binding.21 Together, these two facts make ADAR1 a nucleocytoplasmic shuttling protein. Whether dsRNA concentration has a significant physiological function in controlling ADAR1 localization is yet unknown. ADAR1S is predominantly localized to the nucleus where it associates dynamically with the nucleolus.22 ADAR1L, however, has an additional classical NES in the N-terminal-most Z-DNA-binding domain that is recognized by the nuclear export receptor CRM1.23 This causes ADAR1L to be localized mostly to the cytoplasm.
Less is known about the subcellular localization of ADAR2. It is primarily nuclear and localizes dynamically in the nucleolus.22,24 This nucleolar localization is likely caused by binding to double-stranded ribosomal RNA as it is dispelled in ADAR2 with non-functional dsRBDs or upon inhibition of ribosomal RNA synthesis. Inhibition of ribosomal RNA synthesis also increases efficiency of ADAR2 pre-mRNA editing, indicating a yet poorly understood biological function of the nucleolar localization in controlling editing efficiency.
Mechanism of Editing
A requirement of editing substrates seems to be perfect base pairing in the residues immediately surrounding the editing site, as is discussed below. In the perfectly double-stranded helix that results, it is hard to imagine the enzyme catalyzing chemistry at the adenosine without first inducing some large-scale conformational change. Therefore, and in view of the enzyme active site being positioned in a pocket on the protein surface, a nucleotide-flipping mechanism has been suggested. Through rotation around sugar-phosphate bonds, the nucleobase can be positioned pointing into the active site hollow.
A hypothesis describing the chemistry performed in this pocket has been formed on the basis of critically conserved deaminase residues25 and their position in crystal structures.16 These conserved residues are two cysteines and a histidine, or sometimes three cysteines, involved in the coordination of a zinc ion, and a glutamate proposed to function as a proton shuttle as outlined in Figure 3.
Figure 3.
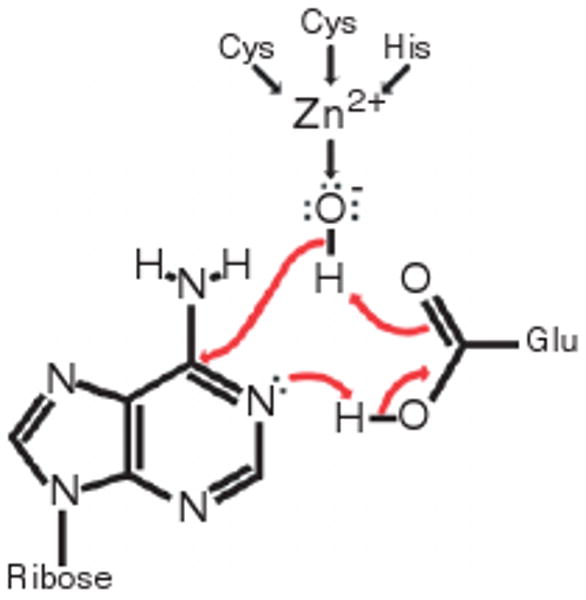
The hypothesized mechanism underlying catalysis by ADAR, based on an ADAR crystal structure16 and the conservation of the four amino acid residues included in the figure.25
Substrate Specificity
A large part of an ADAR's interaction with the substrate is through its dsRBDs. However, the crystal structure of this interaction15 shows that only a small minority of specific interactions is with nucleobase-distinguishing functional groups and the specificity of the overall interaction therefore relies more on the RNA secondary structure. The main requirement appears to be a double-stranded region of at least 15–20 base pairs, although editing efficiency drops already when the size decreases below 100 base pairs.26
Excepting some residues near the duplex end, a perfectly double-stranded RNA can therefore be edited at any adenosine. This leads to editing of about half of adenosine residues before the destabilization of the duplex by IU base pairs prevents further editing.27 Although such hyperediting is of physiological importance, a more interesting phenomenon is the site-selective editing which takes place in dsRNAs with imperfect base pairing, bulges, and loops.
