Abstract
SB-3CT, (4-phenoxyphenylsulfonyl)methylthiirane, is a potent, mechanism-based inhibitor of the gelatinase sub-class of the matrix metalloproteinase (MMP) family of zinc proteases. The gelatinase MMPs are unusual in that there are several examples where both enantiomers of a racemic inhibitor have comparable inhibitory abilities. SB-3CT is one such example. Here, the inhibition mechanism of the MMP2 gelatinase by the (S)-SB-3CT enantiomer and its oxirane analogue is examined computationally, and compared to the mechanism of (R)-SB-3CT. Inhibition of MMP2 by (R)-SB-3CT was shown previously to involve enzyme-catalyzed C–H deprotonation adjacent to the sulfone, with concomitant opening by β-elimination of the sulfur of the three-membered thiirane ring. Similarly to the R enantiomer, (S)-SB-3CT was docked into the active site of MMP2, followed by molecular dynamics simulation to prepare the complex for combined quantum mechanics and molecular mechanics (QM/MM) calculations. QM/MM calculations with B3LYP/6-311+G(d,p) for the QM part (46 atoms) and the AMBER force field for the MM part were used to compare the reaction of (S)-SB-3CT and its oxirane analogue in the active site of MMP2 (9208 atoms). These calculations show that the barrier for the proton abstraction coupled ring opening reaction of (S)-SB-3CT in the MMP2 active site is 4.4 kcal/mol lower than its oxirane analogue, and the ring opening reaction energy of (S)-SB-3CT is only 1.6 kcal/mol less exothermic than its oxirane analogue. Calculations also show that the protonation of the ring-opened products by water is thermodynamically much more favorable for the alkoxide obtained from the oxirane, than for the thiolate obtained from the thiirane. In contrast to (R)-SB-3CT and the R-oxirane analogue, the double bonds of the ring-opened products of (S)-SB-3CT and its S-oxirane analogue have the cis-configuration. Vibrational frequency and intrinsic reaction path calculations on a reduced size QM/MM model (2747 atoms) provide additional insight into the mechanism. These calculations yield 5.9 and 6.7 for the deuterium kinetic isotope effect for C–H bond cleavage in the transition state for the R and S enantiomers of SB-3CT, in good agreement with the experimental results.
The matrix metalloproteinases (MMPs) are key proteolytic regulators of the integrity of the extracellular matrix. MMPs are implicated in embryonic development,1–3 tissue remodeling and repair,4–6 neurophathic pain processes,7 cancer,8–11 and other diseases.12–15 MMP2 (Gelatinases A), one of these zinc-dependent proteolytic enzymes, digests type IV collagens.16 The structure and function of this protein have been studied extensively for the purpose of selective inhibitor design.17–29 One of these inhibitors, (4-phenoxyphenylsulfonyl)methylthiirane (SB-3CT), selectively inhibits MMP2 with high potency.30,31 The inhibition mechanism of MMP2 by (R)-SB-3CT is coupled deprotonation of the methylene group juxtaposed between the sulfone and the thiirane, and the opening of the thiirane ring.31,32 This reaction creates a thiolate anion which strongly coordinates with the zinc at the active site.
It is remarkable that the R and S enantiomers of SB-3CT display similar potency as inhibitors of MMP2, even though they are expected to have rather different binding modes in the active site.33 The kinetic parameters for the R enantiomer are Kon = 2.2 ± 0.5 × 104 M−1 s−1, Koff = 5.3 ± 0.5 × 10−4 s−1 and Ki = 24 ± 6 nM, and the corresponding values for the S enantiomer are Kon = 1.7 ± 0.4 × 104 M−1 s−1, Koff = 4.0 ± 0.3 × 10−4 s−1 and Ki = 23 ± 6 nM. Their similar potency suggests that both enantiomers have a similar inhibitory mechanism, despite this anticipated difference in their binding modes. In this study we used methods similar to our previous study of MMP2·(R)-SB-3CT32 to investigate these binding modes, and to compare the inhibition mechanism of (S)-SB-3CT (3) and its oxirane analogue (4) (Scheme 1). The MMP2·(S)-SB-3CT complex was constructed by docking and molecular dynamics studies. The details of the deprotonation/ring opening mechanism for inhibition were examined by combined quantum mechanics and molecular mechanics (QM/MM) methods, and compared to (R)-SB-3CT. Vibrational frequencies, intrinsic reaction paths and kinetic isotope effects were calculated for both the R and S enantiomers of SB-3CT.
