Abstract
In previous studies we found expression of the protein colligin 2 (heat shock protein 47 (HSP47), SERPINH1) in glioma neovasculature while not in normal brain tissue. Generally, the regulation of heat shock gene expression in eukaryotes is mediated by heat shock factors (HSF). In mammals, three heat shock transcription factors, HSF-1, -2, and -4, have been isolated. Here we investigated the relation between the expression of colligin 2 and these heat shock factors at the mRNA level using real-time reverse transcriptase PCR (qRT-PCR) in different grades of astrocytic tumorigenesis, viz., low-grade glioma and glioblastoma. Endometrium samples, representing physiological angiogenesis, were included as controls. Since colligin 2 is a chaperon for collagens, the gene expression of collagen I (COL1A1) was also investigated. The blood vessel density of the samples was monitored by expression of the endothelial marker CD31 (PECAM1). Because NG2-immunopositive pericytic cells are involved in glioma neovascularization, the expression of NG2 (CSPG4) was also measured.
We demonstrate overexpression of HSF2 in both stages of glial tumorigenesis (reaching significance only in low-grade glioma) and also minor elevated levels of HSF1 as compared to normal brain. There were no differences in expression of HSF4 between low-grade glioma and normal brain while HSF4 was downregulated in glioblastoma. In the endometrium samples, none of the HSFs were upregulated. In the low-grade gliomas SERPINH appeared to be slightly overexpressed with a parallel 4-fold upregulation of COL1A1, while in glioblastoma there was over 5-fold overexpression of SERPINH1 and more than 150-fold overexpression of COL1A1. In both the lowgrade gliomas and the glioblastomas overexpression of CSPG4 was found and overexpression of PECAM1 was only found in the latter. Our data suggest that the upregulated expression of colligin 2 in glioma is accompanied by upregulation of COL1A1, CSPG4, HSF2 and to a lesser extent, HSF1. Further studies will unravel the association of these factors with colligin 2 expression, possibly leading to keys for therapeutic intervention.
Keywords: colligin 2, heat shock factor 2, glioblastoma blood vessels
A wide range of physiological and pathological stresses trigger the heat shock gene transcription. Cells respond to elevated temperatures and to chemical and physiological stresses by an increase in the synthesis of heat shock proteins. HSP are a highly conserved family of proteins which function as molecular chaperones or proteases.1,2 Molecular chaperones form a class of proteins that control the proper folding of nascent polypeptides into the correct 3D structure. During stress responses the role of HSPs is critical in preventing the appearance of intermediates that lead to misfolding or otherwise damaging molecules.3 HSPs assist in the recovery from stress either by repairing damaged proteins (protein refolding), or protein degrading, thus restoring protein homeostasis and promoting cell survival. The regulation of heat shock proteins is mediated by heat shock transcription factors (HSF). Under normal conditions, HSFs reside in the cytoplasm, but are activated upon stress and relocalize to the cell nuclei.4 Activated HSFs form a trimmer with high-affinity binding to DNA; it binds to heat shock elements (HSE) in the promoters of the heat shock genes.4 The activation results in the expression of heat shock proteins (HSPs). In vertebrates and plants, there are at least four members of the HSF gene family (HSF1–4),1 while in human cells, three HSFs (HSF-1, -2, and -4) have been characterized.2,5 The expression of HSF1 and HSF2 is ubiquitous. However, the factors that induce their activation differ. While HSF1 is activated by heat shock and other forms of stress, HSF2 activity has been associated with development and differentiation. The expression of HSF4 appears to be tissuespecific and is restricted to heart, skeletal muscle or brain.5 The simultaneous expression of the different HSFs in particular tissues would enable differential responses to various forms of stress. In the context of tumors, HSPs may be tumor-specific and therefore, they may well become therapeutic targets.6–8
In previous studies, we found specific overexpression of colligin 2 in glioma neovasculature as compared to the normal vasculature of the brain.9,10 Here we investigate whether there is a correlation between the expression of colligin 2 and any of the HSF genes (HSF1, HSF2 and HSF4) at the mRNA level in low- and high-grade glioma. We isolated RNA from human frozen samples by using RNeasy Micro kit (Qiagen BV, Venlo, The Netherlands). We measured the relative transcription levels of colligin 2 (SERPINH1), HSF1, 2 and 4 by real time RT-PCR in four glioblastoma (GBM) samples, four samples of low-grade glioma (LGG), four samples of proliferating endometrium and four samples of normal brain tissue. Prior to isolation all tissues were assessed by a qualified pathologist to ensure the origin and quality of the tissues. The blood vessel density of the samples was monitored by expression of the endothelial marker CD31 (PECAM1).11 Because NG2-immunopositive pericytic cells are involved in glioma neovascularization, the expression of NG2 (CSPG4) was also measured. The expression sites of the HSFs were visualized by confocal microscopy. NormFinder with the Datan Framework GenEx Pro package version 4.3.2 was used to calculate the most stable mRNA set for our four tissue groups.12 Therefore, a set of four reference genes were used for data normalization (GUSB, HMBS, HPRT1 and NOXA1) (see also our supplementary file). For statistical testing the mean values were used. Comparisons between groups were made by using the Kruskal Wallis test.
