Abstract
Aim
To test the contribution of programmed cell death 4 (PDCD4) tumour suppressor gene in Barrett's carcinogenesis.
Methods
PDCD4 immunohistochemical expression was assessed in 88 biopsy samples obtained from histologically proven long-segment Barrett's mucosa (BM; 25 non-intestinal columnar metaplasia, 25 intestinal metaplasia (IM), 16 low-grade intraepithelial neoplasia (LG-IEN), 12 high-grade IEN (HG-IEN) and 10 Barrett's adenocarcinoma (BAc)). As controls, 25 additional samples of native oesophageal mucosa (N) were obtained from patients with dyspepsia. To further support the data, the expression levels of miR-21, an important PDCD4 expression regulator, in 14 N, 5 HG-IEN and 11 BAc samples were determined by quantitative real-time PCR analysis.
Results
PDCD4 immunostaining decreased progressively and significantly with the progression of the phenotypic changes occurring during Barrett's carcinogenesis (p<0.001). Normal basal squamous epithelial layers featured strong PDCD4 nuclear immunoreaction (mostly coexisting with weak–moderate cytoplasmic staining). Non-intestinal columnar metaplasia and intestinal metaplasia preserved a strong nuclear immunostaining; conversely, a significant decrease in PDCD4 nuclear expression was seen in dysplastic (LG-IEN and HG-IEN) and neoplastic lesions. Weak–moderate cytoplasmic immunostaining was evident in cases of LG-IEN, while HG-IEN and BAc samples showed weak cytoplasmic or no protein expression. As expected, miR-21 expression was significantly upregulated in HG-IEN and BAc samples, consistently with PDCD4 dysregulation.
Conclusions
These data support a significant role for PDCD4 downregulation in the progression of BM to BAc, and confirm miR-21 as a negative regulator of PDCD4 in vivo. Further efforts are needed to validate PDCD4 as a potential prognostic marker in patients with Barrett's oesophagus.
Keywords: Barrett's oesophagus, immunohistochemistry, oesophageal cancer, PDCD4, tumour markers, tumour suppressor gene
Introduction
In many western countries, the incidence of Barrett's adenocarcinoma (BAc) has dramatically increased over the last two decades. Barrett's oesophagus (BE) is defined as the replacement of squamous oesophageal epithelium by columnar intestinalised mucosa (Barrett's mucosa, BM) and this metaplastic transformation is recognised as the cancerisation field in which BAc develops.1–3
Many investigators have focused on which histopathological characteristics of BE have malignant potential, and which factors promote Barrett's carcinogenesis. The histological diagnosis of dysplasia (defined as low-grade or high-grade intraepithelial neoplasia (IEN or NiN)) is currently considered the only biomarker for defining high-risk BE populations.4–6 The identification of additional molecular markers of cancer progression might support strategies for cancer secondary prevention and/or provide a biological rationale for targeted therapies.
A growing number of reports have pointed to PDCD4 (programmed cell death 4) as a new tumour suppressor gene.7–13 PDCD4 is a 64 kDa protein involved in the apoptotic machinery, which suppresses cell transformation, tumorigenesis and invasion.7–13 Different mechanisms have been implicated in the control of the steady-state and subcellular location of PDCD4. Among others, the oncogenic microRNA miR-21 (hsa-miR-21) has been shown to specifically target the PDCD4 3′-UTR, which negatively regulates PDCD4 expression.14–19
PDCD4 expression is significantly downregulated in various human cancers, as well as in cancer cell lines, and this has been associated with a poor patient prognosis.19–24 PDCD4 protein levels have been found to be inversely correlated with miR-21 expression in oesophageal squamous cell carcinoma cell lines,18 and we have shown that PDCD4 expression is significantly downregulated in oesophageal cancers (adenocarcinoma and squamous cell carcinoma histotypes) and it predicts patient outcome.25
To test the role of PDCD4 in contributing to oesophageal carcinogenesis, we investigated PDCD4 immunohistochemical expression in Barrett's carcinogenesis. We also examined miR-21 expression levels in high-grade IEN (HG-IEN) and BAc samples by quantitative real-time PCR (qRT-PCR analysis).