ADAR1 treats loops of more than six nucleotides as equivalents of helix ends.28 One can therefore envision a scenario where short stretches of double-strandedness between such interrupts accurately positions the enzyme on the dsRNA. This is, however, not enough to explain all observed selectivity and the importance of other RNA traits, such as smaller bulges, is less well understood.
Some further degree of selectivity may be provided by the deaminase domain which interacts more intimately with the nucleobases. This domain must induce conformational changes around the editing site to make the site available to the catalytic machinery and it is therefore unsurprising that the immediately adjacent nucleotides affect editing efficiency. The 5′-nearest neighbor preference is U = A > C > G for ADAR129 and U = A > C = G for ADAR213. ADAR2 also has a 3′-nearest neighbor preference of U = G > C = A13 although ADAR1 appears to have no such inclinations.29 Both enzymes also have the preference C > U > A = G30,31 for the nucleotide opposing the editing site.
Adherence of a substrate to these neighbor preferences is not an absolute requirement. However, the importance of the deaminase domain is demonstrated by chimeric ADARs containing the dsRBDs of ADAR1 and the deaminase domain of ADAR2 or vice versa. These are more efficient at editing the natural substrate of the ADAR from which they got their deaminase domain. This could be due both to nearest-neighbor preferences and some yet poorly understood affinity of the substrate-binding groove on the ADAR surface for specific RNA secondary structures.30
Recently, the emergence of high-throughput sequencing has allowed for investigation into ADAR specificity in ways previously not possible. The recent discovery32 that editing at sites separated by a multiple of 12 nucleotides is positively coupled indicates that ADARs bind the substrates in register. If so, the suitability of an adenosine residue as an ADAR substrate is improved by the presence of a second suitable adenosine 12 nucleotides removed on the same side of the helix. ADAR specificity still largely remains an unsolved problem into which more investigation is needed. However, the value of understanding editing site selectivity to grasping the phenomenon of A-to-I editing as a whole should not be underestimated and it is therefore encouraging to see new technology such as next-generation sequencing providing new paths to insight.
mRNA as Adar Substrate
Many mRNAs can achieve the double-stranded structure required for ADAR editing because their introns contain a sequence, termed the editing-site-complementary sequence (ECS), able to base pair with the exonic regions.33 This creates stretches of imperfectly basepaired dsRNA in the precursor mRNA (pre-mRNA) suitable for specific editing by ADAR, as is shown in Figure 4. This mandates that the editing takes place cotranscriptionally, before splicing has happened. Consequently, ECSs tend to lie downstream of their base pairing partner exons, possibly allowing for base pairing to take place even before all of the intron has been transcribed. This provides a time window in which the pre-mRNA can be acted upon by ADAR but not yet by the splicing machinery.
Figure 4.
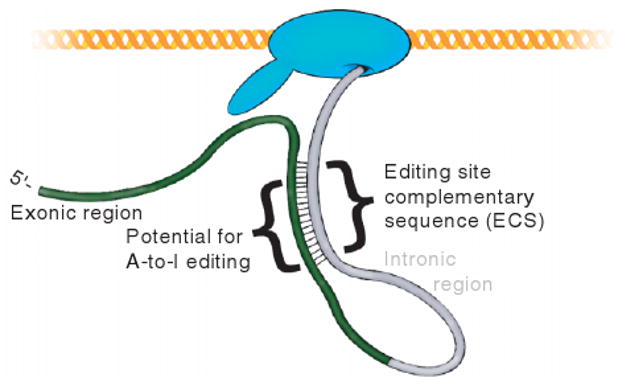
Exonic regions of a messenger RNA can be edited cotranscriptionally because they often base pair with downstream intronic regions before splicing takes place. This forms the double-stranded structure required for editing by ADAR. The complementary intronic regions are termed editing site complementary sequences.
Codon Modifications
During translation, inosine is decoded by the translation machinery as if it were a guanosine and editing in exonic regions can therefore cause recoding events.5 As shown in Figure 5, these various recoding events include the creation of a start codon from an isoleucine codon or the change of a stop codon to a tryptophan codon. A few dozen cases of such events are known, of which the human glutamate receptor5 (GluR) and serotonin receptor subtype 2C34 (5-HT2CR) are archetypical examples. With few exceptions, the edited proteins are neurotransmitter receptors and ion channels in nervous tissue.
