Abstract
In contrast to rodents, adipose tissue serves as the major site of lipogenesis and storage reservoir for excess dietary energy in cattle. Research in rodents shows that adding corn oil (57% C18:2 n-6) to the diet alters lipogenesis enhancing deposition of omega-6 fatty acids. This study examines changes in lipogenic gene expression of subcutaneous adipose tissue from eighteen steers fed increasing levels of dietary corn oil [0 (NONE), 0.31 kg/d (MED) and 0.62 kg/d (HI)] using two platforms, qPCR and microarray. The results show that MED level of oil supplementation up-regulates gene expression of key lipogenic enzymes but that as oil supplementation reaches HI level mRNA encoding lipogenic enzymes responsible for de novo synthesis and desaturation are down-regulated. Changes in specific lipogenic mRNA levels are correlated with changes in tissue fatty acid composition where de novo and desatured fatty acids were reduced with the highest level of oil supplementation.
Keywords: bovine, omega-6 fatty acids, gene expression, microarray, qPCR
Introduction
Fat accumulation in bovine adipose tissues is the result of combined adipocyte hyperplasia and hypertrophy. However, Robelin1 showed that 70% of fat accumulation during postnatal growth is due to an increase in adipocyte hypertrophy alone. Fatty acids used for fat accumulation and adipocyte hypertrophy originate from either de novo fatty acid synthesis or dietary sources. In rodents, dietary polyunsaturated fatty acids (PUFA; both omega-6 and omega-3) are potent inhibitors of hepatic de novo lipogenesis.2–4 Jeffcoat and James2 reported that supplementing corn oil to weanling rats at levels from 0.5 to 10% of diet (w/w) reduced FASN activity to about 30% of the level for a fat-free diet. Further studies showed diets rich in PUFA fed to adult and weanling rats reduced hepatic abundance of FASN message by 2-fold3 and 5.8-fold.4 The majority of research to date examining changes in lipogenic gene expression and dietary factors regulating gene expression has been conducted in rodents; however, several differences exist among murine and bovine lipogenesis including location of and primary substrate utilized for fatty acid synthesis.5–8 Therefore, our objective was to examine changes in expression of genes involved in lipogenesis (LPL, ACC, FASN, and SCD) and proposed transcription factors or co-activators (PPARγ, C/EBPα, NF-Y, STAT5, Spot-14, SREBP1) in bovine adipose tissues from animals supplemented with increasing levels corn oil (57% C18:2 n-6).
Materials and Methods
Eighteen Angus steers (438 ± 4 kg body weight) were randomly assigned to 1 of 3 supplementation treatments. The 3 treatments consisted of 3 corn oil (57% linoleic acid, 28% oleic acid; 11% palmitic acid) supplementation levels: 0 (NONE), 0.31 kg/d (MED) and 0.62 kg/d (HI). Pelleted cottonseed hulls were used as a carrier for the oil supplement and were fed at equal amounts to all steers regardless of treatment throughout the experiment. Additional information on dietary conditions and animal performance are available in Pavan et al (2007).9 After 116 d on treatment, animals were euthanized and a sample of subcutaneous fat from the tail head region was removed from each carcass. Approximately, 10 g of subcutaneous fat was immediately frozen in liquid nitrogen and stored at −80 °C for subsequent RNA extraction.
Initial attempts to isolate high quality RNA from subcutaneous adipose tissue using TRIzol (Invitrogen, Town, CA) procedure found low 260/280 ratios (≤1.6) and yields (<10 ug/g). Therefore, we developed a method (Table 1) that would yield high quality RNA in appropriate amounts for qRT-PCR and microarray from subcutaneous adipose tissue. Snapfrozen adipose samples (2 g) were homogenized in 5 vol of TRIzol and centrifuged at 2600 x g, 2 °C for 30 min. to remove cellular debri and excess lipid. Chloroform (0.2 ml/ml of TRIzol used) was added, mixed, and centrifuged at 2600 x g, 2 °C for 30 min. Isopropanol (0.57x vol of aqueous layer) was added to the clear aqueous layer, and this phase was added to PureYield™ RNA Midiprep columns (Promega, Madison, WI). RNA purification was used completed following the manufacturer’s directions. tcRNA was quantified using Quant-iT™ RiboGreen RNA Reagent (Invitrogen, Town, CA). Integrity of the isolated RNAs was confirmed by visualization of 18S and 28S ribosomal bands of individual samples subjected to denaturing slab gel electrophoresis in 1.2% agarose gel and 260/280 absorbance ratios. All tcRNA samples used in qRT-PCR analysis exhibited 260/280 ratios of >1.8 and RNA yield averaged 32 ug/g adipose tissue.
Table 1.
Protocol developed for high quality RNA isolation from high fat, subcutaneous adipose tissues.
|
For microarray analysis, tcRNA was pooled by treatment for NONE and HI on an equal weight basis for all individual samples in a treatment group. The Affymetrix GeneChip Bovine Genome array was used for hybridization of the extracted mRNAs according to the Affymetrix protocol at Medical College of Georgia, Genomics facility. Four GeneChips were analyzed per treatment group. The GeneChip Bovine Genome Array contained 24,027 probe sets, representing over 23,000 transcripts. The array was scanned and the gene expression data was generated using the Affymetrix software. Statistical analysis was performed for each gene separately using the following simple linear model: yijk = μi + Tij + eijk, where yijk is the base-2 logarithm of background adjusted and normalized average difference of signal intensity for gene i; μi is the overall mean expression of gene i, Tij is dietary supplement j(j = 1,2) on gene i, and eijk is the residual term. F-ratios were computed for all possible contrasts between the treatment levels using the contrast statement in SAS. In order to control experiment wise error rates, a false discovery rate (FDR) criteria was employed. The Benjamini Hochberg method10 was used to identify genes differentially expressed at a given FDR level. The process is summarized in the following steps: 1) after selecting a significance level, αe, genes were sorted by P-values from the most to the least significant, 2) threshold values were calculated for each test as: thresholdi = i*αe/R for i = 1 to R, where R is the total number of tests (also the total number of genes) and i is the sorted position of the test, 3) each P-value was then compared to its respective threshold, starting with the most significant P-value, until a P-value greater than the threshold value was encountered. All following contrast was then considered to be insignificant. For this study an FDR of 0.05 was used to identify potentially differentially expressed genes. Least square estimates of both treatments for each single gene was calculated using the LSMEANS statement of SAS. The fold-change for each gene was calculated by taking the ratio of the least square estimates for all the two treatment comparisons to determine the magnitude of change in gene activity corresponding to each diet supplement.