Materials and methods
cDNA microarray analysis
The Oncomine database and gene microarray analysis tool, a repository for published cDNA microarray data (http://www.oncomine.org/),26 27 was explored (on 15 December 2009) for PDCD4 mRNA expression in non-neoplastic oesophageal tissues, BM and primary BAc. Oncomine algorithms were used to perform a statistical analysis of the differences in PDCD4 expression, since it allows for multiple comparisons among different studies.26–28 Only studies with analytical results with a p<0.05 were considered.
Patients
The cases in the present study were retrospectively collected from the files of the Veneto Region's multicentre Barrett's Oesophagus Registry (EBRA; Padova Unit),29 selecting cases of histologically proven, long-segment BE. A total of 88 biopsy samples of oesophageal mucosa were obtained from different patients with BE, that is, 25 with non-intestinal columnar metaplasia (cardiac-type columnar metaplasia), 25 with intestinal metaplasia (Barrett's mucosa), 16 with low-grade intraepithelial neoplasia (LG-IEN), 12 with high-grade IEN (HG-IEN), and 10 with BAc. Another 25 native oesophageal mucosa samples (N) were obtained from patients with dyspepsia who had no gastro-oesophageal disease at endoscopy/histology (from the files at the Surgical Pathology Unit of the University of Padova). For the qRT-PCR study, 2 mm cores were obtained from paraffin blocks selected from 14 consecutive patients who had undergone oesophagectomy at the Department of Gastroenterological and Surgical Sciences, University of Padova, for HG-IEN and/or BAc, giving a total of 14 N, 5 HG-IEN and 11 BAc samples. None of the patients had received neoadjuvant therapy. Written consent was obtained from all the patients involved in the study.
Histological and immunohistochemical study
All biopsy specimens were immediately fixed in 10% buffered formalin and embedded in paraffin wax. Serial histological sections 4–6 μm thick were obtained from each paraffin wax block and stained with H&E and Alcian-PAS. The original diagnosis was confirmed on the histological evidence in all cases. Immunohistochemical staining was done automatically (Ventana Benchmark XT system; Ventana, Touchstone, Arizona, USA)30 for PDCD4 (1:100; catalogue no. HPA001032; Atlas Antibodies, Stockholm, Sweden) according to the manufacturer's instructions. Sections were lightly counterstained with haematoxylin. Appropriate positive and negative controls were run concurrently. In cancer samples, the presence of positive stromal and inflammatory cells served as an internal control. PDCD4 expression was jointly scored by two pathologists (MF and MR) who were unaware of the patients' clinical history. Consistent with Mudduluru et al,22 nuclear and cytoplasmic immunoreactions were considered. Nuclear PDCD4 staining results were classified in four groups according to the percentage of positively stained nuclei: score 0, none; score 1, ≤30%; score 2, 30–70%; score 3, ≥70%. Cytoplasmic PDCD4 expression was scored by staining intensity (score 0, none; score 1, weak; score 2, intermediate; score 3, strong). A total score was also calculated as the sum of the nuclear and cytoplasmic scores, and cases were divided into four groups, defined as: negative (total score 0), weak staining (total score 1 or 2), intermediate staining (total score 3 or 4), and strong staining (total score 5 or 6).
Quantitative real-time PCR
Tissue cores were deparaffinised with xylene at 50°C for 3 min. Total RNA extraction was done using the RecoverAll kit (Ambion, Austin, Texas, USA) according to the manufacturer's instructions. The NCodeTM miRNA qRT-PCR method (Invitrogen, Carlsbad, California, USA) was used to detect and quantify mature miR-21 (primer sequence: 5′-CGGTAGCTTATCAGACTGATGTTGA-3′) on real-time PCR instruments according to the manufacturer's instructions (Applied Biosystems, Foster City, California, USA). Normalisation was done with the small nuclear RNA U6B (RNU6B; Invitrogen). PCR reactions were run in triplicate, including no-template controls. The data were analysed using the comparative CT method.
