Abstract
A promising approach to developing a vaccine against O111 strains of diarrheagenic Escherichia coli that exhibit different mechanisms of virulence is to target either the core or the polysaccharide chain (O antigen) of their lipopolysaccharide (LPS). However, due to structural variations found in both these LPS components, to use them as antigen targets for vaccination, it is necessary to formulate a vaccine able to induce a humoral immune response that can recognize all different variants found in E. coli O111 strains. In this study, it was demonstrated that, despite differences in composition of oligosaccharide repeat units between O111ab and O111ac LPS subtypes, antibodies against one O111 subtype can recognize and inhibit the adhesion to human epithelial cells of all categories of O111 E. coli (enteropathogenic E. coli [EPEC], enterohemorrhagic E. coli [EHEC], and enteroaggregative E. coli [EAEC]) strains regardless of the nature of their flagellar antigens, mechanisms of virulence, or O111 polysaccharide subtypes. These antibodies were also able to increase the clearance of different strains of O111 E. coli by macrophages. PCR analyses of the pathways involved in O111 LPS core biosynthesis showed that all EAEC strains have core type R2, whereas typical EPEC and EHEC have core type R3. In contrast, atypical EPEC strains have core types R2 and R3. In summary, the results presented herein indicate that the O111 polysaccharide and LPS core types R2 and R3 are antigen targets for panspecific immunotherapy against all categories of O111 E. coli.
Pathogenic strains of O111 Escherichia coli exist as three distinct categories of diarrheagenic organisms, namely, enteropathogenic E. coli (EPEC; typical and atypical), enterohemorrhagic E. coli (EHEC), and enteroaggregative E. coli (EAEC) (7). In developing countries, diarrhea induced by these pathogens is a serious illness that inflicts a huge health and economic burden on the population (46, 48). Despite the fact that sanitation and clean water can markedly reduce the cases of diarrhea in areas of endemicity, surveillance studies have demonstrated that in Latin America alone more than 80% of the population has no access to sewage systems or treated water (44). Different serotypes of Shiga toxin-producing E. coli pathogens (O111:H−, O111:H8, and O111:H2) are also a public health problem in developed countries worldwide, where they have been responsible for outbreaks of bloody diarrhea and cases of hemolytic-uremic syndrome (HUS) (4, 12, 14, 21, 28, 32, 35, 55). One of the worst outbreaks of O111 E. coli happened in August 2008 in Oklahoma, where 341 people become ill, 70 people were hospitalized, 17 people developed HUS, and 1 person died (5, 8). In addition, other pathogens such as Salmonella enterica subsp. enterica serovar Adelaide and Salmonella enterica subsp. enterica serovar 50:z:e,n,x also have the same lipopolysaccharide (LPS) polysaccharide structure as that found in O111 E. coli (29).
Because of the impact that O111 E. coli strains have on public health, a lot of effort has been devoted to developing a safe, cheap, and effective vaccine to prevent diarrheagenic diseases caused by these pathogens.
The best approach to constructing a vaccine capable of protecting against a wide range of different strains of O111 E. coli is to target the LPS polysaccharide chain (O antigen), since 75% of the outer membrane of all Gram-negative bacteria is covered by LPS (38, 50). This approach is supported by the fact that conjugated vaccines against polysaccharides have been used successfully against polysaccharide-encapsulated organisms such as Streptococcus pneumoniae and Haemophilus influenzae type b in clinical practice (42). However, to use the O111 polysaccharide chain as an antigen target for the construction of a universal vaccine against enteric O111 E. coli pathogens, the antigenic variation of O111 subtypes between different E. coli strains has to be taken into account (7, 33, 59). In addition, although the O111 polysaccharides that compose their capsules are identical to the ones present on their external membranes (17, 53, 54), it has been demonstrated by Goldman and coworkers that the capsules of O111 bacteria are poorly recognized by antibodies raised against O111 LPS derived from the bacterial membrane (17), indicating that immunization with capsulated bacteria induces antibody responses different from those induced by immunization with noncapsulated bacteria.
In addition, the O111 E. coli strains can be either naked or capsulated, although the O111 polysaccharides that compose their capsules are identical to the ones present on their external membranes, except for the absence of a lipid A core (17, 53, 54).
The LPS core can also be targeted for vaccination or immunotherapy (11, 19, 39). It is not considered a virulence factor, although its involvement in bacterial adhesion has been reported (24). Structural variations are also found in the external part of the LPS core (37), and they have to be considered in order to generate antibodies capable of identifying all antigenic variants encountered within O111 bacteria.