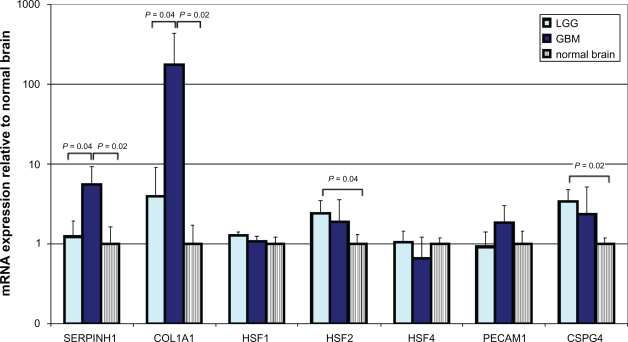
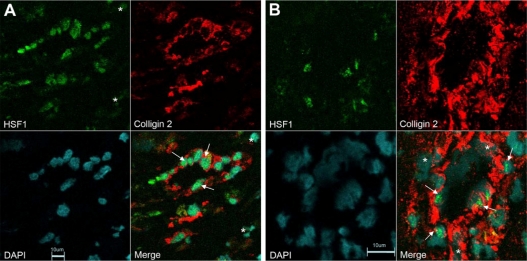
qRT-PCR revealed a significant 5.6-fold increase in mRNA levels for colligin 2 in glioblastoma and mildly elevated levels in low-grade glioma (Fig. 1). In parallel, significant increases in mRNA levels of collagen type I was found in the gliomas. Of the heat shock factors, HSF2 was overexpressed in glioma which reached significance in low-grade glioma. There was only minor overexpression of HSF1 while HSF4 was underexpressed in GBM (Fig. 1). Different types of blood vessels in glioblastoma showed a coexpression of both HSF1 and colligin 2. Endothelial cells of small blood vessels that expressed HSF1 were also immunopositive for colligin 2 (Fig. 2A). Additionally, some cells that compose the vessel wall of hypertrophied vessels co-expressed colligin 2 and HSF1 whiule others showed colligin 2 expression only (Fig. 2B). In GBM both PECAM1 (1.8-fold) and CSPG4 (2.3-fold) were overexpressed while only CSPG4 was overexpressed (3.4-fold) in LGG. We also observed 2.2-fold upregulation of the expression of colligin 2 mRNA in endometrium samples as compared to normal controls (not significant; P = 0.0833) with significant 55-fold upregulation of the expression of collagen I mRNA (P = 0.0209). Remarkably, none of the HSFs was upregulated in endometrium as compared to normal brain controls (HSF1: P = 0.1489; HSF2: P = 0.2482; HSF4: P = 0.2482).
Figure 1.
mRNA expression of colligin 2, HSF1, 2 and 3, collagen 1, CD31 and NG2 in low- and high-grade glioma and normal control brain. Data in this figure are the average ± SD of one representative experiment with 4 tissues in each group. Expression data are presented relative to the average mRNA expression levels measured in total RNA isolated from normal brain tissues (n = 4). Prior to isolation, all tissues were assessed by a qualified pathologist to ensure the origin and quality of the tissues. Total RNA was isolated with the RNeasy Micro kit (Qiagen BV, Venlo, The Netherlands). cDNA was prepared by use of the RevertAid H Minus First Strand cDNA synthesis kit (Fermentas, St Leon-Rot, Germany). The resulting cDNA preparations were analyzed by real-time PCR with TaqMan gene expression assays and TaqMan Universal PCR Master Mix (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). PCRs were performed in a 20 μL reaction volume in an Applied BioSystems 7900HT Fast Real-Time PCR system. Negative controls included minus RT and H2O-only samples, which showed to be negative in all cases. The most stable mRNA set for our 4 tissue groups were calculated with NormFinder19 with the Datan Framework GenEx Pro package version 4.3.2 and was shown to be a combination of GUSB, HMBS, HPRT1 and NOXA1. Expression of GUSB, HMBS, HPRT1 and NOXA1 was therefore used as a reference to control sample loading and RNA quality, as described previously.20
Differences in mRNA concentrations were determined by the non parametric Kruskal-Wallis test with P < 0.05 being considered statistically significant. All statistical tests were two-sided.