Figure 5.
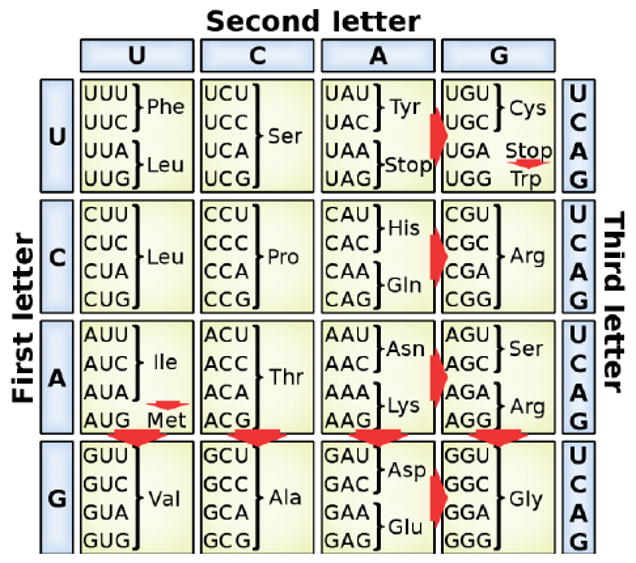
Summary of recoding events that can be caused by A-to-I editing, including the creation of a Met start codon and the destruction of stop codons.
Four GluR subunits form, in the membrane of post-synaptic cells' dendrites, a single channel which allows passage of cations upon binding of L-glutamate released by the pre-synaptic cell. In this way, the receptors mediate fast neurotransmission. A glutamine codon in this receptor constitutes the first known instance of A-to-I editing affecting the amino acid incorporated into a protein.5 An imperfectly inverted repeat in the immediate downstream intron forms a double-stranded segment around this codon and ADAR2 edits the genomically encoded CAG to form a CIG coding for arginine. Despite high sequence similarity, the extent of the editing varies from 0% to 100% for different homologs of the GluR subunit. Incorporation of a single edited subunit into the channel prevents passage of Ca2+ ions with important physiological consequences. Homozygous Adar2-null mice never incorporate the critical arginine residue into their receptors, resulting in excess influx of Ca2+ that causes epileptic seizures. The mutant mice die within a few weeks of birth.35
The 5-HT2CR receptor transcript contains five potential editing sites within three codons that can be edited in various combinations decreasing G protein coupling to different extents.34 Again, a downstream ECS is required for editing which in one codon is carried out by ADAR1, in a second by ADAR2, and the third by both.
Splice Site Modifications
Upon formation of hairpins in a pre-mRNA, intronic and splice site positions can be edited as easily as exonic positions. Because editing removes an adenosine residue and creates a residue probably interpreted as guanosine by the splicing machinery, editing has the potential to create instances of the canonical GU 5′ splice sites, to destroy adenosine branch points, and to both create and destroy instances of the canonical AG 3′ splice sites. These scenarios are illustrated in Figure 6. However, editing of splice sites has proved difficult to identify and only a few cases are yet known. Among them, the self-editing of ADAR2 is of especial interest. ADAR2 edits its own transcript at an intronic AA, creating an alternative 3′-splice acceptor site incorporating an additional 47 nucleotides into the mature mRNA. This causes a reading frame shift and produces a non-functional protein, providing a negative-feedback mechanism for the expression of functional ADAR2.36
Figure 6.

Summary of splice site modifications possible upon editing of canonical splice site and branch point motifs. The original splice pattern (a) can be modified by the creation of a new 5′ splice site (b), by the creation of a new 3′ splice site (c), by the destruction of a 3′ splice site (d), or by the destruction of the branch point(e).
There may be specific reasons for the lack of observations of the above scenarios. Scenario (d), for an example, requires transcription of the entire intron before editing can take place, at which point the editing activity may be preempted by splicing. In general, however, the lack of observed splice site modifications by A-to-I editing seems to be more grounded in our current lack of efficient means to screen for such sites.