Another element of the humoral immune response involved in clearance of pathogens is the complement system, which, independently of antibody, can be activated by pathogens in the initial stages of infection and, by itself, can kill pathogens directly. However, it is not effective in recognizing or eliminating all bacteria in samples (3, 30, 43, 45). The complement system can also promote bacterial uptake and destruction by macrophages by interacting with both the pathogen and the complement receptors present on the macrophage membrane. However, when complement activation is not enough to promote bacterial killing by macrophages, antibodies are required (25, 26, 34).
To investigate whether the O111 LPS polysaccharide of E. coli is a good antigen candidate for the formulation of a universal vaccine capable of preventing infection by O111 pathogens, electrophoretic, molecular, serological, and immunological analyses were conducted in order to determine whether antibodies against O111 polysaccharides can recognize O111 EHEC, EPEC, and EAEC, can inhibit their adhesion to human epithelial cells, and can stimulate their clearance by macrophages.
In addition, the compositions of the cores of 73 samples of all categories of O111 bacteria were characterized by PCR analysis of the enzymes responsible for the biosynthesis of all five types of LPS core: R1, R2, R3, R4, and K12.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 1. The prototype EPEC strain E2348/69 was kindly donated by James B. Kaper (University of Maryland School of Medicine, Baltimore, MD) (23). The study is based on the analysis of 74 E. coli strains of the O111 serogroup, most of which were originally isolated from children with diarrhea in Brazil and whose virulence properties were characterized by Campos et al. (6, 7). Stocks derived from the E. coli collection of the Instituto Butantan, Laboratory of Bacteriology, São Paulo, Brazil, were utilized in this work. For comparative purposes, we included two E. coli strains of the O111 serogroup isolated from a cat and a dog in Brazil (36).
TABLE 1.
Strains and categories of diarrheagenic E. coli
| Serotype | Pathotypea | Reference |
|---|---|---|
| E2348/69-O127:H6 | t-EPEC | 23 |
| O111ab:H2 | t-EPEC | 6 |
| O111ab:H− | t-EPEC | 6 |
| O111ab:H9 | a-EPEC | 6 |
| O111ab:H− | a-EPEC | 6 |
| O111ac:H9 | a-EPEC | 7 |
| O111ab:H25 | a-EPEC | 6 |
| O111ac:H8 | EHEC | 7 |
| O111ac:H− | EHEC | 6 |
| O111ab:H12 | EAEC | 6 |
| O111ab:H10 | EAEC | 7 |
| O111ab:H21 | EAEC | 7 |
| O111ac:H4 | EAEC | 6 |
| O111:H25 (2) | EPEC | 36 |
t-EPEC, typical EPEC; a-EPEC, atypical EPEC.
Cell lines.
The HEp-2 cell line used in this study was obtained from the Instituto Adolfo Lutz, São Paulo, Brazil; it was previously acquired from the American Type Culture Collection (CCL 2). Cell line J774A.1 (murine macrophage) was purchased from the European Collection of Cell Cultures (ECACC) and kindly donated by Roger R. C. New. Both cell lines were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% calf serum, 1 mM l-glutamine, and 50 IU/ml penicillin-streptomycin.
Extraction and purification of LPS by gas chromatography (GC).
LPS (20% [wt/vol] of bacterial mass) derived either from O111ac:H8 EHEC or typical O111ab:H2 EPEC was extracted using phenol-EDTA-TEA buffer (0.25 M EDTA and 5% phenol adjusted to pH 6.9 with triethylamine). The cell suspensions were incubated at 37°C with constant agitation for 1 h and centrifuged at 10,000 × g for 1 h. The supernatants were collected and dialyzed in dialysis tubing with a 2,000-molecular-weight (MW) cutoff against running water for 3 days and for 1 day against deionized water. After dialysis, the samples were concentrated using a rotary evaporator, clarified by centrifugation at 5,000 × g for 20 min, and lyophilized. For purification, the LPS extracts were solubilized in distilled water and centrifuged at 45,000 × g for 6 h at 4°C. The pellets containing purified LPS were solubilized in distilled water and lyophilized.
Derivatization prior to GC analysis.