Scheme 1.
MMP2 inhibition mechanisms by SB-3CT (3, X = S) and its oxirane analogue (4, X = O)
Computational Methods
As in the previous study of the R isomer of SB-3CT, the initial structures of the MMP2 complex with (S)-SB-3CT and its oxirane analogue were built by docking and molecular dynamics (MD) methods.32 A two-layer ONIOM method34–41 was used for the QM/MM study of the inhibition mechanism of 3 and 4. The QM region (46 atoms) consists of the zinc ion, the three imidazole rings from His403, His407 and His413, the CH2CO2− part of the Glu404 side chain, the thiirane and the SO2CH2 group of the inhibitor, and one water molecule. The B3LYP/6-31G(d) level of density functional theory (DFT) described the QM part of the system, and the Amber force field42 described the MM part of the system. QM/MM geometry optimization was carried out with a mechanical embedding scheme. The QM part and all residues and solvent molecules in the MM part within 6 Å of the QM part were fully optimized (936 atoms), while the remaining atoms were held fixed. Similar cut-offs have been used previously in QM/MM studies of enzymatic systems to avoid spurious changes in the energy due to remote fluctuation in the geometry.43,44 Because the MMP2 active site is rather open to the solvent, a smaller cut-off of 6 Å was used. The partial charges for the reactive system were refined by alternating between QM/MM geometry optimization and RESP45,46 charge fitting.32 With mechanical embedding, the electrostatic interactions between the QM and MM regions are calculated using these partial charges. Single point calculations with electronic embedding41 were used for the final QM/MM energies calculated at the ONIOM(B3LYP/6-311+G(d,p):AMBER) level of theory. When calculated with the same basis set, the mean absolute difference in the relative energies with electronic embedding vs mechanical embedding is 3 kcal/mol (see Table S1 for details). All ONIOM calculations were performed with the development version of Gaussian.47 The ONIOM toolkit48 facilitated the QM/MM calculations.
Results and Discussion
The MMP2· (R)-SB-3CT and MMP2·(S)-SB-3CT complexes are compared in Fig. 1. These complexes are obtained by docking, followed by MD simulation and QM/MM geometry optimization. Similar to (R)-SB-3CT in the MMP2 active site,32 the phenoxyphenyl side chain of (S)-SB-3CT fits into the S1′ pocket, and the hydrogen bond (1.83 Å) between the backbone NH of Leu191 to the pro-S oxygen of the sulfone is preserved. The second oxygen of the sulfone is exposed to solvent. In contrast to (R)-SB-3CT, the plane of the thiirane ring of (S)-SB-3CT points away from the zinc. The Zn-S distance in MMP2·(S)-SB-3CT (4.55 Å) is significantly longer than in MMP2·(R)-SB-3CT (2.91 Å). As a consequence, a water molecule coordinates with the zinc in the (S)-SB-3CT complex (Zn-O distance is 2.11 Å) but not in the (R)-SB-3CT complex (3.58 Å). The MMP2 complex of the oxirane analogue of (S)-SB-3CT is generated by replacement of the sulfur of the thiirane with oxygen. The QM part and adjacent regions were examined visually (see Fig. S1 for a superposition of the reactants and transition states for 3 and 4) to ensure that the reactant complex and the transition state have similar hydrogen-bonding patterns with the solvent water molecules, to avoid spurious differences in barrier heights caused by minor changes in the solvent.
Fig. 1.