Abbreviations: LGG, low grade glioma; GBM, glioblastoma; SERPINH1, mRNA coding for colligin 2; COL1A1, mRNA coding for collagen 1; HSF, mRNA coding for heat shock factor; PECAM1, mRNA coding for CD31; CSPG4, mRNA coding for NG2; RT-PCR, reverse transcriptase–polymerase chain reaction; cDNA, complementary DNA.
Figure 2.
Expression of HSF1 and colligin 2 in blood vessels of glioblastoma. A) Double immunolabeling for HSF1 and colligin 2 in small blood vessels of glioblastoma. The endothelial cells express both colligin 2 = red and HSF1 = green (arrows). Colligin 2 is exclusively expressed in the blood vessels while HSF1 is expressed in some cells that surround the blood vessel (asterix). B) Double immunolabeling for HSF1 and colligin 2 in hypertrophied blood vessels of glioblastoma. Some of the endothelial cells and the pericytes in the blood vessels co-express colligin 2 = red and HSF1 = green (arrows). Some cells in the blood vessel wall express colligin 2 exclusively (asterix). The co-expression of colligin 2 and HSF1 in blood vessels of glioblastoma depends on the blood vessel type and in the activity level of a blood vessel. We used normal brain samples that do not express colligin 2 as a negative control. For the quality control we used adjacent slides from the same glioblastoma samples stained with the secondary antibodies only. We obtained negative controls for both single and double staining.
Several studies showed the transcription of HSP genes requires the activation and translocation to the nucleus of heat shock factors (HSF’s).13 It has been shown that HSF1 induces the mRNA expression of colligin 2 in rats14 and in young zebrafish.15 This is the first report on the parallel upregulation of colligin 2 and heat shock factors in human glioma. Heat shock responses appear to be implicated in a broad range of pathological conditions including heat shock, oxidative stress, ischemia and reperfusion, inflammation, tissue damage, exposure to heavy metals and infection.1 In addition, tumor neovascularization is associated with the heat shock response. Mammals have three different HSFs which are considered to be functionally distinct: HSF1 is essential for the heat shock response and is also required for developmental processes; HSF2 and HSF4 are important for differentiation and development.16 Although deletion of HSF1 in mammalian cells still allows a basal expression of HSPs, it leads to the abrogation of induction of the response to a variety of stresses.17,18 The genes encoding HSF1 and HSF2 are constitutively expressed in most cell lines and tissues under normal growth conditions and both factors are kept in a latent, non-DNA-binding state, indicating that the DNA-binding activity of both HSF1 and HSF2 is negatively regulated.19 Despite differences in expression, HSF1 and HSF2 both act as positive activators of transcription for all their functions. In contrast, HSF4 lacks activity of a transcriptional activator.20 HSF4 is highly expressed in the lens and in brain. It may well be a HSE binding trimmer because it lacks an inhibitory domain of trimerization. HSF4 constitutively binds to DNA and regulates the expression of HSP in the absence of stress.21 HSF4 contains two alternative splice variants: HSF4a and HSF4b.22 HSF4a isoform acts as a repressor of HSF122 by competitively binding to the heat shock element (HSE). This is the first observation of downregulation of HSF4 in glioblastoma and is suggestive of a role of HSF4 in the regulation of the expression of HSPs including colligin 2. High-resolution chromatin immunoprecipitation on microarray (ChIP-chip) screens have successfully been used for identifying direct target genes for many transcription factors.23 This approach has also been used for searching target genes for HSF216 and may be used for further unravelling the relation between HSF2, HSF1, HSF4 and colligin 2. In the proliferating endometrium samples we found overexpression of colligin 2 and collagen type I. In contrast to the glioma samples, no associated upregulation of any of the three HSFs was observed in these samples (data not shown; please see the supplementary file). This may well be an important difference between physiological and neoplastic angiogenesis. Obviously, if there is a causal relation between the upregulated HSFs found and the expression of colligin 2, the HSFs may become important for the design of antiangiogenic therapy.