Interaction with microRNA Pathway
MicroRNAs (miRNAs) are transcribed from the genome as primary miRNAs (pri-miRNAs) that fold up into a number of double-stranded stem regions. These are then cut to form individual hairpins of approximately 70 nucleotides in length by the enzyme Drosha with help from the dsRBD-containing DGCR8. Each hairpin is exported to the cytoplasm where it is cut by the enzyme Dicer with help from the dsRBD-containing TRBP to form RNA duplexes of approximately 21 nucleotides per strand. One strand is loaded into the RNA-induced silencing complex (RISC) while the other is degraded. RISC degrades or sequesters RNA molecules and its specificity is determined by ability to base pair with its loaded RNA, something the cell makes use of to control gene expression.37 The miRNA biogenesis pathway is summarized in Figure 7.
Figure 7.
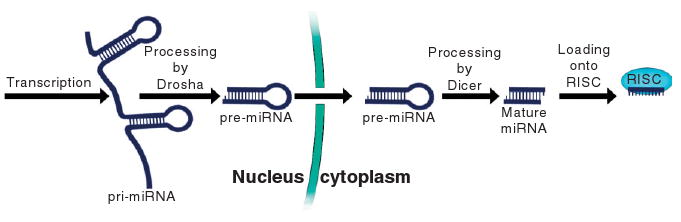
The microRNA biogenesis pathway begins with the newly transcribed pri-miRNA and ends in the RISC-loaded mature microRNA. The loaded microRNA directs RISC to its targets which it degrades. Many instances of editing of pri-miRNA are known and there is also evidence that editing could take place at the pre-miRNA stage.
Both components of the miRNA biogenesis pathway and ADARs make use of dsRBDs and the consequent overlap of substrate pools provides the ground for much crosstalk between the two pathways, as is shown for one miRNA in Figure 8. A number of pri-miRNAs are known to be edited by ADARs, resulting in prevention of Drosha-DGCR8 processing,38 prevention of Dicer-TRBP processing,39 or in a RISC-loaded edited miRNA that can target a different set of transcripts compared to its unedited counterpart.40 Because each miRNA can degrade a large set of mRNAs, controlling the extent of editing of a single miRNA could be used by the cell to control large-scale expression patterns. Finally, the possibility remains that editing may prevent loading onto the RISC complex or affect which strand is chosen for loading. Strand selection depends on the relative stability of the two duplex ends,37 something which could be affected by editing. In vitro evidence also suggests the possibility of editing at the pre-miRNA stage, although whether such editing is of significance in vivo is still uncertain.39
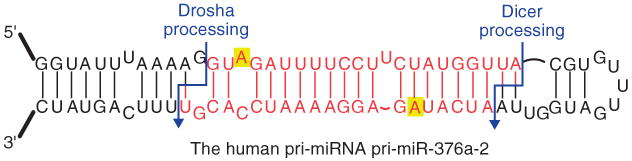
Figure 8.
The human microRNA pri-miR-376a-2 is an example of an edited microRNA. It forms one of the stem structures of the larger pri-miRNA and this stem structure allows for the editing by ADAR, which is known to take place at the sites highlighted in yellow. In this case, editing results in the mature miRNA, shown in red, targeting a new set of messenger RNAs.40.
Interaction with Small Interfering RNA Pathway
Small interfering RNAs (siRNAs) begin as long dsRNAs in the cytoplasm which are cut into duplexes of approximately 23 nt per strand by the enzyme Dicer with help from the dsRBD-containing TRBP. One strand of each duplex is then loaded onto the RISC described above for miRNAs. As for miRNAs and RNA editing, the involvement of dsRBDs in both pathways lays the ground for crosstalk. Cytoplasmic ADAR1L has the ability to compete with Dicer for the long dsRNA substrate. In vitro editing of the long dsRNA has been shown to severely decrease the efficacy of Dicer cleavage, suppressing mature siRNA production and the RNA interference phenomenon they would normally effect.41 Any siRNA that is still generated may have A-to-I changes preventing it from efficiently targeting the original dsRNA. ADAR1L can also bind tightly to any mature siRNA generated, competing with RISC.42 These phenomena are outlined in Figure 9.