Structures of (R)-SB-3CT and (S)-SB-3CT in the MMP2 active site optimized at the ONIOM(B3LYP/6-31G(d):AMBER) level of theory. Residues of MMP2·(R)-SB-3CT and MMP2·(S)-SB-3CT are shown in ball-and-stick representation with atom colored according to atom types (H, C, N, O, S, Zn, shown in white, cyan, blue, red, yellow, and grey, respectively). The same color scheme is used in all the figures.
(a) Deprotonation and Ring Opening of the Inhibitor
The reactant, transition state (TS), and product structures for the inhibition of MMP2 by (S)-SB-3CT are shown in the top two rows of Fig. 2 and selected geometrical parameters are given in Fig. S2. The respective structures for the oxirane analogue are shown at the bottom of these Figures. In both reactant structures, the zinc at the MMP2 active site is coordinated with Glu404, three histidines, and one water molecule. Depending on how the Glu404 coordinates with the zinc, two local minima are identified for the reactant complex of (S)-SB-3CT·MMP2 and of its oxirane analogue (3-R and 3-R′; 4-R and 4-R′ in Fig. 2). In 3-R, the oxygen of the Glu404 that will accept the transferring proton of (S)-SB-3CT is coordinated (2.05 Å) to the zinc, while the other Glu404 oxygen is not (3.29 Å). For 3-R′, the –CO2− group of Glu404 is shifted so that the acceptor oxygen is farther from the zinc (3.60 Å) and the other oxygen is coordinated to the zinc (1.96 Å). 3-R′ is only 2.6 kcal/mol higher in energy than 3-R. Likewise, the oxirane analogue 4-R′ is 3.9 kcal/mol higher than 4-R. In both 3-R and 3-R′, the sulfur of the thiirane is significantly further from the zinc (by 4.55 and 4.11 Å, respectively) than the distance between the oxygen of oxirane and the zinc in 4-R and 4-R′ (2.32 and 2.26 Å, respectively). The energy of MMP2·(S)-SB-3CT increases by 4.9 kcal/mol if the sulfur is constrained to be closer to the zinc, similar to the distance in MMP2·(R)-SB-3CT (2.86 Å).
Fig. 2.
Reactants, transition states and products for (S)-SB-3CT (3) and its oxirane analogue (4) in the MMP2 active site optimized at the ONIOM(B3LYP/6-31G(d):AMBER) level of theory. Energies (in kcal/mol) were calculated at the ONIOM(B3LYP/6-311+G(d,p):AMBER) using electronic embedding with the reactant complexes used as the reference states. 3-P1 and 4-P1 are the cis isomers of the unprotonated ring opening products. 3-P1′ and 4-P1′ are the trans isomers. In 3-P2 and 4-P2, the cis ring opening products are protonated by the water molecule, and the resulting hydroxide anion coordinates with the zinc.
In the TS identified in the QM/MM calculations, 3-TS (Fig. 2 and Fig. S2), the transferring proton is 1.50 Å from the donor carbon and 1.18 Å from the acceptor oxygen, indicating the TS is a little earlier compared to MMP2·(R)-SB-3CT (C–H and H–O distances of 1.57 Å and 1.14 Å, respectively). The breaking C–S bond of the thiirane is elongated to 2.04 Å in 3-TS. The Glu404 side chain moves away from the zinc in order to abstract the proton. The TS of the oxirane analogue (4-TS) is similar to the thiirane system (C–H and H–O distances are 1.58 Å and 1.13 Å), but a little later than the (R)-oxirane TS (C–H and H–O distances are 1.43 Å and 1.24 Å). In the transition states, the thiirane sulfur and oxirane oxygen are strongly coordinated to the zinc (Zn–S and Zn–O distances are 2.38 Å and 1.94 Å, respectively), but the glutamate is not.