PCR primers that span intron/exon junctions were designed using Primer3 software. Primer sequences for both end-point and real time quantitative PCR efficiencies are presented in Table 2. Primer sets were evaluated first by using end-point PCR. Pooled tcRNA (1 ug) was reverse-transcribed in a 20 ul reaction volume using oligo dT and SuperScript™ III reverse transcriptase (Invitrogen) in a 2-step RT-PCR reaction. PCR was conducted using GoTaq (Promega) and 100 pM of each respective primer. Products were subjected to slab gel electrophoresis and visualized by ethidium bromide staining and fluorescence. Further all products were purified and subjected to di-deoxy sequencing at the Clemson University Genomics Institute to verify identity. For primer efficiency, a standard curve based on the original mass of tcRNA in the RT reaction was generated (50, 10, 2, 0.4, 0.08, and 0.016 ng per reaction) ran in triplicate, and subjected to qPCR by using the QuantiTect® SYBR® Green PCR kit (Qiagen) on an Eppendorf® Mastercycler ® ep realplex (Eppendorf). Primer efficiency was calculated by regression analyses using the Eppendorf Mastercycler Software and all efficiencies were between 0.85–1.07. For all qPCR conducted the thermal cycling conditions included: DNA polymerase activation at 95 °C for 15 min., 40 PCR cycles for 15s at 94 °C, 30s at 60 °C, and 30s at 72 °C in the presence of 100 nM of each primer combination.
Table 2.
Primer sequences (5’ to 3’) for quantitative real-time PCR.
| Gene | Forward | Reverse | Efficiency |
|---|---|---|---|
| β-Actin | CTCTTCCAGCCTTCCTTCCT | GGGCAGTGATCTCTTTCTGC | 0.85 |
| ACC | AGCTGAATTTTCGCAGCAAT | GGTTTTCTCCCCAGGAAAAG | 1.07 |
| C/EBPα | TGGACAAGAACAGCAACGAG | GGTCATTGTCACTGGTCAGC | 0.95 |
| FASN | GCATCGCTGGCTACTCCTAC | GTGTAGGCCATCACGAAGGT | 0.93 |
| GAPDH | GGGTCATCATCTCTGCACCT | GGTCATAAGTCCCTCCACGA | 0.93 |
| LPL | GGGTTTTGAGCAAGGGTACA | GCCACAATGACCTTTCCAGT | 0.97 |
| NF-Y | ATGCAGGATCCAAATCAAGC | AAAAGGGCAGAATGTGATCG | 1.05 |
| PPARγ | AGGATGGGGTCCTCATATCC | GCGTTGAACTTCACAGCAAA | 0.86 |
| SCD | TTATTCCGTTATGCCCTTGG | GGTAGTTGTGGAAGCCCTCA | 0.95 |
| Spot14 | CCTCACCCATCTTACCCTGA | CAAGCTAGCAAACTGCACCA | 1.05 |
| SREBP1 | CTGGAGAAGCTGGACTGAGG | GCTTTCCCAAGACTCAGCAC | 0.86 |
| STAT5 | TGGGAAAGATGGGAACTGAG | ACCAACAAGTCTGGGTCAGG | 1.00 |
For real time quantitative PCR (qPCR), 1 μg of tcRNA per steer was reverse transcribed (RT) and 2 ng of RT reaction subjected to qPCR by using the QuantiTect® SYBR® Green PCR kit (Qiagen) on an Eppendorf ® Mastercycler® ep realplex (Eppendorf). Each 96-well plate with all samples, primer set of interest, and housekeeping gene was replicated. For all qPCR conducted the thermal cycling conditions included: DNA polymerase activation at 95 °C for 15 min., 40 PCR cycles for 15s at 94 °C, 30s at 60 °C, and 30s at 72 °C in the presence of 100 pM of each primer combination. Melting curves were generated at the conclusion of amplification to verify presence of a single product. Two genes, GAPDH and β-actin, were evaluated as housekeeping genes for data normalization. To determine the appropriate housekeeping gene to be used to normalize the data, the CT for GAPDH and β-actin, and all target genes per sample were entered into the BESTKEEPER program (http://www.gene-quantification.info/). The program determines the most stable housekeeping gene to be used for normalization by repeated pair-wise correlation analysis. Both GAPDH and β-actin in the analysis exhibited a coefficient of correlation of 0.99 (P < 0.001) and would be suitable for data normalization.
The transcript levels for each gene were calculated at cycle threshold values (CT) at which each fluorescent signal was first detected above background. For the analysis of relative gene expression, all CT values for each sample/primer pair combination and the respective primer efficiency were analyzed using the REST-2008 program (http://www.gene-quantification.de/rest-2008.html) for analysis and data normalized using GAPDH. The software calculates relative gene expression using Pair-wise Fixed Reallocation Randomization Test and relative expression determined at the 95% confidence interval. The fold-change in gene expression for MED or HI versus NONE were calculated using X−ΔΔCT method if upregulated or −1 divided by X−ΔΔCT if downregulated, where X = primer efficiency for each gene of interest, according to Pfaffl et al (2002)11 and these results are presented in the manuscript.