Figure 9.
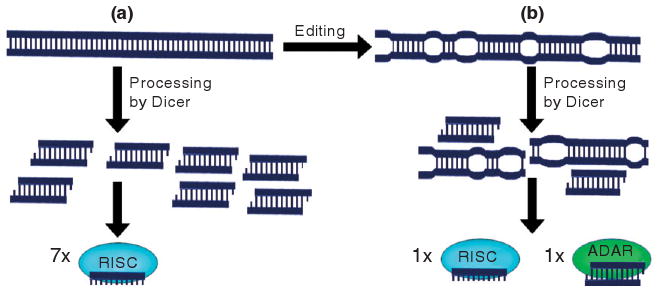
(a) In the absence of ADAR editing, long double-stranded RNAs are efficiently processed by Dicer into duplexes suitable for loading onto RISC. (b) ADAR antagonizes this pathway by editing the double-stranded RNA41, thereby unwinding it and making it less suitable as a Dicer substrate, and by sequestering the RNA duplexes generated so as to make them unavailable to RISC42.
A-to-I Editing of Viral RNAs
RNA viruses form perfectly complementary dsRNA on at least one point in their lifecycle, and this allows for hyperediting–editing at up to about half their adenosine residues. This makes it possible for ADARs to play an important part in the defense against such RNA viruses. Because most RNA viruses localize to the cell cytoplasm, the cytoplasmic ADAR1L is likely the main ADAR player in antiviral defense, a fact consistent with its expression being induced by interferons upon cellular stresses that include viral infections.
Viral hyperediting has been observed in defective measles viruses causing persistent infection.43 The measles virus is a negative-sense single-stranded RNA virus which replicates in the cytoplasm. Upon transcription of its genes, the transcript is believed to occasionally base pair with the gene itself, allowing for indiscriminate editing by ADAR.44 This leads to A-to-G mutations in the negative-sense genome and U-to-C mutations in the positive-sense antigenome at as much as half of adenosine residues for certain genes. Similar hyperediting is also observed for the human respiratory syncytial45 and lymphocytic choriomeningitis viruses.46
Viruses have also been found to take advantage of ADAR1 editing. The hepatitis delta virus (HDV) is a single-stranded, closed circular RNA of about 1700 nucleotides which replicates via an anti-genome. Two sides of the circle have a high degree of complementarity, causing the circle to fold into a rod-shaped structure where approximately 70% of the nucleotides are base paired, forming a dsRNA structure which is selectively edited. The sole HDV protein comes in two forms: a shorter form required for replication and a C-terminus extended form required for packing. The extended form results when the UAG stop codon is edited in the anti-genome to form a UGG tryptophan codon.47 This change is subsequently passed to the genome from which the extended protein's mRNA is transcribed. HDV therefore makes use of editing by ADAR as a trigger for the switch from replication to the assembly of viral particles. It has been shown that IFN-alpha-stimulated upregulation of ADAR1L makes ADAR1L the main editor of HDV whereas in unstimulated cells, ADAR1S is mostly responsible for this editing.48 Human immunodeficiency virus type 1 (HIV-1) also makes use of ADAR1 to aid in its replication in manners both dependent and independent of editing49,50 and measles virus has been shown to make use of ADAR1 as a pro-viral and anti-apoptotic host factor.51 It is clear that ADARs can variously play for both the side of the host and the virus.
Identification of A-TO-I Editing Sites
Identification of ADAR sites has always been difficult due to the lack of a signature sequence motif, and editing sites found before the early 2000s were mostly chance discoveries. At that time, the small number of known editing sites compared to a large number of sites predicted to exist from high mRNA inosine content,52 and the discrepancy between the broad expression pattern of ADARs and the brain-specific expression pattern of the known substrates began to fuel bioinformatics approaches to identifying new editing sites. By aligning cDNA or expressed sequence tags with genomic sequences, attempts were aimed at identifying A/G variation in the prior two that does not exist in the genome.31,53 Although these studies increased the number of known editing sites by over two orders of magnitude, the vast majority of new sites were found in intronic or intergenic regions rather than translated exons. Most sites clustered to high-copy-number repeats, usually Alu repeats, a topic which deserves a section of its own below.