(b) Vibrational Frequencies and Reaction Path Following
While the full frequency calculations at the current level of theory are not feasible for the whole system (9208 atoms), the two lowest vibrational modes can be calculated. Only one imaginary frequency is found for 3-TS and 4-TS (1018i and 540i cm−1, respectively), verifying that these structures are transition states. Two reduced size (partial) models were built from the full thiirane and oxirane TS complexes by extracting all the QM atoms (46 atoms), the MM atoms which are allowed to move during geometry optimization (936 atoms) and enough frozen MM atoms to surround the first two parts, for a total of 2747 atoms. When the transition states of the reduced systems were re-optimized, the key parameters changed very little (RMSD = 0.04 Å, C-H and O-H distances involving the transferring proton changed by less than 0.01 Å). The fact that these changes are quite small suggests that the reduced systems are good models of the full protein-inhibitor complexes. Using the optimized transition states of these reduced size models, the full frequency analysis and calculation of the intrinsic reaction coordinate (IRC) are both practical with the latest version of the code.49 The frequency analysis for the partial models of the TSs gives imaginary frequencies close to the full system for both the thiirane (1031i cm−1) and oxirane (601i cm−1). As a part of the present work, similar calculations were carried out for the TS structure for the inhibition of MMP2 by (R)-SB-3CT.32 The imaginary frequencies are 767i and 642i for the full and reduced size thiirane systems, respectively. For its oxirane analogue, the imaginary frequencies are 1217i and 1234i for the full and reduced size systems, respectively.
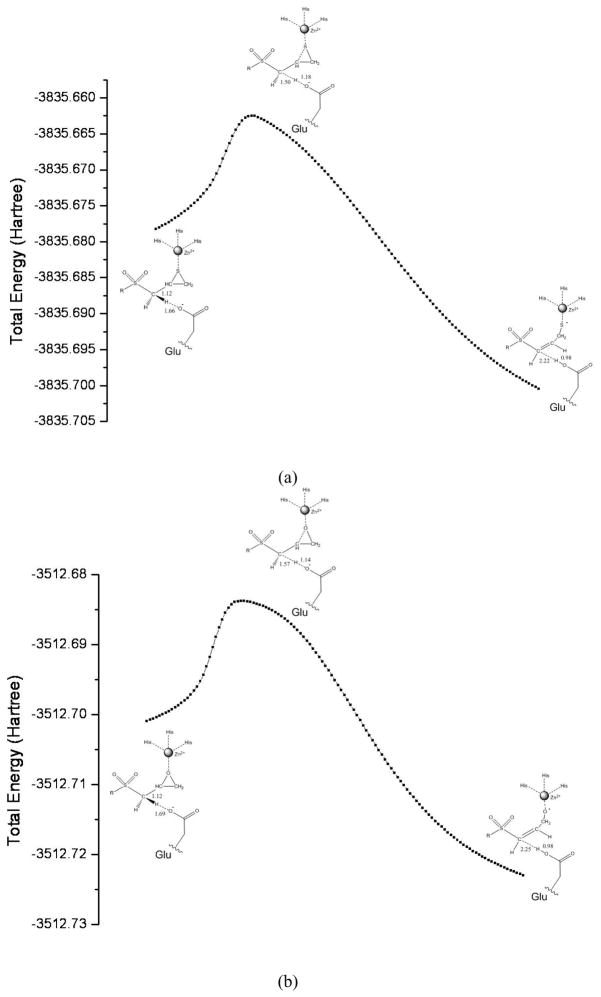
The intrinsic reaction coordinate (IRC) was calculated using the reduced size model to generate the reaction path from the reactants to the products through the TSs. The profiles of the IRC paths, and the key structures, are shown in Fig. 3 (see Fig. S3 for R enantiomer). The reaction path calculations clearly demonstrate that there are no additional barriers between the TSs and the reactants, and TSs and the products, for both MMP2·(S)-SB-3CT and its oxirane analogue. The reaction path shows that the R enantiomer of SB-3CT and its oxirane analogue open to the trans products. The corresponding reaction paths for the S enantiomer of the thiirane and oxirane lead to the cis products, 3-P1 and 4-P1. The cis products are 6.9 and 5.4 kcal/mol higherthan the trans products, 3-P1′ and 4-P1′, part because of less favorable interactions with the active site. The different stereochemical outcome for the two enantiomers demonstrates that stereoelectronic control exists in the transition state. Animations of the normal mode corresponding to the imaginary frequencies and the IRC paths for MMP2·(S)-SB-3CT and its oxirane analogue are provided in the Supporting Information.