Statistical analysis
Kruskal–Wallis test, Student t test and Pearson's correlation tests were used to analyse differences and correlations between groups; p values <0.05 were considered significant. All the statistical analyses were done with Stata software (Stata Corporation, College Station, Texas, USA).
Results
The PDCD4 gene is downregulated in Barrett's carcinogenesis
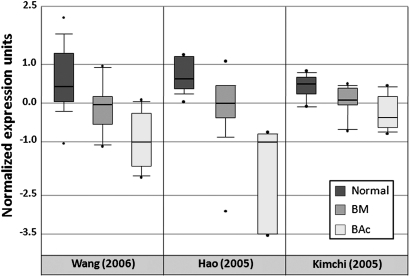
PDCD4 gene expression was analysed by checking different publicly available BE microarray studies using the Oncomine database and gene microarray data analysis tools.26 27 The analysis considered the mRNA expression levels for each of the studies involved, and the significance of the expression of the gene across the studies considered was also taken into account. In the three independent data sets of human Barrett's oesophageal carcinogenesis considered, PDCD4 mRNA expression levels decreased significantly when primary BAc was compared versus BE and versus native squamous epithelium (Pearson's correlation p=1.98×10−4)31–33 (figure 1).
Figure 1.
Expression array analysis of multiple available Barrett's oesophageal adenocarcinoma microarray data sets was performed for PDCD4, calculating the statistical significance, using the Oncomine database and gene microarray data analysis tool.31–33 PDCD4 expression in normal tissues (Normal), Barrett's mucosa (BM) and esophageal adenocarcinomas (BAc) is shown. Class analysis: Wang_Esophagus (correlation=−0.614; p=1.3×10−6; 24 normal, 19 BM, 9 BAc)32; Hao_Esophagus (correlation=−0.656; p=8.4 1×10−5; 15 normal, 14 BM, 5 BAc)33; Kimchi_Esophagus (correlation=−0.665; p=4.0×10−4; 8 normal, 8 BM, 8 BAc).31
PDCD4 expression is downregulated in BAc
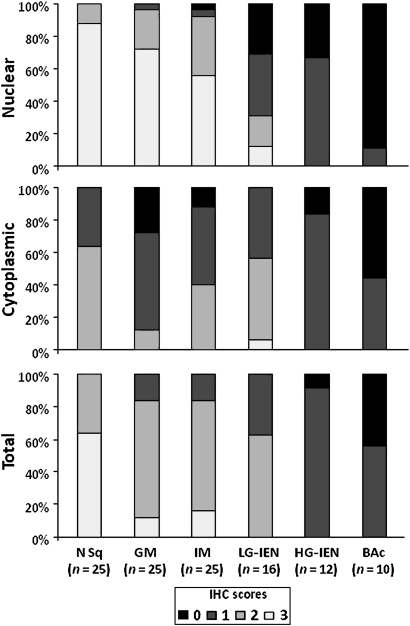
The normal basal squamous epithelial layer (proliferative zone) consistently featured strong PDCD4 nuclear expression (mostly coexisting with weak-moderate cytoplasmic staining; figures 2 and 3). In normal and cancer tissue samples, non-cancerous and non-epithelial cells (fibroblasts, lymphocytes, smooth muscle cells and endothelia) always featured PDCD4 nuclear expression (this finding was considered as a positive internal control and it did not interfere with the histological assessment).19 22 34 35 PDCD4 nuclear downregulation was consistently found in dysplastic and neoplastic lesions (Kruskal–Wallis p<0.001). In particular, moderate–strong nuclear staining was always associated with non-intestinal columnar metaplasia and intestinal metaplasia (scoring 2/3 in 24/25 and 23/25 cases, respectively), whereas intraepithelial (LG and HG) and invasive neoplasia showed a significantly lower PDCD4 nuclear expression (scores 2/3: 5/16, 0/12 and 0/10, respectively). Weak–moderate cytoplasmic staining was apparent in LG-IEN (scores 1/2: 15/16); HG-IEN and BAc samples showed weak cytoplasmic or no protein expression (score 1: 10/12 and 4/10, respectively). Overall, PDCD4 levels decreased progressively and significantly with the dedifferentiation of the lesions considered (Kruskal–Wallis p<0.001).