Results
RNA quality and yield from the TRIzol procedure without clean up were unacceptable for qRT-PCR or microarray analysis. After development of a new method for RNA extraction from high fat, subcutaneous adipose tissues (Table 1), all tcRNA samples used in these analyses exhibited 260/280 ratios of >1.8 and RNA yield averaged 32 μg/g adipose tissue (range of 15–50 ug/g).
Results from the Affymetrix GeneChip Bovine Genome array show that several genes involved in lipogenesis were altered with omega-6 fatty acid supplementation (Table 3). Carnitine palmitoytransferase II, fatty acid synthase (FASN), isocitrate dehydrogenase 1, hormone-sensitive lipase, glycerol-3-phosphate acyltransferase, diacylglycerol O-acyltransferase, 1-acylglycerol-3-phosphate O-acyltransferase 1, signal transducer and activator of transcription 5B (STAT5), and stearoyl-CoA desaturase (SCD) were all down regulated (P < 0.05) with HI levels of oil supplementation compared to NONE. Expression of CCAAT/enhancer binding protein (C/EBPα), C/EBPΔ, isocitrate dehydrogenase 3, lipoprotein lipase (LPL), long-chain fatty acid acyl elongase, and peroxisome proliferators activated receptor γ (PPARγ) mRNA were up-regulated (P < 0.05) with HI oil supplementation compared to NONE. Additional genes associated with fatty acid synthesis and metabolism that were identified to be differentially expressed in subcutaneous adipose tissues from steers supplemented with HI corn oil levels compared to NONE is shown in supplementary Table 1.
Table 3.
Microarray results for gene expression in subcutaneous adipose tissues from steers supplemented with HI corn oil levels compared to NONE.*
| Gene product | Gene symbol | P-value | Fold-change | Up or down regulated |
|---|---|---|---|---|
| Genes involved in lipogenesis | ||||
| Acetyl-CoA carboxylase | ACC | 0.17 | ||
| Carnitine palmitoyltransferase II | CPT2 | <0.01 | 2.63 | Down |
| Fatty acid synthase | FASN | <0.01 | 2.73 | Down |
| Isocitrate dehyrogenase 2 | IDH2 | 0.06 | 1.52 | Up |
| Isocitrate dehydrogenase 1 | IDH1 | <0.01 | 6.81 | Down |
| Lipase, hormone-sensitive | HSL | <0.01 | 7.46 | Down |
| Lipoprotein lipase | LPL | <0.01 | 3.70 | Up |
| Long-chain fatty acyl elongase | ELOVL | 0.04 | 4.96 | Up |
| Stearoyl-CoA desaturase | SCD | 0.001 | 7.78 | Down |
| Glycerol-3-phosphate acyltransferase mitochondrial | GPAT | <0.01 | 24.96 | Down |
| Putative diacylglycerol O-acyltransferase | DGAT2 | <0.01 | 5.40 | Down |
| 1-Acylglycerol-3-phosphate O-acyltransferase 1 | AGPAT1 | <0.01 | 7.96 | Down |
| Solute carrier family 25 (citrate transporter) | SLC25 A | 0.002 | 3.31 | Down |
| Transcription factors/co-activators involved in fat metabolism | ||||
| CCAAT/enhancer binding protein-α | C/EBPα | <0.01 | 14.31 | Up |
| CCAAT/enhancer binding protein-Δ | C/EBPΔ | <0.01 | 4.37 | Up |
| Peroxisome proliferators activated receptor γ | PPAR-γ | <0.01 | 34.99 | Up |
| Signal transducer and activator of transcription 5B | STAT5b | <0.01 | 2.54 | Down |
| Sterol regulatory element binding protein | SREBP | 0.23 | ||
Note:
If the FDR corrected P-value is not significant then the fold-change and directionality of the expression of those genes are not provided.
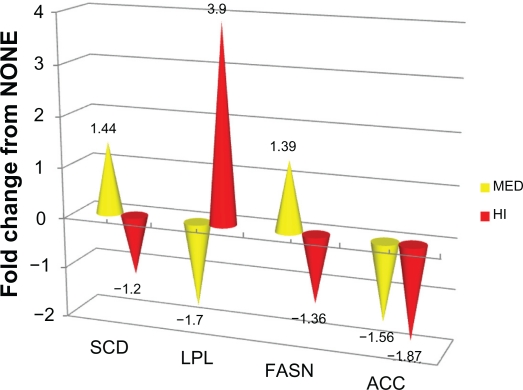
For qPCR analyses, expression of SCD mRNA was biomodal being up regulated (P < 0.05) in the MED group and down regulated (P < 0.05) in the HI group compared to NONE (Table 4; Figs. 1 and 2). Relative message levels for ACC mRNA were decreased (P < 0.05) compared to NONE for both oil treatments. At MED oil supplementation levels, FASN mRNA tended to be up-regulated (P = 0.08) compared to NONE. However as for expression levels of ACC and SCD, FASN message decreased (P = 0.05) in HI compared to NONE. Among the lipogenic genes examined, only LPL message levels increased (P < 0.05) by 3.9-fold in the HI treatment compared to NONE. At MED corn oil supplementation levels, LPL expression was down-regulated (P < 0.05) compared to NONE.