Fig. 3.
IRC profiles for (S)-SB-3CT (a) and its oxirane analogue (b) in the MMP2 active site using the partial model at the ONIOM(B3LYP/6-31G(d):AMBER) level of theory. Key bond lengths are in Angstroms.
(c) Thermodynamics
The energy profiles for (S)-SB-3CT (3) and its oxirane analogue (4) in the MMP2 active site are shown in Fig. 4 and summarized in Table 1. The ring opening barriers for the S-thiirane and S-oxirane in the active site of MMP2 are 16.1 and 20.5 kcal/mol, respectively. The thiolate ring-opened product from the thiirane (3-P1), and the alkoxide ring-opened product from the oxirane (4-P1), both show tight coordination with the zinc in these product complexes. The protonated Glu404 moves significantly away from the zinc. The water molecule in both 3-P1 and 4-P1 moves away from the zinc to distances of 3.16 Å and 3.37 Å, respectively. The reaction energies for thiirane and oxirane are −13.3 and −14.9 kcal/mol, respectively.
Fig. 4.

Energy profiles for (S)-SB-3CT (3) and its oxirane analogue (4) in the MMP2 active site. Relative energies (in kcal/mol) were calculated at the ONIOM(B3LYP/6-311+G(d,p):AMBER) using electronic embedding with the reactant complexes used as reference states.
Table 1.
QM/MM calculations of the energetics for the ring-opening reactions of inhibitions in the active site of MMP2a
| Inhibitor | Barrier Height | Reaction Enthalpy |
||
|---|---|---|---|---|
| P1 (unprotonated cis product) | P1′ (unprotonated trans product) | P2 (protonated cis product) | ||
| (S)-SB-3CT (3)b | 16.1 | −13.3 | −20.2 | 8.7 |
| (S)-Oxirane Analogue (4)b | 20.5 | −14.9 | −20.3 | −11.2 |
ONIOM(B3LYP/6-311+G(d,p):AMBER)//ONIOM(B3LYP/6-31G(d):AMBER) with electronic embedding; energies in kcal/mol.
See Fig. 2.
(d) Product Protonation by Water
Since solvent has substantial access to the active site of MMP2, the ring-opened products can be protonated by solvent. Proton transfer from a nearby water to 3-P1 and 4-P1 gives structures 3-P2 and 4-P2, respectively (Fig. 2 and Fig. S2). Proton transfer from the water molecule generates a hydroxide ion which coordinates with the zinc, replacing the thiolate and alkoxide as zinc ligands. For the thiirane system, the protonated product complex 3-P2 is 8.7 kcal/mol endothermic compared to the reactant complex 3-R. For the oxirane system, by contrast, the protonated product complex 4-P2 is 11.2 kcal/mol exothermic compared to the reactant complex 4-R. A similar result was observed in the study of the R enantiomer.32 This difference between the thiirane and oxirane may explain why both the R and S enantiomers of SB-3CT are slow binding inhibitors of MMP2, while their oxirane analogues are linear competitive inhibitors.30
(e) Kinetic Isotope Effect (KIE) Calculations using the QM/MM model
In our earlier studies, the KIE calculation for the R-thiirane could only be carried out for the solution model.32 However, recent improvements in computer code and resources have made it feasible to calculate the full set of frequencies for QM/MM systems. Using the frequency calculations for the 2747 atom QM/MM models, deuterium KIEs were calculated for both the MMP2·(R)-SB-3CT and MMP2·(S)-SB-3CT complexes. The Wigner tunneling correction50 contributes a factor of 1.15 for R-thiirane and 1.27 for S-thiirane. The calculated KIE (kH/kD) is 5.9 for the inhibition of MMP2 by (R)-SB-3CT, and 6.7 for (S)-SB-3CT. These numbers agree well with the experimental result for the racemic mixture (kH/kD = 5.0).31