LPS (500 μg) was methanolyzed with 0.5 M HCl in methanol by incubation for 18 h at 37°C. The methanolyzed products were extracted with hexane, and the methanolic phase was neutralized by addition of silver carbonate. The methyl glycosides were N acetylated with acetic anhydride and then trimethylsilylated by the addition of bis(trimethylsilyl)-trifluoracetamide (BSTFA; BSTFA/pyridine ratio, 1:1 [vol/vol]) and incubation at room temperature for 1 h. The trimethylsilylated methylglycosides were analyzed by gas-liquid chromatography coupled with mass spectrometry (GC-MS) (52).
Bacterial suspension for the production of serum against capsulated O111 E. coli.
To generate IgG antibodies against O111 capsular polysaccharide, rabbits were immunized independently with either capsulated O111ab:H2 (EPEC) or O111ac:H− (EHEC) strains. For immunization, bacterial colonies grown overnight in LB agar were homogenized in 0.5 ml of 0.5% Formol saline solution (85%) to fix the capsulated material. The fixed capsulated bacterial suspensions were then centrifuged in an Eppendorf 5804 centrifuge (rotor number F 34-6-38) for 20 min at 5,000 × g, and the supernatants were discarded. The pellets containing the capsulated bacteria were resuspended in saline to achieve a concentration of 9 × 108 cells/ml on the McFarland scale.
Bacterial suspension for the production of serum against naked O111 E. coli.
To generate serum against the O111 LPS polysaccharide present on the bacterial outer membrane, the O111ab:H2 (EPEC) or O111ac:H− (EHEC) strain was utilized. For immunization, bacteria were treated as described above with the exception that, after being formalinized, they were heated at 100°C for 2 h to release the pseudocapsule. Subsequently, the bacterial suspensions were centrifuged in an Eppendorf 5804 centrifuge (rotor number F 34-6-38) for 20 min at 5,000 × g, and the supernatants containing the pseudocapsule were discarded. The pellets containing naked bacteria were resuspended in saline until they achieved a concentration of 9 × 108 cells/ml on the McFarland scale.
Immunization protocol.
New Zealand White male rabbits (60 days old) were immunized intravenously with either naked or capsulated formalinized bacterial suspensions to obtain serum against membrane O111 polysaccharide (O antigen) or serum against capsular O111 polysaccharide (OK antigen), respectively. For this, increasing volumes, 0.5, 1, 2, 3, and 4 ml of the bacterial samples described above, were used for each successive immunization, with an interval of 4 days between each. Blood samples were collected 7 days after the last immunization, and the sera obtained by centrifugation at 500 × g for 10 min in an Eppendorf 5804 R centrifuge were stored at −20°C until use.
LPS extracts for immunoelectrophoresis.
LPS extracts were prepared according to the methodology described by Hitchcock and Brown (22) with a few modifications. Bacterial samples were grown in 3 ml of LB broth at 37°C for 18 h. After incubation, 1 ml of each culture was added to 5 ml of LB broth and kept in agitation at 37°C until an optical density of 0.4 at 530 nm was achieved. Subsequently, 1.5 ml of each culture was centrifuged in a Hitachi CR21E centrifuge (rotor 46) at 17.3 × g for 5 min. The pellets were resuspended in 50 μl of lysis buffer (0.5 M Tris-HCl [pH 6.8]-4% SDS-2 ml mercaptoethanol-0.05% bromophenol blue in double-distilled water to a final volume of 100 ml) and incubated for 10 min at 100°C. After the samples were cooled down, 20 μl of proteinase K solution (2.5 mg/ml) was added to each of the samples, which were then incubated in a water bath at 60°C for 1 h. After incubation, the samples were kept at 4°C until use.
Immunoblotting analysis.
Extracts of LPS derived from different strains of pathogenic O111 E. coli were solubilized in a nonreducing sample buffer and run on a 15% (22) SDS-PAGE gel (31). Gels were stained with silver or blotted onto nitrocellulose. After transfer, the membranes were blocked with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 2 h at room temperature. The membranes were then washed three times with PBS and incubated for 2 h at room temperature with rabbit serum against either membrane or capsular LPS O111 polysaccharides from O111ab:H2 EPEC or O111ac:H− EHEC diluted 1/10 in PBS-1% BSA. After incubation, the membranes were washed 3 times with PBS plus 0.05% Tween 20 for 10 min and incubated with a goat anti-rabbit IgG-alkaline phosphatase conjugate diluted 1/5,000 in PBS-BSA for 1 h at room temperature. The membranes were washed once more, and the reaction was developed using as the substrate 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium tablets (Sigma) dissolved in 10 ml of distilled water.