Table 4.
qPCR results for differential gene expression in subcutaneous adipose tissues from steers supplemented with MED and HI corn oil levels compared to NONE.
|
Corn oil treatment level |
MED |
HI |
|||||
|---|---|---|---|---|---|---|---|
| Gene | Gene symbol | P-level | Fold-change | Up or down | P-level | Fold-change | Up or down |
| Acetyl-CoA carboxylase | ACC | 0.03 | 1.56 | Down | 0.01 | 1.87 | Down |
| Fatty acid synthase | FASN | 0.08 | 1.39 | Up | 0.04 | 1.36 | Down |
| Lipoprotein lipase | LPL | 0.02 | 1.70 | Down | 0.02 | 3.9 | Up |
| Stearoyl-CoA desaturase | SCD | 0.01 | 1.44 | Up | 0.02 | 1.20 | Down |
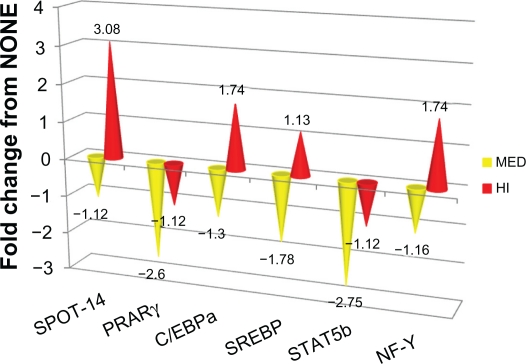
| CCAAT/enhancer binding protein-α | C/EBPα | 0.13 | 1.30 | Down | 0.06 | 1.63 | Down |
| Nuclear factor Y | NF-Y | 0.17 | 1.16 | Down | 0.02 | 1.74 | Up |
| Peroxisome proliferators activated receptor γ | PPARγ | 0.01 | 2.60 | Down | 0.42 | 1.12 | Down |
| Signal transducer and activator of transcription 5B | STAT5b | 0.03 | 2.75 | Down | 0.67 | 1.12 | Down |
| Spot-14 | Spot-14 | 0.08 | 1.12 | Down | 0.01 | 3.08 | Up |
| Sterol regulatory element binding protein | SREBP | 0.01 | 1.78 | Down | 0.26 | 1.13 | Up |
Figure 1.
Fold change in mRNA expression for lipogenic genes (ACC, FASN, LPL, SCD) for MED and HI versus NONE.
Figure 2.
Fold change in mRNA expression for transcription factors or co-activators (PPARγ, SREBP1, STAT5, C/EBPα, SPOT 14, NF-Y) for MED and HI versus NONE.
The relative expression of transcription factors or co-activators thought to be involved in fat metabolism12 is presented in Table 4. At MED supplementation level, the transcription factors of PPARγ, SREBP1, and STAT5 mRNA were down-regulated (P < 0.05) compared to NONE. Message levels of C/EBPα and NF-Y did not differ (P > 0.05) in MED group compared to NONE. At the HI corn oil level, PPARγ, STAT5 and SREBP-1 mRNA expression did not differ (P > 0.05) compared to NONE. C/EBPα mRNA expression level tended (P = 0.06) to be reduced with HI compared to NONE. NF-Y and Spot-14 message levels were up-regulated (P < 0.05) 1.7- and 3-fold, respectively, for HI group compared to NONE.
The correspondence between the two methods of gene expression analysis is presented in Table 5. Four out of eight genes showed similar expression changes. There was agreement between microarray and qPCR for FASN, LPL, SCD, and SREBP in which significance and direction were similar between the two platforms. In contrast, there was a lack of agreement between the platforms for changes in ACC, PPARγ, C/EBPα, and STAT5 mRNA expression with oil supplementation. For ACC, mRNA expression was unchanged (P > 0.05) using microarray analysis but was downregulated (P < 0.05) with qPCR analysis. For PPARγ and C/EBPα, mRNA expression was up-regulated (P < 0.05) in microarray analysis but unchanged (P > 0.05) according to qPCR. For STAT5, message levels were down-regulated (P < 0.05) for microarray but unchanged (P > 0.05) according to qPCR.
Table 5.
Comparison among microarray and qPCR results for HI compared to NONE.*
| Gene Symbol | MicroarrayP-value | qPCRP-value |
|---|---|---|
| ACC | 0.167 | <0.01* (1.87) ⇓ |
| FASN | <0.01* (2.73) ⇓ | 0.04* (1.36) ⇓ |
| LPL | <0.01* (3.70) ⇑ | 0.02* (1.58) ⇑ |
| SCD | <0.01* (7.78) ⇓ | 0.02* (1.20) ⇓ |
| PPARγ | <0.01* (34.99) ⇑ | 0.420 |
| SREBP | 0.229 | 0.26 |
| STAT5 | <0.01* (2.54) ⇓ | 0.67 (1.12) ⇓ |
| C/EBPα | <0.01* (14.31) ⇑ | 0.06 |
Note:
Indicates statistical significance; Fold change is shown with in the brackets; If the p-value is not statistically significant then the fold-change and direction of expression changes of those genes are not shown.
Discussion
Extraction of high quality RNA from fatty tissues is especially difficult compared to other tissues. Utilization of typical extraction methods, ie, TRIzol procedure, for fatty tissues results in poor RNA yields, unacceptable RNA quality and DNA carryover.13 Thus, extracted RNA from TRIzol procedure is unacceptable for use in qPCR or microarray without additional purification procedures that remove protein and/or DNA contaminants not effectively removed due to high lipid levels in adipose tissue. In addition, the low yield of RNA from fatty tissues requires the use of larger tissue samples sizes to purify adequate quantities for gene expression analyses including northern blotting, protection assays, qPCR, and microarray analyses. Therefore, we developed a new protocol for the extraction of high quality RNA from high fat, subcutaneous adipose tissues utilizing the chemistry of two commercially available RNA purification reagants. This protocol produced tcRNA samples with high quality (260/280 ratios of > 1.8) and adequate RNA yield (avg = 32 μg/g adipose tissue with range of 15–50 ug/g).