Figure 2.
Immunohistochemical (IHC) staining scores for PDCD4. Nuclear PDCD4 immunostaining was significantly weaker in high-grade intraepithelial neoplasia (HG-IEN) and Barrett's adenocarcinoma (BAc) lesions. Cytoplasmic PDCD4 expression was stronger in low-grade intraepithelial neoplasia (LG-IEN), but weaker in HG-IEN and BAc. The combined (total) PDCD4 staining scores were significantly lower in preneoplastic/neoplastic lesions (LG-IEN, HG-IEN and BAc) than in normal (N) and metaplastic tissues (non-intestinal columnar metaplasia (GM); intestinal metaplasia (IM)) (p<0.001).
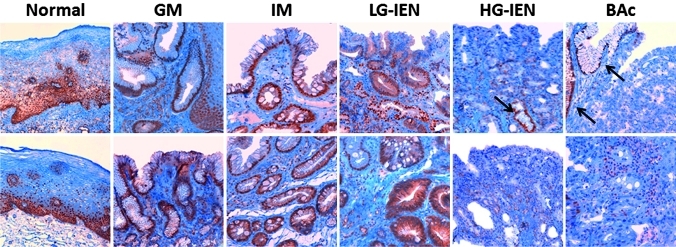
Figure 3.
Representative PDCD4 immunostainings of non-cancer oesophageal tissues and preneoplastic/neoplastic lesions in the Barrett's carcinogenic cascade. In native squamous oesophageal epithelium, normal oesophageal basal cells feature strong nuclear and weak–moderate cytoplasmic staining. Non-intestinal columnar metaplasia (GM) and intestinal metaplasia (IM) preserved a strong nuclear immunostaining. Weak–moderate cytoplasmic immunostaining was seen in cases of in low-grade intraepithelial neoplasia (LG-IEN), whereas in high-grade intraepithelial neoplasia (HG-IEN) and Barrett's adenocarcinoma (BAc) samples showed weak cytoplasmic or no protein expression. Note the presence in the HG-IEN and BAc examples of PDCD4-nuclear positive residual GM/IM glands (black arrows) (original magnifications ×20 and ×40).
miR-21 is overexpressed in HG-IEN and BAc
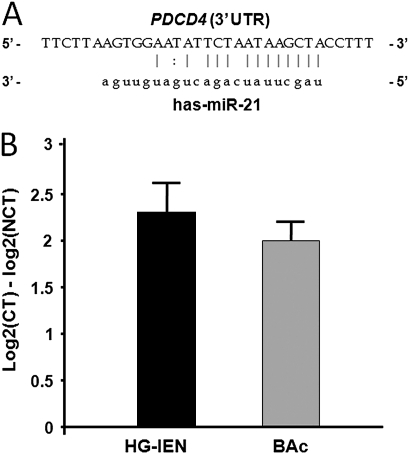
It has been demonstrated that PDCD4 is regulated directly by the oncomiR miR-21, with a 100% match sequence at nt228-249 (figure 4A).14–19 The PDCD4 3′-UTR miR-21 target sequence is highly conserved among different species,14 so we investigated miR-21 expression in preneoplastic and neoplastic Barrett's lesions. When we determined miR-21 expression levels in 14 N, 5 HG-IEN and 11 BAc samples (figure 4B), a significant miR-21 upregulation was found in the HG-IEN (with a 2.3-fold increase; t test p<0.001) and BAc (with a twofold increase; t test p<0.001).