Dot blot technique.
Strips of nitrocellulose membrane were coated with live, naked, capsulated, or free-capsule samples of the following strains of E. coli: O111ab/H12 (EAEC), O111ab:H2 (typical EPEC), O111ab:H9 (atypical EPEC), O111:H25 (atypical EPEC derived from cats and dogs), and O127:H6 (typical EPEC). Live bacterial samples were previously grown in tryptic soy broth (TSB) at 37°C for 18 h before being added to the strips. Capsulated and naked samples were obtained as described previously. To obtain free-capsule samples, live bacterial suspensions were heated at 100°C for 10 min and centrifuged at 5,000 × g for 20 min. The pellets were discarded, and the supernatants containing free capsules were kept for dot blot analysis. Briefly, 100 μl of each live, naked, capsulated, or free-capsule sample of the O111 E. coli strains described above was used to directly coat strips of nitrocellulose membranes. Subsequently, the strips were washed three times with PBS-0.05% Tween 20 for 10 min each. After being washed, the strips were incubated with 1% BSA in PBS for 1 h at room temperature. After incubation, the strips were washed and incubated with rabbit serum against O111ac:H− EHEC diluted 1/500 in PBS-1% BSA for 2 h at room temperature. After subsequent washing, the strips were incubated for 1 h at room temperature with goat anti-rabbit IgG conjugate labeled with alkaline phosphatase diluted 1/5,000 in PBS-1% BSA. The strips were washed once more, and the enzymatic reaction was developed using as the substrate 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium tablets (Sigma) dissolved in 10 ml of distilled water.
Agglutination assay.
The titer of the antibodies against O111ac:H− EHEC generated in rabbits was determined by the test tube agglutination method as described by Ewing (13). Briefly, bacterial samples derived from humans, dogs, and cats were grown in TSB at 37°C for 18 h. The following day, rabbit serum against O111ac:H− EHEC was diluted 1/50 in 3 ml of bacterial culture. The tube was homogenized, and serial doubling dilutions in tubes, each containing 3 ml of bacterial sample, were performed for titration. Subsequently, the tubes were incubated at 37°C, and the reading was performed after 30, 60, and 120 min of incubation. The titer was determined as the last serum dilution which visually induced agglutination.
Inhibition of bacterial adhesion to epithelial cells.
HEp-2 cells were grown to 70% confluence on circular coverslips in wells of 24-well tissue culture plates. In parallel, 40-μl samples of O111ab:H2 (typical EPEC) and O127:H6 (atypical EPEC) at a concentration of 105/ml were incubated for 1 h at 37°C with 1 ml of rabbit serum against O111ac:H− (EHEC) diluted 1/10 in 1 ml of DMEM containing 1% d-mannose and 2% bovine fetal serum without antibiotic. Subsequently, the samples were added in triplicate to the wells containing the HEp-2 cells and incubated for 3 h at 37°C in 5% CO2. As a positive control for bacterial adhesion, the cells were incubated only with bacteria in the absence of rabbit antibodies against O111ac:H− EHEC. After incubation, the monolayers were washed six times with sterile PBS and then fixed with 100% methanol for 10 min, stained for 5 min with May-Grunwald stain (Merck) diluted 1:2 in Sorensen buffer, and finally stained for 20 min with Giemsa stain (Merck) diluted 1:3 in Sorensen buffer. The excess stain was discarded, and the coverslips with the stained cells were affixed to microscope slides for visualization by light microscopy (eyepiece, ×10; objective, ×100).
Phagocytosis.
J774A.1 macrophages at a concentration of 105 cells/ml in DMEM containing 10% fetal bovine serum were seeded in 24-well cell culture plates (1 ml/well) and incubated for 48 h at 37°C in a 5% CO2 incubator. In parallel, 40-μl bacterial samples containing 107 cells were incubated for 1 h at 37°C with rabbit anti-O111ac:H− serum diluted 1/10 in 1 ml of DMEM without antibiotic and containing 10% noninactivated or inactivated fetal bovine serum purchased from Cultilab. After incubation, the samples were added in triplicate to the cells. Control samples in which antibody was omitted were also set up. The cells were incubated for 3 h at 37°C in a 5% CO2 incubator. After incubation, the cells were washed 6 times with sterile PBS. The cells were then treated with 50 μg/ml of gentamicin for 30 min and subsequently washed 6 times with PBS. After being washed, the macrophages were lysed by incubating each well with 1 ml of Triton X-100 (Merck) diluted 1/10 in PBS for 10 min at room temperature. After incubation, 400 μl of PBS was added to each well and the lysate was resuspended. Subsequently, 100 μl of each lysate was 10-fold serially diluted in saline starting with a dilution of 1 in 10. One hundred microliters of each dilution was then added in triplicate to an LB agar plate and spread with a loop. The plates were then incubated overnight at 37°C, and the number of CFU was determined.