Comparative genomics approaches have also been successful in identifying new editing sites. One study54 was based on the observation of a local maximum in conservation surrounding a known Drosophila exonic editing site. This local maximum was explained by assuming that the editing conferred a selective advantage to the fly and therefore a selective pressure to maintain complementarity between the editing site sequence and the ECS. This prompted the search for similar exonic local maxima in conservation as potential signatures of ADAR editing sites, resulting in the identification of over a dozen new exonic editing sites.
More recently, high-throughput sequencing has allowed for the sequencing of nearly 60,000 genomic locations and the corresponding cDNA.55 The locations were chosen for reported A/G variation in the cDNA without any corresponding reports of SNPs in the genomic DNA, excluding sites located in repetitive regions. With high specificity and selectivity, this survey reported 239 new editing sites, the majority previously unreported. Because this study did not include features such as conservation, coding potential, or RNA secondary structure in predicting editing sites, the editing sites identified may allow for an assessment of the distribution of editing sites in the transcriptome. The data show a clear tendency of depletion of editing sites in coding regions and miRNAs, indicating that ADAR editing has had deleterious effects during evolution whenever unwanted editing sites arose.
Editing of Repeat RNA
The approximately 300-base pair Alu element is a short interspersed element (SINE) unique to primates and more than a million copies are interspersed throughout various locations of the human genome, including the introns and UTRs of 75%56 of all known genes. The abundance of Alu elements in the primate genome has been linked to the fact that the frequency of A-to-I editing in humans is at least an order of magnitude higher than in the mouse, rat, chicken, or fly genomes, because transcripts containing two adjacent oppositely oriented Alu copies will base pair to form a hairpin structure suitable as an ADAR substrate.57 The total amount of SINEs is similar in primate and rodent genomes; however, the dominance of a single SINE in primates makes it far more likely that two adjacent, oppositely oriented SINEs will base pair to form a dsRNA structure.
The high level of editing in Alu elements might suggest for them a function in keeping editing of more physiologically important sequences in check by competing for ADAR active sites, possibly controlling the extent of editing through their concentration in a biologically significant way. Editing of Alu repeats also raises the possibility of ADARs having a role in controlling transposable RNA elements by altering their sequences.
Physiological Significance
Various phenotypes have been linked to improper A-to-I editing patterns in a number of species, including human. In line with editing controlling a number of pathways and with edited mRNA being mostly confined to the central nervous system, these phenotypes take on a variety of forms but are generally related to neural function.
The best understood of such phenotypes is based on homozygous Adar2-null mice, whose GluR receptors are underedited, allowing for increased influx of Ca2+ which leads to neuronal death, epilepsy seizures, and death a few weeks after birth.35 The same phenotype is also observed in mice with mutations surrounding the editing site or lacking the required ECS, both preventing editing at the Q/R site.58
Lack of ADAR1 function in mice causes defective hematopoiesis, widespread apoptosis, and liver disintegration, causing death at the embryonic stage.59 Inactivation of the single ADAR gene in Drosophila melanogaster causes tremors, lack of coordination, mating defects, and neurodegeneration.60
In humans, abnormal 5-HT2CR editing patterns in the prefrontal cortex have been linked to suicidal depression61 and heterozygosity for the functional null mutation of ADAR1 results in dyschromatosis symmetrica hereditearia, a human pigmentary genodermatosis.62 Underediting at the Q/R site has also been proposed as responsible for the death of motor neurons in sporadic amyotrophic lateral sclerosis (ALS) 63 and apoptotic death of neurons during ischemia caused by cardiac arrest and disruption of the blood flow to the brain.64 Known mammalian A-to-I editing sites cannot yet explain all of these phenotypes, and it is therefore hoped that discoveries of new editing sites and a better understanding of the A-to-I editing phenomenon may lead to breakthroughs in understanding these human ailments.