Lipogenic gene expression was altered due to dietary lipid in this study. Stearoyl-CoA desaturase catalyzes the rate-limiting step in monounsaturated fatty acid (MUFA) synthesis and conversion of trans-11 vaccenic acid (TVA) to cis-9 trans-11 conjugated linoleic acid (CLA), a potent anticarcinogen found in ruminant products.14 Oil supplementation level influenced the mRNA expression of SCD in subcutaneous adipose tissues. At MED levels, SCD mRNA was up-regulated; however at HI levels, SCD mRNA was down-regulated. Because the majority of cis-9 trans-11 CLA is produced via desaturation by SCD (80%),15 the down-regulation of SCD mRNA expression with HI would result in a lower conversion of TVA to CLA even though dietary linoleic acid supply was increasing. Indeed, a curvilinear response in CLA concentration with increasing corn oil supplementation was identified.9 Similarly, Duckett et al (2009)16 and Waters et al (2009)17 also reported down-regulation of SCD expression in adipose tissue of steers supplemented with corn oil or soybean oil.
Research in rodents has shown that omega-6 supplementation in vitro or in vivo down regulated SCD-1 mRNA. Rodents have several SCD isoforms, with SCD-1 being highly expressed in lipogenic tissues. 18 Jeffcoat and James (1978)2 found that SCD activity was diminished by 60% in the first 18 h after feeding a PUFA diet (corn oil) to rats. Addition of arachidonic acid to 3T3-L1 adipocytes in culture reduced SCD activity by 60% and SCD-1 mRNA by 80% within 6–12 h after treatment.19 Additional research in vitro showed that addition of individual PUFA (linoleic, linolenic and eicosapentaeinoic acid) also down-regulated SCD-1 mRNA; whereas, stearic and oleic acids did not alter SCD-1 mRNA level. Similarly, Ntambi (1992) found reductions in SCD-1 mRNA abundance after feeding triglycerides containing linoleic, linolenic and arachidonic acids but not with triglycerides containing palmitic, stearic or oleic acid compared to a fat-free diet. In the rodent studies, dietary PUFA supplementation was at higher levels (10% of diet) than in this study. Total dietary fatty acid content in this study was lower (HI = 8% of diet) than those in rodent studies (10%) but higher than recommended for optimal growth performance in high concentrate fed cattle (4%–6% of diet).20 In addition, dietary PUFA would be biohydrogenated in ruminant animals such that much smaller proportions of PUFA would reach the tissues and may explain the lower response in changes in gene expression to PUFA supplementation observed in this beef study versus rodent studies.
Both ACC and FASN are key enzymes regulating de novo fatty acid synthesis and palmitic acid production. These results indicate that at HI supplementation levels (8% fatty acids) both ACC and FASN were down regulated, which would result in lower de novo fatty acid synthesis and palmitic acid production. Indeed, tissue (L.M. and s.c.) concentrations of palmitic acid were lower in steers grazing endophyte free tall fescue with HI levels of oil supplementation.9 This reduction in palmitic acid content with oil supplementation was an interesting result from this research project as this fatty acid is considered to be hypercholesterolemic.21 Thus, oil supplementation appears to down-regulate de novo fatty acid synthesis to reduce the concentration of the end product, palmitic acid, a hypercholesterolemic fatty acid. Dietary polyunsaturated fatty acids (PUFA; both omega-6 and omega-3) are potent inhibitors of hepatic de novo lipogenesis in rodents.2–4 Wilson et al (1990)22 showed that PUFA feeding to rats resulted in a dose-dependent reduction of hepatic fatty acid synthesis in vivo. Jeffcoat and James (1978)2 reported that supplementing corn oil to weanling rats at levels from 0.5% to 10% of diet (w/w) reduced FASN activity to about 30% of the level for a fat-free diet. Further studies showed diets rich in PUFA fed to adult and weanling rats reduced hepatic abundance of FASN by 2-fold3 and 5.8-fold4 in vivo.
Lipoprotein lipase (LPL) is the enzyme responsible for the uptake of dietary fatty acids. These results suggest that HI levels of corn oil supplementation up-regulate LPL expression to uptake greater amounts of available dietary fatty acids. Similarly, Waylan et al (2004)23 reported that feeding flaxseed at 5% of diet also increased gene expression of LPL in muscle tissues.
Morrison and Farmer (2000)12 proposed several transcription factors that are believed to be involved in fat metabolism and these were examined in this study. Our qPCR results show that STAT5 was downregulated at MED oil supplementation level but unchanged at HI oil supplementation level. Hogans and Stephens (2005)24 found that STAT5 directly represses the expression of FASN in adipocytes. These results agree with those in this study as STAT5 was down-regulated and FASN up-regulated at the MED oil supplementation level. SREBP-1 contains a sterol regulatory element (SRE) and PUFA supplementation in rodents down regulates this transcription factor in rodents25 and chickens.26 Current research shows that FASN27 and SCD26 promoters contains recognition sites for SREBP-1, upstream stimulatory factor, stimulatory protein 1, and NF-Y, which are involved in the regulation by PUFA. Teran-Garcia et al (2007)27 has shown PUFA decrease in vivo binding of NF-Y and SREBP-1c to the proximal promoter of hepatic FASN, Spot 14 and SCD. These results suggest that PUFA regulate the binding of NF-Y and SREBP-1c to coordinately target genes involved in lipid metabolism. Waters et al (2008)17 also observed a decreased expression of SREBP-1c and no change in PPARα with 4% soybean oil and 2% fish oil addition to high concentrate diet in steers. PPARγ and C/EBPα are transcription factors known to be involved in the differentiation of adipocytes. Data suggests that the majority of adipocyte differentiation in beef animals occurs prior to 8 month of age.28 In addition, accumulation of fat in older beef animals appears to be primarily due to hypertrophy of the fat cell instead of fat cell proliferation/differentiation.1 Taken with our current data, the low levels of PPARγ and C/EBPα would support little adipocyte proliferation and greater adipocyte hypertrophy occurring for lipid accumulation in these steers at the time of this study.