Figure 4.
miR-21 is overexpressed in high-grade intraepithelial neoplasia (HG-IEN) and Barrett's adenocarcinoma (BAc). (A) Predicted base complementarity of miR-21 to 3′-UTR binding site of PDCD4, as predicted by the Sanger miR-database (miRBase, http://www.mirbase.org/). (B) Altered miR-21 expression between cancerous tissue (CT; HG-IEN and BAc) and non-cancerous tissue (NCT) in BAc patients by qRT-PCR analysis. miR-21 was significantly upregulated in HG-IEN and BAc samples (p<0.001). (Values are means±SD).
Discussion
The progression of BM to BAc passes through well-established histological changes, in which dysplasia is considered the most advanced phenotypic change before invasive cancer sets in.1 2 36 However, not all dysplastic lesions progress to cancer, and a better understanding of the molecular biology of this disease may lead to improvements in the diagnosis, treatment and prognosis of patients with BE.37
PDCD4 has been characterised as a new tumour suppressor gene, which is downregulated in several human malignancies.19–24 Its overexpression inhibits neoplastic transformation in both cancer cell lines and in vivo models.10 38 39
Low PDCD4 protein levels (assessed by western blot and qRT-PCR analyses) have been observed in oesophageal squamous cell carcinoma cell lines.18 In a large series of oesophageal tumours, we recently found that PDCD4 protein was completely lost or significantly reduced in almost 60% of the tumour samples considered.25 As in the lung cancer setting,20 no significant difference in PDCD4 dysregulation was observed between the two main histotypes of oesophageal cancer (adenocarcinoma (Barrett related and non-Barrett related) versus squamous cell carcinoma). Moreover, nuclear PDCD4 expression was associated with longer disease-free and overall survival rates.25
Hence we performed this study, which explored the immunohistochemical expression of PDCD4 in a series of Barrett-related lesions. PDCD4 protein expression was consistently lost/reduced in almost all HG-IEN and BAc tissue samples. In fact, the combined (nuclear and/or cytoplasmic) PDCD4 expression decreased significantly as the lesions considered became more dedifferentiated.
In particular, PDCD4 nuclear expression was significantly downregulated in preneoplastic–neoplastic tissue samples by comparison with normal metaplastic tissues. On the other hand, LG-IEN showed a weak–moderate cytoplasmic immunostaining. These data strongly suggest that the intracellular localisation of PDCD4 (as assessed by immunohistochemistry) is a useful additional diagnostic tool in the clinical and biological characterisation of BE-related lesions. A similar nuclear-to-cytoplasmic shift was documented by Mudduluru et al22 in colorectal carcinogenesis.
The biological interpretation of the subcellular expression of the protein remains inconsistent. In vitro studies have shown that, under normal growth conditions, PDCD4 is located mainly in the nucleus, moving to the cytoplasm on serum withdrawal.7 The nuclear-to-cytoplasm shift documented in preneoplastic/neoplastic lesions could theoretically result from epigenetic (and/or genetic) gene dysregulation.
In oesophageal squamous carcinoma cell lines, Hiyoshi et al recently demonstrated that PDCD4 gene expression is regulated by miR-21.18 We observed a significant upregulation of miR-21 in HG-IEN and BAc. miR-21 is overexpressed in different human cancers, being causally linked to cell proliferation, apoptosis and migration.14–19 miR-21 was also found to be upregulated in both oesophageal adenocarcinoma and squamous cell carcinoma,18 40 41 suggesting a major oncogenic function for miR-21 (acquired early in both of these oncogenic pathways).
In conclusion, our findings consistently point to PDCD4 being a promising biomarker in the identification of cancer-prone BM-related lesions. The association of PDCD4 with more traditional immunohistochemistry markers (and p53 in particular) may be useful in the routine histological assessment of BM-related lesions. In addition, the evidence of PDCD4 and miR-21 involvement in Barrett's carcinogenesis represents a solid biological rationale for innovative targeted therapy.
Take-home messages.