PCR amplification of core-type-specific fragments from chromosomal waa loci.
Chromosomal DNA was prepared by using DNA preparation kits (Qiagen, CA; DNA minikit). The waa gene, which encodes the enzymes responsible for the biosynthesis of the LPS cores R1, R2, R3, R4, and K12 from E. coli (1), was PCR amplified by using the primers described in Table 2. The PCR was performed in a 50-μl (total volume) mixture by using Taq polymerase. The PCR was run in 35 cycles as follows: 94°C for 1 min, 94°C for 20 s, 50°C for 30 s, 72°C for 2 min 15 s, and 72°C for 2 min. The PCR products were analyzed by electrophoresis on agarose gel (0.7%).
TABLE 2.
Sequences of oligonucleotide primers related to the biosynthesis of LPS core
| Primer name | Sequence (5′-3′) | Size (bp) | Type of core |
|---|---|---|---|
| R1C3 | GGG ATG CGA ACA GAA TTA GT | ||
| R1K15 | TTC CTG GCA AGA GAG ATA AG | 651 | R1 |
| R2C4 | GAT CGA CGC CGG AAT TTT TT | ||
| R2K9 | AGC TCC ATC ATC AAG TGA GA | 1,141 | R2 |
| R3C2 | GGC CAA AAC ACT ATC TCT CA | ||
| R3K13 | GTG CCT AGT TTA TAC TTG AA | 1,785 | R3 |
| R4C4 | TGC CAT ACT TTA TTC ATC A | ||
| R4K14 | TGG AAT GAT GTG GCG TTT AT | 699 | R4 |
| K12-1 | TTC GCC ATT TCG TGC TAC TT | ||
| K12-2a | TAA TGA TAA TTG GAA TGC TGC | 916 | K12 |
Statistical analysis.
Results were expressed as means ± standard deviations (SD). Single-criterion analysis of variance (ANOVA) followed by Bonferroni's test was used to analyze data using SigmaStat 3.0 software. Values for which P was <0.05 were considered statistically significant.
RESULTS
Gas chromatography analysis.
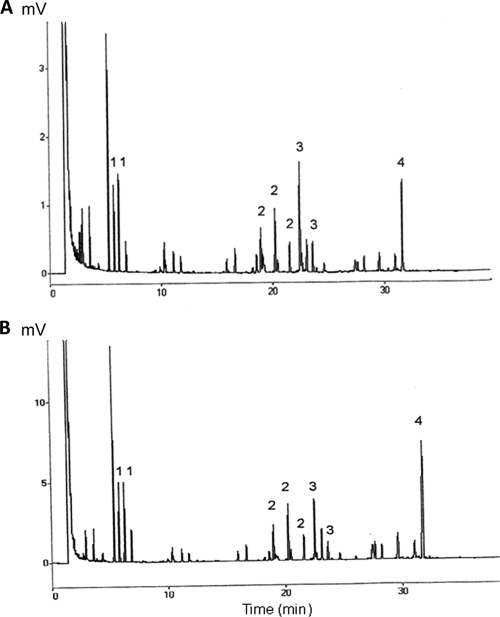
In order to determine whether there was any difference between the number of oligosaccharide units present in O111ab and O111ac polysaccharide subtypes, O111ab:H2 EPEC and O111ac:H8 EHEC samples were analyzed by gas-liquid chromatography to determine the molar ratio of 3,6-dideoxyhexose, galactose, glucose, and N-acetylglucosamine present in each strain. The results showed that, quantitatively, both samples have the same chromatographic profile (Fig. 1). However, the molar ratios for colitose, galactose, glucose, and N-acetylglucosamine in each sample were, respectively, 2.0:2.4:1.0:2.0 for O111ac:H8 (EHEC) and 1.3:1.5:1.0:2.2 for O111ab:H2 (EPEC), indicating that the numbers of colitose and galactose oligosaccharide units present in the O111ac:H8 sample are almost twice those detected in the O111ab:H2 sample (Table 3).