Spot-14 is a nuclear protein that is responsive to thyroid hormone (T3) and is involved in regulation of lipid synthesis in rat hepatocytes.29 The function of Spot-14 is unknown but a high correlation between fatty acid synthase (FASN) and Spot-14 indicate that it is involved in lipid metabolism.3 Zhu et al30 suggested that Spot-14 protein is involved in allosteric regulation of de novo fatty acid synthesis. Zhu et al1 has also shown that Spot-14 knock-out mice had increased hepatic lipogenesis, which shows that Spot-14 is not required but may alter lipogenic rates. In this study, HI level of oil supplementation increased Spot-14 expression and tended to decrease FASN expression, which resulted in a reduction of palmitic acid, a product of de novo lipogenesis, in s.c. adipose tissues.
The correspondence between microarray and qRT-PCR for four lipogenic genes and four transcription factors of interest was within the acceptable limits. Joseph et al32 also reported a similar agreement among the two gene expression platforms. Hausman et al33 reported significant correlations and linear regression between microarray and RT-PCR; however they noticed that changes related to animal age were only detected by RT-PCR and not by microarray analysis. Even though both the platforms allow assay of gene expression, microarrays are infamous for their associated error both as technical and biological error as well as their sequence-specific effects which makes expression prediction difficult for at least some of the genes.34 This difference in the correspondence across the two platforms might also be accounted due to the pooling of RNA for microarray analysis where as individual RNA samples were used for qRT-PCR. Also, the qRT-PCR primer is designed based on the sequence database, which is derived from multiple sequence alignment of ESTs. As a result there is a high chance that Affymetrix may target one gene, while the sequence database may infact represent an entire gene family with less specificity and therefore will interrogate neither the Affymetrix gene nor the database gene and thus give results that differ from both platforms.35
The results from this study show that the MED level of oil supplementation (4.94% total fatty acids in diet) up-regulates gene expression of key lipogenic enzymes (FASN, SCD, or LPL) but that as oil supplementation reaches HI level (7.99% total fatty acids in diet) genes encoding lipogenic enzymes responsible for de novo fatty acid synthesis and MUFA synthesis are down-regulated. These results agree with changes in tissue fatty acid composition in which palmitic (C16:0) acid concentration, a product of de novo fatty acid synthesis, and CLA cis-9 trans-11 isomer, a product of desaturation, were reduced with the highest level of oil supplementation.
Supplementary Material
S Table 1.
Additional genes associated with fatty acid synthesis and metabolism that were identified to be differentially expressed in subcutaneous adipose tissues from steers supplemented with HI corn oil levels compared to NONE.
| Gene | Gene symbol | FDR corrected P-value | Pathway involved |
|---|---|---|---|
| Malic enzyme 3, NADP(+)-dependent, mitochondrial isoform 2 | ME3 | <0.01 | Esterification pathway |
| Malate dehydrogenase 2, mitochondrial | MDH2 | <0.01 | Esterification pathway |
| Pyruvate dehydrogenase phosphatase | PDP | 0.01 | Esterification pathway |
| Pyruvate dehydrogenase kinase isoform 4 | PDK4 | <0.01 | Esterification pathway |
| Pyruvate dehydrogenase (lipoamide) beta | PDHB | <0.01 | Esterification pathway |
| Adipose differentiation-related protein | ADFP | <0.01 | Esterification pathway |
| Propionyl-CoA carboxylase beta | PCCB | <0.01 | Proprionyl-CoA metabolism |
| Methylmalonyl-coA mutase | MCM | <0.01 | Proprionyl-CoA metabolism |
| Methylmalonyl-coA Epimerase | MCE | 0.04 | Proprionyl-CoA metabolism |
| Pyruvate Kinase | PK | <0.01 | Proprionyl-CoA metabolism |
| Succinate dehydrogenase complex, subunit C, integral membrane protein | SDHC | <0.01 | Proprionyl-CoA metabolism |
| Succinate dehydrogenase complex, subunit D, integral membrane protein | SDHD | <0.01 | Proprionyl-CoA metabolism |
| Aconitase 2, mitochondrial | ACO2 | <0.01 | Proprionyl-CoA metabolism |
| Phosphoenolpyruvate Carboxykinase 2 (mitochondrial) | PCK2 | <0.01 | Glycolysis/gluconeogenesis |
| Fructose 2,6-bisphosphatase | Fru-2,6-P2 | <0.01 | Glycolysis/gluconeogenesis |
| Glucose-6-phosphate dehydrogenase | G6PD | 0.027 | Pentose pathway |
| 6-Phosphogluconate dehydrogenase | 6PGDH | <0.01 | Pentose pathway |
| Sterol regulatory element binding protein cleavage-activating protein (SREBP cleavage-activating protein) | SCAP | 0.02 | Pentose pathway |
| Superoxide Dismutase 2, Mitochondrial | SOD2 | <0.01 | Pentose pathway |
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Robelin J. Growth of Adipose Tissues in Cattle—Partitioning between Depots, Chemical-Composition and Cellularity—a Review. Livestock Production Science. 1986;14:349–64. [Google Scholar]