Programmed cell death 4 (PDCD4) is a 64 kDa protein that is downregulated in different human cancers, including oesophageal tumours.
The oncogenic microRNA miR-21 (hsa-miR-21) has been demonstrated to specifically target the PDCD4 3′-UTR, negatively regulating PDCD4 expression.
PDCD4 expression is significantly downregulated during Barrett's carcinogenesis.
PDCD4 and miR-21 are novel molecular targets of Barrett's carcinogenesis.
Acknowledgments
The authors are grateful to Valentina Ferri, Vanni Lazzarin and Cristiano Lanza for their technical assistance.
Footnotes
Funding: We would like to acknowledge the continuous support of the G Berlucchi Foundation.
Competing interests: None.
Patient consent: Obtained.
Contributors: All authors of this research paper participated directly in the planning, execution, and analysis of the study.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Sampliner RE. Update guidelines for the diagnosis, surveillance, and therapy of Barrett's esophagus. Am J Gastroenterol 2002;97:1888–95 [DOI] [PubMed] [Google Scholar]
- 2.Enzinger PC, Mayer RJ. Esophageal cancer. NEJM 2003;349:2241–52 [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225–49 [DOI] [PubMed] [Google Scholar]
- 4.Jankowski JA, Wright NA, Meltzer SJ, et al. Molecular evolution of the metaplasia–dysplasia–adenocarcinoma sequence in the esophagus. Am J Pathol 1999;154:965–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jankowski JA, Harrison RF, Perry I, et al. Barrett's metaplasia. Lancet 2000;356:2079–85 [DOI] [PubMed] [Google Scholar]
- 6.Conio M, Blanchi S, Lapertosa G, et al. Long-term endoscopic surveillance of patients with Barrett's esophagus. Incidence of dysplasia and adenocarcinoma: a prospective study. Am J Gastroenterol 2003;98:1931–9 [DOI] [PubMed] [Google Scholar]
- 7.Bohm M, Sawicka K, Siebrasse JP, et al. The transformation suppressor protein PDCD4 shuttles between nucleus and cytoplasm and binds RNA. Oncogene 2003;22:4905–10 [DOI] [PubMed] [Google Scholar]
- 8.Afonja O, Juste D, Das S, et al. Induction of PDCD4 tumour suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene 2004;23:8135–45 [DOI] [PubMed] [Google Scholar]
- 9.Bitomsky N, Bohm M, Klempnauer KH. Transformation suppressor protein PDCD4 interferes with JNK-mediated phosphorylation of c-Jun and recruitment of the coactivator p300 by c-Jun. Oncogene 2004;23:7484–93 [DOI] [PubMed] [Google Scholar]
- 10.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res 2005;65:6034–41 [DOI] [PubMed] [Google Scholar]
- 11.Dorrello NV, Peschiaroli A, Guardavaccaro D, et al. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006;314:467–71 [DOI] [PubMed] [Google Scholar]
- 12.Bitomsky N, Wethkamp N, Marikkannu R, et al. siRNA-mediated knockdown of PDCD4 expression causes upregulation of p21(Waf1/Cip1) expression. Oncogene 2008;27:4820–9 [DOI] [PubMed] [Google Scholar]
- 13.Carayol N, Katsoulidis E, Sassano A, et al. Suppression of programmed cell death 4 (PDCD4) protein expression by BCR-ABL-regulated engagement of the mTOR/p70 S6 kinase pathway. J Biol Chem 2008;283:8601–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008;27:2128–36 [DOI] [PubMed] [Google Scholar]
- 15.Frankel LB, Christoffersen NR, Jacobsen A, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 2008;283:1026–33 [DOI] [PubMed] [Google Scholar]
- 16.Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008;27:4373–9 [DOI] [PubMed] [Google Scholar]
- 17.Talotta F, Cimmino A, Matarazzo MR, et al. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene 2009;28:73–84 [DOI] [PubMed] [Google Scholar]
- 18.Hiyoshi Y, Kamohara H, Karashima R, et al. Micro-RNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res 2009;15:1915–22 [DOI] [PubMed] [Google Scholar]