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst. 2000;92(19):1564–72. doi: 10.1093/jnci/92.19.1564. [DOI] [PubMed] [Google Scholar]
- 2.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–69. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 3.Sistonen L, Sarge KD, Phillips B, Abravaya K, Morimoto RI. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol. 1992;12(9):4104–11. doi: 10.1128/mcb.12.9.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13(3):1392–407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12(24):3788–96. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 6.Ma WW, Adjei AA. Novel agents on the horizon for cancer therapy. CA Cancer J Clin. 2009;59(2):111–37. doi: 10.3322/caac.20003. [DOI] [PubMed] [Google Scholar]
- 7.Fuller KJ, Issels RD, Slosman DO, Guillet JG, Soussi T, Polla BS. Cancer and the heat shock response. Eur J Cancer. 1994;30A(12):1884–91. doi: 10.1016/0959-8049(94)00362-9. [DOI] [PubMed] [Google Scholar]
- 8.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mustafa D, van der Weiden M, Zheng P, Nigg A, Luider TM, Kros JM. Expression Sites of Colligin 2 in Glioma Blood Vessels. Brain Pathol. 2008 doi: 10.1111/j.1750-3639.2008.00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mustafa DA, Burgers PC, Dekker LJ, et al. Identification of glioma neovascularization-related proteins by using MALDI-FTMS and nano-LC fractionation to microdissected tumor vessels. Mol Cell Proteomics. 2007;6(7):1147–57. doi: 10.1074/mcp.M600295-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Folkerth RD. Descriptive analysis and quantification of angiogenesis in human brain tumors. J Neurooncol. 2000;50:165–72. doi: 10.1023/a:1006499824379. [DOI] [PubMed] [Google Scholar]
- 12.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 13.Santoro MG. Heat shock factors and the control of the stress response. Biochem Pharmacol. 2000;59:55–63. doi: 10.1016/s0006-2952(99)00299-3. [DOI] [PubMed] [Google Scholar]
- 14.Heydari AR, You S, Takahashi R, Gutsmann-Conrad A, Sarge KD, Richardson A. Age-related alterations in the activation of heat shock transcription factor 1 in rat hepatocytes. Exp Cell Res. 2000;256:83–93. doi: 10.1006/excr.2000.4808. [DOI] [PubMed] [Google Scholar]
- 15.Jill M, Murtha Evan T. Keller: Characterization of the heat shock response in mature zebrafish (Danio rerio) Experimental Gerontology. 2003;38:683–91. doi: 10.1016/s0531-5565(03)00067-6. [DOI] [PubMed] [Google Scholar]
- 16.Akerfelt M, Henriksson E, Laiho A, et al. Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc Natl Acad Sci U S A. 2008;105(32):11224–9. doi: 10.1073/pnas.0800620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao X, Zuo X, Davis AA, et al. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. Embo J. 1999;18(21):5943–52. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodson ML, Park-Sarge OK, Sarge KD. Tissue-dependent expression of heat shock factor 2 isoforms with distinct transcriptional activities. Mol Cell Biol. 1995;15(10):5288–93. doi: 10.1128/mcb.15.10.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sistonen L, Sarge KD, Morimoto RI. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol Cell Biol. 1994;14(3):2087–99. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17(1):469–81. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. Faseb J. 2001;15(7):1118–31. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 22.Frejtag W, Zhang Y, Dai R, Anderson MG, Mivechi NF. Heat shock factor-4 (HSF-4a) represses basal transcription through interaction with TFIIF. J Biol Chem. 2001;276(18):14685–94. doi: 10.1074/jbc.M009224200. [DOI] [PubMed] [Google Scholar]
- 23.van Steensel B. Mapping of genetic and epigenetic regulatory networks using microarrays. Nat Genet. 2005;37(Suppl):S18–24. doi: 10.1038/ng1559. [DOI] [PubMed] [Google Scholar]; Sieuwerts AM, Meijer-van Gelder ME, Timmermans M, et al. How ADAM-9 and ADAM-11 differentially from estrogen receptor predict response to tamoxifen treatment in patients with recurrent breast cancer: a retrospective study. Clin Cancer Res. 2005;11(20):7311–21. doi: 10.1158/1078-0432.CCR-05-0560. [DOI] [PubMed] [Google Scholar]