Conclusion
Since its discovery more than two decades ago, A-to-I editing catalyzed by ADARs has become integral to our understanding of a broad array of cellular functions. Beyond causing recoding events, A-to-I editing also has other roles that include modifying splice sites, controlling RNA interference, and playing a role in antiviral defense. In hindsight, this is not surprising. Unlike some other forms of RNA editing which make stricter requirements of their potential substrates, ADARs can affect any RNA molecule able to achieve a double-stranded structure. This makes an understanding of A-to-I editing important to our apprehension of many other phenomena. With the recognition that the human genome makes a very limited set of transcripts, it has become clear that the cell must make use of other phenomena to widen the functionality of the transcriptome. RNA editing is such a phenomenon, allowing for the expression of two related yet distinct gene products without the need to encode them both in the genome.
Furthering our understanding of A-to-I editing catalyzed by ADARs therefore holds the promise for both understanding other biological phenomena with which A-to-I editing interacts and for ameliorating or curing a number of human ailments directly connected to A-to-I editing. Consequently, it is encouraging to see recent research improving our understanding of ADAR substrate specificity, increasing our library of known editing sites, and furthering our understanding of the interaction of ADARs with other cellular pathways. With many aspects and consequences of ADAR A-to-I editing being discerned but not yet fully understood, research in this field promises to remain a valuable source for new knowledge concerning important biological phenomena.
Acknowledgments
This work was supported by grants from the National Institutes of Health, the Ellison Medical Foundation, and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
References
- 1.Bass BL, Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- 2.Rebagliati MR, Melton DA. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell. 1987;48:599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- 3.Wagner RW, Nishikura K. Cell cycle expression of RNA duplex unwindase activity in mammalian cells. Mol Cell Biol. 1988;8:770–777. doi: 10.1128/mcb.8.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 5.Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-J. [DOI] [PubMed] [Google Scholar]
- 6.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. PNAS. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, et al. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 9.Jin Y, Zhang W, Li Q. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life. 2009;61:572–578. doi: 10.1002/iub.207. [DOI] [PubMed] [Google Scholar]
- 10.Patterson JB, Thomis DC, Hans SL, Samuel CE. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology. 1995;210:508–511. doi: 10.1006/viro.1995.1370. [DOI] [PubMed] [Google Scholar]
- 11.Maas S, Gommans WM. Novel exon of mammalian ADAR2 extends open reading frame. PLoS ONE. 2009;4:e4225. doi: 10.1371/journal.pone.0004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho DS, Yang W, Lee JT, Shiekhattar R, Murray JM, et al. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem. 2003;208:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminase ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 14.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, et al. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryter JM, Schultz SC. Molecular basis of doublestranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macbeth MR, Schubert HL, VanDemark AP, Lingam AT, Hill CP, et al. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 1995;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, et al. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. PNAS. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown BA, Lowenhaupt K, Wilbert CM, Hanlon EB, Rich A., 2nd The zalpha domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. PNAS. 2000;97:13532–13536. doi: 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maas S, Gerber AP, Rich A. Identification and characterization of a human tRNA-specific adenosine deaminase related to the ADAR family of pre-mRNA editing enzymes. PNAS. 1999;16:8895–8900. doi: 10.1073/pnas.96.16.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckmann CR, Neunteufl A, Pfaffstetter L, Jantsch MF. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol Biol Cell. 2001;12:1911–1924. doi: 10.1091/mbc.12.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fritz J, Strehblow A, Taschner A, Schopoff S, Pasierbek P, et al. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol Cell Biol. 2009;29:1487–1497. doi: 10.1128/MCB.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desterro JMP, Keegan LP, Lafarga M, Berciano MT, O'Connell M, et al. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 23.Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol Cell Biol. 2001;21:7862–7871. doi: 10.1128/MCB.21.22.7862–7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. PNAS. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mian IS, Moser MJ, Holley WR, Chatterjee A. Statistical modelling and phylogenetic analysis of a deaminase domain. J Comp Biol. 1998;5:57–72. doi: 10.1089/cmb.1998.5.57. [DOI] [PubMed] [Google Scholar]