FIG. 1.
Gas chromatography analysis of O111ab and O111ac polysaccharides of E. coli. O111ab and O111ac polysaccharides derived from EHEC O111ac:H8 EHEC (A) and O111ab:H2 EPEC (B) were extracted with phenol-EDTA-TEA. Peaks 1, 2, 3, and 4 are 3,6-dideoxyhexose, galactose, glucose, and N-acetylglucosamine, respectively.
TABLE 3.
Oligosaccharide units of 0111ac:H8 EHEC and O111ab:H2 EPEC quantified by gas chromatography
| Saccharide | GC peak | Oligosaccharide units for: |
|
|---|---|---|---|
| EHEC O111ac:H8 | EPEC O111ab:H2 | ||
| 3,6-Dideoxyhexose | 1 | 2.0 | 1.3 |
| Galactose | 2 | 2.4 | 1.5 |
| Glucose | 3 | 1.0 | 1.0 |
| N-Acetylglucosamine | 4 | 2.0 | 2.2 |
Immunoblotting analysis.
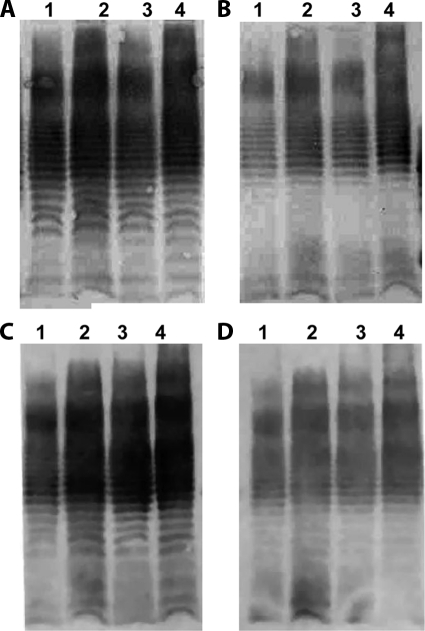
Immunoblotting analysis was performed to determine whether antibodies against capsulated or naked bacteria were able to recognize extracts of O111 LPS derived from different categories of O111 E. coli, regardless of their O111ab or O111ac subtype. Accordingly, LPS extracts of O111ab:H2 (typical EPEC), O111ab:H12 (EAEC), O111ac:H− (EHEC), and O111ac:H9 (atypical EPEC) strains were run on polyacrylamide gel, transferred to nitrocellulose membranes, and incubated with antibodies against O111ac:H− (EHEC) or O111ab:H2 (typical EPEC) derived from the immunization with either naked or capsulated bacteria. The results showed that antibodies against O111ab:H2 (typical EPEC) or O111ac:H− (EHEC) bacteria derived from immunization with either naked or capsulated bacteria recognize the same fractions of LPS extracts in all categories tested regardless their O111ab or O111ac subtype or origin (capsule or membrane) (Fig. 2).
FIG. 2.
Recognition of O111ab and O111ac LPS extracts by antibodies against O111ab and O111ac subtypes generated by the immunization of rabbits with naked or capsulated bacteria. LPS extract of O111ab:H2 (typical EPEC) (lanes 1), O111ab:H12 (EAEC) (lanes 2), O111ac:H− (EHEC) (lanes 3), and O111ac:H9 (atypical EPEC) (lanes 4) were run on polyacrylamide gel, transferred to nitrocellulose membranes, and incubated with antibodies against O111ab OK polysaccharide (A), O111ab membrane polysaccharide (B), O111ac OK polysaccharide (C), or O111ac membrane polysaccharide (D). The results were developed using 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate for goat anti-mouse IgG labeled with alkaline phosphatase.
Dot blot analysis.
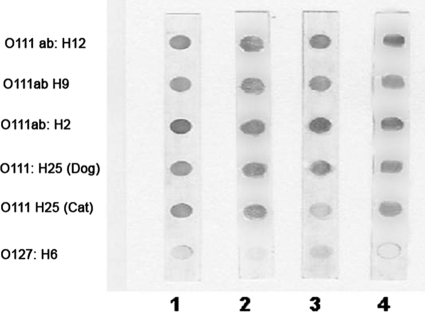
In order to verify the ability of antibodies against capsulated O111ac:H− EHEC to recognize live, naked, capsulated, or free-capsule samples of different categories of diarrheagenic O111 E. coli, the dot blot technique was utilized.