- 2.Jeffcoat R, James AT. The control of stearoyl-CoA desaturase by dietary linoleic acid. FEBS Lett. 1978;85:114–8. doi: 10.1016/0014-5793(78)81260-5. [DOI] [PubMed] [Google Scholar]
- 3.Clarke SD, Armstrong MK, Jump DB. Dietary polyunsaturated fats uniquely suppress rat liver fatty acid synthase and S14 mRNA content. J Nutr. 1990;120:225–31. doi: 10.1093/jn/120.2.225. [DOI] [PubMed] [Google Scholar]
- 4.Jump DB, Clarke SD, Thelen A, Liimatta M. Coordinate regulation of glycolytic and lipogenic gene expression by polyunsaturated fatty acids. J Lipid Res. 1994;35:1076–84. [PubMed] [Google Scholar]
- 5.Ingle DL, Bauman DE, Garrigus US. Lipogenesis in the ruminant: in vivo site of fatty acid synthesis in sheep. J Nutr. 1972;102:617–23. doi: 10.1093/jn/102.5.617. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin RL, Reichl JR, Louis S, Smith NE, Yang YT, et al. Effects of age, pregnancy, and lactation on rat, guinea pig, and cow adipose enzyme activities and cow adipose metabolism. J Dairy Sci. 1973;56:340–9. doi: 10.3168/jds.S0022-0302(73)85177-X. [DOI] [PubMed] [Google Scholar]
- 7.Ingle DL, Bauman DE, Garrigus US. Lipogenesis in the ruminant: in vitro study of tissue sites, carbon source and reducing equivalent generation for fatty acid synthesis. J Nutr. 1972;102:609–16. doi: 10.1093/jn/102.5.609. [DOI] [PubMed] [Google Scholar]
- 8.Yang YT, Baldwin RL. Preparation and metabolism of isolated cells from bovine adipose tissue. J Dairy Sci. 1973;56:350–65. doi: 10.3168/jds.S0022-0302(73)85178-1. [DOI] [PubMed] [Google Scholar]
- 9.Pavan E, Duckett SK. Corn oil supplementation to steers grazing endophytefree tall fescue. II. Effects on longissimus muscle and subcutaneous adipose fatty acid composition and stearoyl-CoA desaturase activity and expression. J Anim Sci. 2007;85:1731–40. doi: 10.2527/jas.2006-732. [DOI] [PubMed] [Google Scholar]
- 10.Hochberg Y, Benjamini Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 11.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison RF, Farmer SR. Hormonal signaling and transcriptional control of adipocyte differentiation. J Nutr. 2000;(130):3116S–21. doi: 10.1093/jn/130.12.3116S. [DOI] [PubMed] [Google Scholar]
- 13.Mandrekar M, Brisco P, Hoffman K, Bitner R. Cleanup of TRIzol reagent-pufieid total TNA using the PureYield™ RNA Midiprep System. Promega Notes. 2006;94:7–11. [Google Scholar]
- 14.IP C, Singh M, Thompson HJ, Scimeca JA. Conjugtaed linoleic acid suppresses mammary carcinogenesis and proliferative activity of the mammary gland in the rat. Cancer Research. 1994;54:1212–5. [PubMed] [Google Scholar]
- 15.Mosley EE, Shafii B, Moate PJ, Mcguire MA. cis-9, trans-11 conjugated Linoleic Acid Is Synthesized Directly from Vaccenic Acid in Lactating Dairy Cattle. Journal of Nutrition. 2006;136:570–5. doi: 10.1093/jn/136.3.570. [DOI] [PubMed] [Google Scholar]
- 16.Duckett SK, Pratt SL, Pavan E. Corn oil or corn grain supplementation to steers grazing endophyte-free tall fescue. II. Effects on subcutaneous fatty acid content and lipogenic gene expression. J Anim Sci. 2009;87:1120–8. doi: 10.2527/jas.2008-1420. [DOI] [PubMed] [Google Scholar]
- 17.Waters SM, Kelly JP, O’Boyle P, Moloney AP, Kenny DA. Effect of level and duration of dietary n-3 polyunsaturated fatty acid supplementation on the transcriptional regulation of Delta(9)-desaturase in muscle of beef cattle. Journal of Animal Science. 2009;87:244–52. doi: 10.2527/jas.2008-1005. [DOI] [PubMed] [Google Scholar]
- 18.Miyazaki M, Jacobson MJ, Man WC, Cohen P, Asilmaz E, et al. Identification and characteriztation of murine SCD4, a novel heart-specific stearoyl-CoA desaturase isoform regulated by leptin and dietary factors. Journal of Biological Chemistry. 2003;278:33904–11. doi: 10.1074/jbc.M304724200. [DOI] [PubMed] [Google Scholar]
- 19.Sessler AM, Kaur N, Palta JP, Ntambi JM. Regulation of stearoyl-CoA desaturase 1 mRNA stability by polyunsaturated fatty acids in 3T3-L1 adipocytes. Journal of Biological Chemistry. 1996;271:29854–8. doi: 10.1074/jbc.271.47.29854. [DOI] [PubMed] [Google Scholar]
- 20.Zinn RA. Effects of excessive supplemental fat on feedlot cattle growth performance and digestive function. The Professional Animal Scientist. 1994;10:66–72. [Google Scholar]
- 21.Ulbricht TL, Southgate DA. Coronary heart disease: seven dietary factors. Lancet. 1991;338:985–92. doi: 10.1016/0140-6736(91)91846-m. [DOI] [PubMed] [Google Scholar]
- 22.Wilson MD, Blake WL, Salati LM, Clarke SD. Potency of Polyunsaturated and Saturated Fats as Short-Term Inhibitors of Hepatic Lipogenesis in Rats. Journal of Nutrition. 1990;120:544–52. doi: 10.1093/jn/120.6.544. [DOI] [PubMed] [Google Scholar]
- 23.Waylan AT, Dunn JD, Johnson BJ, Kayser JP, Sissom EK. Effect of flax supplementation and growth promotants on lipoprotein lipase and glycogenin messenger RNA concentrations in finishing cattle. Journal of Animal Science. 2004;82:1868–75. doi: 10.2527/2004.8261868x. [DOI] [PubMed] [Google Scholar]