- 26.Nishikura K, Yoo C, Kim U, Murray JM, Estes PA, et al. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991;10:3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner RW, Smith JE, Cooperman BS, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. PNAS. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehmann KA, Bass BL. The Importance of internal loops within RNA substrates of ADAR1. J Mol Biol. 1999;291:1–13. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- 29.Polson AG, Bass BL. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong SK, Shuji S, Lazinski DW. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ensterö M, Daniel C, Wahlstedt H, Major F, Öhman M. Recognition and coupling of A-to-I edited sites are determined by the tertiary structure of the RNA. Nucleic Acids Res. 2009;37:6916–6926. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higuchi M, Single FN, K{ö}hler M, Sommer B, Sprengel R, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 34.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 35.Higuchi M, Stefan M, Single FN, Hartner J, Rozov A, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 36.Reuter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNAediting. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 37.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature Reviews Molecular Cell Biology. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scadden AD, Smith CW. RNAi is antagonized by A–>I hyper-editing. EMBO Rep. 2001;2:1107–1111. doi: 10.1093/embo-reports/kve244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang W, Wang Q, Howell KL, Lee JT, Cho DS, et al. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, et al. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bass BL, Weintraub H, Cattaneo R, Billeter MA. Biased hypermutation of viral RNA genomes could be due to unwinding/modification of double-stranded RNA. Cell. 1989;56:331–331. doi: 10.1016/0092-8674(89)90234-1. [DOI] [PubMed] [Google Scholar]
- 45.Rueda P, García-Barreno B, Melero JA. Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A-G substitutions (hypermutations) Virology. 1994;198:653–662. doi: 10.1006/viro.1994.1077. [DOI] [PubMed] [Google Scholar]
- 46.Zahn RC, Schelp I, Utermöhlen O, von Laer D. A-to-G hypermutation in the genome of lymphocytic choriomeningitis virus. J Virol. 2007:457–464. doi: 10.1128/JVI.00067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polson AG, Bass BL, Casey JL. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 48.Hartwig D, Schütte C, Warnecke J, Dorn I, Hennig H, et al. The large form of ADAR 1 is responsible for enhanced hepatitis delta virus RNA editing in interferon-alpha-stimulated host cells. J Vir Hep. 2005;13:150–157. doi: 10.1111/j.1365-2893.2005.00663.x. [DOI] [PubMed] [Google Scholar]
- 49.Doria M, Neri F, Gallo A, Farace MG, Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 2009;37:5848–5858. doi: 10.1093/nar/gkp604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phuphuakrat A, Kraiwong R, Boonarkart C, Lauhakirti D, Lee TH, et al. Double stranded RNA adenosine deaminases enhance HIV-1 expression of human immunodeficiency virus type 1 proteins. J Virol. 2008;82:10864–10872. doi: 10.1128/JVI.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toth AM, Li Z, Cattaneo R, Samuel CE. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem. 2009;284:29350–29356. doi: 10.1074/jbc.M109.045146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paul MS, Bass BL. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 55.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 56.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20:813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 57.Eisenberg E, Nemzer S, Kinar Y, Sorek R, Rechavi G, et al. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. PNAS. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 60.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 61.Yang W, Wang Q, Kanes SJ, Murray JM, Nishikura K. Altered RNA editing of serotonin 5-HT2C receptor induced by interferon: implications for depression associated with cytokine therapy. Mol Brain Res. 2004;124:70–78. doi: 10.1016/j.molbrainres.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 62.Miyamura Y, Suzuki T, Kono M, Inagaki K, Ito S, et al. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet. 2003;73:693–699. doi: 10.1086/378209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801–801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 64.Peng PL, Zhong XF, Tu WH, Soundarapandian MM, Molner P, et al. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49:719–733. doi: 10.1016/j.neuron.2006.01.025. [DOI] [PubMed] [Google Scholar]