- 19.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol 2009;219:214–21 [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Knosel T, Kristiansen G, et al. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol 2003;200:640–6 [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Ozaki I, Mizuta T, et al. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene 2006;25:6101–12 [DOI] [PubMed] [Google Scholar]
- 22.Mudduluru G, Medved F, Grobholz R, et al. Loss of programmed cell death 4 expression marks adenoma–carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer 2007;110:1697–707 [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Sun Z, Yang HS. Downregulation of tumor suppressor PDCD4 promotes invasion and activates both beta-catenin/Tcf and AP-1-dependent transcription in colon carcinoma cells. Oncogene 2008;27:1527–35 [DOI] [PubMed] [Google Scholar]
- 24.Yamamichi N, Shimomura R, Inada K, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res 2009;15:4009–16 [DOI] [PubMed] [Google Scholar]
- 25.Fassan M, Cagol M, Pennelli G, et al. Programmed cell death 4 (PDCD4) protein in esophageal cancer. Oncol Rep 2010;24:135–9 [DOI] [PubMed] [Google Scholar]
- 26.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhodes DR, Yu J, Shanker K, et al. Large-scale metaanalysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A 2004;101:9309–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin W, Rhodes DR, Ingold C, et al. Dysregulation of the annexin family protein family is associated with prostate cancer progression. Am J Pathol 2003;162:255–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaninotto G, Minnei F, Guirroli E, et al. The Veneto Region's Barrett's Oesophagus Registry: aims, methods, preliminary results. Dig Liver Dis 2007;39:18–25 [DOI] [PubMed] [Google Scholar]
- 30.Rugge M, Fassan M, Clemente R, et al. Bronchopulmonary carcinoid: phenotype and long-term outcome in a single-institution series of Italian patients. Clin Cancer Res 2008;14:149–54 [DOI] [PubMed] [Google Scholar]
- 31.Kimchi ET, Posner MC, Park JO, et al. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res 2005;65:3146–54 [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Zhan M, Yin J, et al. Transcriptional profiling suggests that Barrett's metaplasia is an early intermediate stage in esophageal adenocarcinogenesis. Oncogene 2006;25:3346–56 [DOI] [PubMed] [Google Scholar]
- 33.Hao Y, Triadafilopoulos G, Sahbaie P, et al. Gene expression profiling reveals stromal genes expressed in common between Barrett's esophagus and adenocarcinoma. Gastroenterology 2006;131:925–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshinaga H, Matsuhashi S, Fujiyama C, et al. Novel human PDCD4 (H731) gene expressed in proliferative cells is expressed in the small duct epithelial cells of the breast as revealed by an anti-H731 antibody. Pathol Int 1999;49:1067–77 [DOI] [PubMed] [Google Scholar]
- 35.Goke R, Barth P, Schmidt A, et al. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21(Waf1/Cip1). Am J Physiol Cell Physiol 2004;287:C1541–6 [DOI] [PubMed] [Google Scholar]
- 36.Paulson TG, Reid BJ. Focus on Barrett's esophagus and esophageal adenocarcinoma. Cancer Cell 2004;6:11–16 [DOI] [PubMed] [Google Scholar]
- 37.Fitzgerald RC. Molecular basis of Barrett's oesophagus and oesophageal adenocarcinoma. Gut 2006;55:1810–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nieves-Alicea R, Colburn NH, Simeone AM, et al. Programmed cell death 4 inhibits breast cancer cell invasion by increasing tissue inhibitor of metalloproteinases-2 expression. Breast Cancer Res Treat 2009;114:203–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang HS, Knies JL, Stark C, et al. PDCD4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene 2003;22:3712–20 [DOI] [PubMed] [Google Scholar]
- 40.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg 2008;135:255–60 [DOI] [PMC free article] [PubMed] [Google Scholar]