Conclusions
The inhibition of MMP2 by (S)-SB-3CT (3) and its oxirane analogue (4) involves the concurrent deprotonation of the inhibitor by the active-site glutamate, opening of the respective three-membered heterocycle, and coordination of the heteroatom anion product to the active-site zinc. For the present QM/MM calculations, the barrier for this ring-opening reaction of (S)-SB-3CT in the MMP2 active site is 16.1 kcal/mol, and is 4.4 kcal/mol lower than the barrier for the oxirane analogue. Abstraction of a proton from the inhibitor by glutamate is the key event in the inhibition reaction, as indicated by the kinetic isotope effects, and is directly coupled with the ring-opening. In the transition state, the heteroatom of the three-membered ring moves closer to the zinc, facilitating completion of the deprotonation and ring opening events for progress toward the product complexes. The reaction enthalpies are quite similar (−13.3 kcal/mol for 3 and −14.9 kcal/mol for 4). Reaction path following calculations show that ring opening of (S)-SB-3CT by MMP2 yields the cis product, while ring opening of (R)-SB-3CT in the MMP2 produces the trans product. The calculations show that the protonation of the alkoxide product from the ring opening of 4 by a water molecule in the active site is exothermic, whereas protonation of the thiolate from ring opening of 3 is endothermic. The calculated KIEs of the reaction agree well with the experimental results, increasing the confidence in the present study. The previous and present studies provide the solid theoretical support for the inhibition mechanism of MMP2.
Supplementary Material
Acknowledgments
This work is supported at Wayne State University by the National Science Foundation (CHE0910858) and at the University of Notre Dame by the National Institute of Health (CA122417). Computer time on Wayne State University computer grid is gratefully acknowledged.
Footnotes
Supporting Information Available
Superposition of the reactant and transition state for 3 and 4, QM/MM geometries for the ring opening reaction of 3 and 4 in the active site of MMP2, IRC profiles for (R)-SB-3CT and its oxirane analogue in the MMP2 active site, ONIOM energetics with mechanical embedding, animations of the imaginary frequency modes and intrinsic reaction paths using the partial QM/MM model for 3-TS and 4-TS. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lei H, Furth EE, Kalluri R, Wakenell P, Kallen CB, Jeffrey JJ, Leboy PS, Strauss JF., 3rd Biol Reprod. 1999;60:183–189. doi: 10.1095/biolreprod60.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Liu CH, Wu PS. Biotechnol Lett. 2006;28:1725–1730. doi: 10.1007/s10529-006-9147-y. [DOI] [PubMed] [Google Scholar]
- 3.Shi YB, Fu L, Hasebe T, Ishizuya-Oka A. Pharmacol Ther. 2007;116:391–400. doi: 10.1016/j.pharmthera.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woessner JF., Jr FASEB J. 1991;5:2145–2154. [PubMed] [Google Scholar]
- 5.Smith MF, Ricke WA, Bakke LJ, Dow MP, Smith GW. Mol Cell Endocrinol. 2002;191:45–56. doi: 10.1016/s0303-7207(02)00054-0. [DOI] [PubMed] [Google Scholar]