- 24.Hogan JC, Stephens JM. The regulation of fatty acid synthase by STAT5A. Diabetes. 2005;54:1968–75. doi: 10.2337/diabetes.54.7.1968. [DOI] [PubMed] [Google Scholar]
- 25.Tabor DE, Kim JB, Spiegelman BM, Edwards PA. Transcriptional activation of the stearoyl-CoA desaturase 2 gene by sterol regulatory element-binding protein/adipocyte determination and differentiation factor 1. Journal of Biological Chemistry. 1998;273:22052–8. doi: 10.1074/jbc.273.34.22052. [DOI] [PubMed] [Google Scholar]
- 26.Lefevre P, Tripon E, Plumelet C, Douaire M, Diot C. Effects of polyunsaturated fatty acids and clofibrate on chicken stearoyl-coA desaturase 1 gene expression. Biochem Biophys Res Commun. 2001;280:25–31. doi: 10.1006/bbrc.2000.4070. [DOI] [PubMed] [Google Scholar]
- 27.Teran-garcia M, Adamson A, Yu W, et al. Polyunsaturated fatty acid suppression of fatty acid synthase (FASN): evidence for dietary modulation of NF-Y binding to the RASN promoter by SREBP-1c. Biochemical Journal. 2007;402:591–600. doi: 10.1042/BJ20061722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hood RL, Allen CE. Cellularity of bovine adipose tissue. J Lipid Res. 1973;14:605–10. [PubMed] [Google Scholar]
- 29.Kinlaw WB, Church JL, Harmon J, Mariash CN. Direct evidence for a role of the “spot 14” protein in the regulation of lipid synthesis. J Biol Chem. 1995;270:16615–8. doi: 10.1074/jbc.270.28.16615. [DOI] [PubMed] [Google Scholar]
- 30.Zhu QH, Anderson GW, Mucha GT, Parks EJ, Metkowski JK, et al. The spot 14 protein is required for de novo lipid synthesis in the lactating mammary gland. Endocrinology. 2005;146:3343–50. doi: 10.1210/en.2005-0204. [DOI] [PubMed] [Google Scholar]
- 31.Zhu QH, Mariash A, Margosian MR, Gopinath S, Fareed MT, et al. Spot 14 gene deletion increases hepatic de novo lipogenesis. Endocrinology. 2001;142:4363–70. doi: 10.1210/endo.142.10.8431. [DOI] [PubMed] [Google Scholar]
- 32.Joseph SJ, Robbins KR, Pavan E, Pratt SL, Duckett SK, et al. Effect of diet supplementation on the expression of bovine genes associated with Fatty Acid synthesis and metabolism. Bioinform Biol Insights. 2010;4:19–31. doi: 10.4137/bbi.s4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hausman GJ, Barb CR, Dean RG. Patterns of gene expression in pig adipose tissue: insulin-like growth factor system proteins, neuropeptide Y (NPY), NPY receptors, neurotrophic factors and other secreted factors. Domest Anim Endocrinol. 2008;35:24–34. doi: 10.1016/j.domaniend.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. Independence and reproducibility across microarray platforms. Nature Methods. 2005;2:337–43. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
- 35.Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. Independence and reproducibility across microarray platforms. Nat Methods. 2005;2:337–44. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S Table 1.
Additional genes associated with fatty acid synthesis and metabolism that were identified to be differentially expressed in subcutaneous adipose tissues from steers supplemented with HI corn oil levels compared to NONE.
| Gene | Gene symbol | FDR corrected P-value | Pathway involved |
|---|---|---|---|
| Malic enzyme 3, NADP(+)-dependent, mitochondrial isoform 2 | ME3 | <0.01 | Esterification pathway |
| Malate dehydrogenase 2, mitochondrial | MDH2 | <0.01 | Esterification pathway |
| Pyruvate dehydrogenase phosphatase | PDP | 0.01 | Esterification pathway |
| Pyruvate dehydrogenase kinase isoform 4 | PDK4 | <0.01 | Esterification pathway |
| Pyruvate dehydrogenase (lipoamide) beta | PDHB | <0.01 | Esterification pathway |
| Adipose differentiation-related protein | ADFP | <0.01 | Esterification pathway |
| Propionyl-CoA carboxylase beta | PCCB | <0.01 | Proprionyl-CoA metabolism |
| Methylmalonyl-coA mutase | MCM | <0.01 | Proprionyl-CoA metabolism |
| Methylmalonyl-coA Epimerase | MCE | 0.04 | Proprionyl-CoA metabolism |
| Pyruvate Kinase | PK | <0.01 | Proprionyl-CoA metabolism |
| Succinate dehydrogenase complex, subunit C, integral membrane protein | SDHC | <0.01 | Proprionyl-CoA metabolism |
| Succinate dehydrogenase complex, subunit D, integral membrane protein | SDHD | <0.01 | Proprionyl-CoA metabolism |
| Aconitase 2, mitochondrial | ACO2 | <0.01 | Proprionyl-CoA metabolism |
| Phosphoenolpyruvate Carboxykinase 2 (mitochondrial) | PCK2 | <0.01 | Glycolysis/gluconeogenesis |
| Fructose 2,6-bisphosphatase | Fru-2,6-P2 | <0.01 | Glycolysis/gluconeogenesis |
| Glucose-6-phosphate dehydrogenase | G6PD | 0.027 | Pentose pathway |
| 6-Phosphogluconate dehydrogenase | 6PGDH | <0.01 | Pentose pathway |
| Sterol regulatory element binding protein cleavage-activating protein (SREBP cleavage-activating protein) | SCAP | 0.02 | Pentose pathway |
| Superoxide Dismutase 2, Mitochondrial | SOD2 | <0.01 | Pentose pathway |