- 6.Homandberg GA, Ummadi V, Kang H. Inflammation Res. 2004;53:534–543. doi: 10.1007/s00011-004-1292-y. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki Y, Xu ZZ, Wang X, Park JY, Zhuang ZY, Tan PH, Gao YJ, Roy K, Corfas G, Lo EH, Ji RR. Nat Med. 2008;14:331–336. doi: 10.1038/nm1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coussens LM, Werb Z. Chem Biol. 1996;3:895–904. doi: 10.1016/s1074-5521(96)90178-7. [DOI] [PubMed] [Google Scholar]
- 9.Vihinen P, Kähäri VM. Int J Cancer. 2002;99:157–166. doi: 10.1002/ijc.10329. [DOI] [PubMed] [Google Scholar]
- 10.Egeblad M, Werb Z. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 11.Noël A, Jost M, Maquoi E. Semin Cell Dev Biol. 2008;19:52–60. doi: 10.1016/j.semcdb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Dollery CM, McEwan JR, Henney AM. Circ Res. 1995;77:863–868. doi: 10.1161/01.res.77.5.863. [DOI] [PubMed] [Google Scholar]
- 13.Luft FC. J Mol Med. 2004;82:781–783. doi: 10.1007/s00109-004-0609-1. [DOI] [PubMed] [Google Scholar]
- 14.Janssens S, Lijnen HR. Cardiovasc Res. 2006;69:585–594. doi: 10.1016/j.cardiores.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Chow AK, Cena J, Schulz R. Br J Pharmacol. 2007;152:189–205. doi: 10.1038/sj.bjp.0707344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liotta LA, Tryggvason K, Garbisa S, Robey PG, Abe S. Biochemistry. 1981;20:100–104. doi: 10.1021/bi00504a017. [DOI] [PubMed] [Google Scholar]
- 17.Morgunova E, Tuuttila A, Bergmann U, Isupov M, Lindqvist Y, Schneider G, Tryggvason K. Science. 1999;284:1667–1670. doi: 10.1126/science.284.5420.1667. [DOI] [PubMed] [Google Scholar]
- 18.Briknarova K, Gehrmann M, Banyai L, Tordai H, Patthy L, Llinas M. J Biol Chem. 2001;276:27613–27621. doi: 10.1074/jbc.M101105200. [DOI] [PubMed] [Google Scholar]
- 19.Feng Y, Likos JJ, Zhu L, Woodward H, Munie G, McDonald JJ, Stevens AM, Howard CP, De Crescenzo GA, Welsch D, Shieh HS, Stallings WC. Biochim Biophys Acta, Proteins Proteomics. 2002;1598:10–23. doi: 10.1016/s0167-4838(02)00307-2. [DOI] [PubMed] [Google Scholar]
- 20.Lukacova V, Zhang Y, Mackov M, Baricic P, Raha S, Calvo J, Balaz S. J Biol Chem. 2004;279:14194–14200. doi: 10.1074/jbc.M313474200. [DOI] [PubMed] [Google Scholar]
- 21.Diaz N, Suarez D, Sordo T. J Phys Chem B. 2006;110:24222–24230. doi: 10.1021/jp0656882. [DOI] [PubMed] [Google Scholar]
- 22.Tochowicz A, Maskos K, Huber R, Oltenfreiter R, Dive V, Yiotakis A, Zanda M, Bode W, Goettig P. J Mol Biol. 2007;371:989–1006. doi: 10.1016/j.jmb.2007.05.068. [DOI] [PubMed] [Google Scholar]
- 23.Diaz N, Suarez D. Proteins: Struct, Funct, Bioinf. 2008;72:50–61. doi: 10.1002/prot.21894. [DOI] [PubMed] [Google Scholar]
- 24.Ikejiri M, Bernardo MM, Bonfil RD, Toth M, Chang M, Fridman R, Mobashery S. J Biol Chem. 2005;280:33992–34002. doi: 10.1074/jbc.M504303200. [DOI] [PubMed] [Google Scholar]
- 25.Fisher JF, Mobashery S. Cancer Metastasis Rev. 2006;25:115–136. doi: 10.1007/s10555-006-7894-9. [DOI] [PubMed] [Google Scholar]
- 26.Khandelwal A, Balaz S. J Comput-Aided Mol Des. 2007;21:131–137. doi: 10.1007/s10822-007-9104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khandelwal A, Balaz S. Proteins: Struct, Funct, Bioinf. 2007;69:326–339. doi: 10.1002/prot.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta SP. Chem Rev. 2007;107:3042–3087. doi: 10.1021/cr030448t. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Lukacova V, Bartus V, Nie X, Sun G, Manivannan E, Ghorpade S, Jin X, Manyem S, Sibi M, Cook G, Balaz S. Chem Biol Drug Des. 2008;72:237–248. doi: 10.1111/j.1747-0285.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown S, Bernardo MM, Li ZH, Kotra LP, Tanaka Y, Fridman R, Mobashery S. J Am Chem Soc. 2000;122:6799–6800. [Google Scholar]
- 31.Forbes C, Shi QC, Fisher JF, Lee M, Hesek D, Llarrull LI, Toth M, Gossing M, Fridman R, Mobashery S. Chem Biol Drug Des. 2009;74:527–534. doi: 10.1111/j.1747-0285.2009.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao P, Fisher JF, Shi QC, Vroven T, Mobashery S, Schlegel HB. Biochemistry. 2009;48:9839–9847. doi: 10.1021/bi901118r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M, Bernardo MM, Meroueh SO, Brown S, Fridman R, Mobashery S. Org Lett. 2005;7:4463–4465. doi: 10.1021/ol0517269. [DOI] [PubMed] [Google Scholar]
- 34.Maseras F, Morokuma K. J Comput Chem. 1995;16:1170–1179. [Google Scholar]
- 35.Svensson M, Humbel S, Froese RDJ, Matsubara T, Sieber S, Morokuma K. J Phys Chem. 1996;100:19357–19363. [Google Scholar]
- 36.Humbel S, Sieber S, Morokuma K. J Chem Phys. 1996;105:1959–1967. [Google Scholar]
- 37.Dapprich S, Komaromi I, Byun KS, Morokuma K, Frisch MJ. J Mol Struc -THEOCHEM. 1999;461–462:1–21. [Google Scholar]
- 38.Vreven T, Morokuma K. J Comput Chem. 2000;21:1419–1432. [Google Scholar]
- 39.Vreven T, Morokuma K, Farkas O, Schlegel HB, Frisch MJ. J Comput Chem. 2003;24:760–769. doi: 10.1002/jcc.10156. [DOI] [PubMed] [Google Scholar]
- 40.Vreven T, Frisch MJ, Kudin KN, Schlegel HB, Morokuma K. Mol Phys. 2006;104:701–714. [Google Scholar]
- 41.Vreven T, Byun KS, Komaromi I, Dapprich S, Montgomery JA, Jr, Morokuma K, Frisch MJ. J Chem Theory Comput. 2006;2:815–826. doi: 10.1021/ct050289g. [DOI] [PubMed] [Google Scholar]
- 42.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 43.Lundberg M, Kawatsu T, Vreven T, Frisch MJ, Morokuma K. J Chem Theory Comput. 2009;5:222–234. doi: 10.1021/ct800457g. [DOI] [PubMed] [Google Scholar]
- 44.Meroueh SO, Fisher JF, Schlegel HB, Mobashery S. J Am Chem Soc. 2005;127:15397–15407. doi: 10.1021/ja051592u. [DOI] [PubMed] [Google Scholar]
- 45.Bayly CI, Cieplak P, Cornell W, Kollman PA. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 46.Cornell WD, Cieplak P, Bayly CI, Kollmann PA. J Am Chem Soc. 1993;115:9620–9631. [Google Scholar]
- 47.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Scalmani G, Mennucci B, Barone V, Petersson GA, Caricato M, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Li X, Hratchian HP, Peralta JE, Izmaylov AF, Kudin KN, Heyd JJ, Brothers E, Staroverov V, Zheng G, Kobayashi R, Normand J, Sonnenberg JL, Iyengar SS, Tomasi J, Cossi M, Rega N, Burant JC, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Chen W, Wong MW, Pople JA. Revision F.02 ed. Gaussian, Inc; Wallingford, CT: 2007. [Google Scholar]
- 48.Tao P, Schlegel HB. J Comput Chem. 2010;31:2363–2369. doi: 10.1002/jcc.21524. [DOI] [PubMed] [Google Scholar]
- 49.Tao P, Fisher JF, Shi Q, Mobashery S, Schlegel HB. J Phys Chem B. 2010;114:1030–1037. doi: 10.1021/jp909327y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wigner E. Z Physik Chem. 1932;B19:203–216. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.