The results obtained in this experiment showed that antibodies against capsulated O111ac:H− EHEC are able to recognize naked, capsulated, and free-capsule samples of all categories of O111 E. coli strains tested, regardless their O111 subtype, flagellar antigen, or mechanism of virulence. However, O127:H6, a strain from an unrelated serogroup, was barely recognized (Fig. 3).
FIG. 3.
Recognition of O111ab:H12 (EAEC), O111ab:H9 (atypical EPEC), O111ab:H2 (typical EPEC), and O111:H25 (atypical EPEC derived from cats and dogs) by rabbit antibodies against formalinized capsulated O111ac:H− (EHEC). Lane 1, live bacteria; lane 2, capsulated bacteria; lane 3, naked bacteria; lane 4, free capsules.
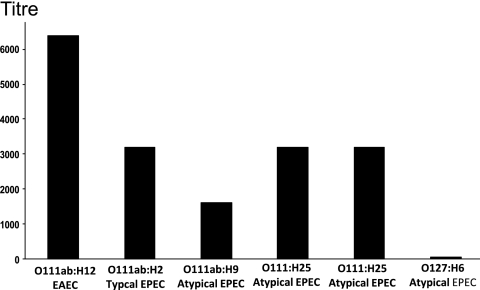
Titer of O111ac:H− antibodies against different categories of O111ab bacterial samples.
The test tube agglutination assay was used to determine the titers of the rabbit anti-O111ac:H− EHEC antibodies against other categories of O111 E. coli strains: typical and atypical EPEC and EAEC. Accordingly, rabbit antibodies against O111ac:H− EHEC were incubated with live samples of the following strains: O111ab:H12 (EAEC), O111ab:H2 (typical EPEC), O111ab:H9 (atypical EPEC), and O111:H25 (atypical EPEC derived from cats and dogs). The O127:H6 bacterial sample derived from typical EPEC was used as a control. The bacterial samples were incubated with different dilutions of rabbit antibodies against O111ac:H− (EHEC). The titer was determined as the last serum dilution which visually still induces agglutination.
The results showed that rabbit antibodies against O111ac:H− EHEC (titer against homologous bacteria, 1/8,000) are able to recognize and aggregate all bacterial samples tested regardless of their category, the nature of their flagellar antigen, or their O111 polysaccharide subtypes. In contrast, they did not recognize a bacterial sample derived from a different serogroup, as demonstrated in the case of the O127:H6 bacterial sample (Fig. 4).
FIG. 4.
Recognition of live O111ab E. coli strains by rabbit antibodies against O111ac:H− EHEC. Different dilutions of rabbit antibodies against O111ac:H− EHEC were incubated with live bacterial samples of other categories of diarrheagenic O111 E. coli (typical and atypical EPEC and EAEC), as determined by the test tube agglutination method (11). The titer was determined as the last serum dilution which visually showed a positive reaction. This experiment was repeated on three subsequent occasions, and similar observations were made.
Inhibition of bacterial adhesion.
To determine whether rabbit antibodies against O111ac:H8 EHEC polysaccharides were able to inhibit the adhesion of other categories of O111 E. coli to human epithelial cells, HEp-2 cells were incubated with an O111ab:H2 bacterial sample in either the presence or absence of antibodies.
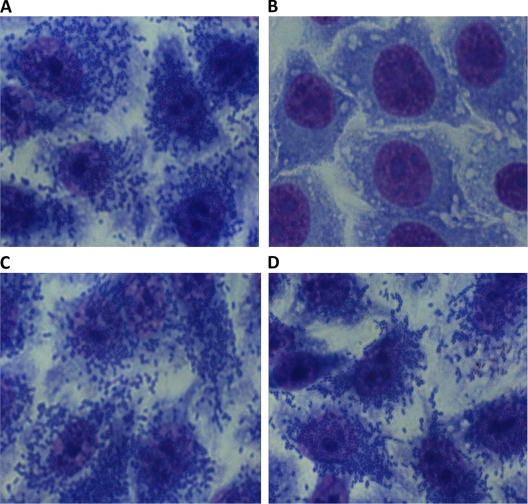
The results showed that antibodies against O111ac:H− EHEC are able to inhibit the adhesion of the O111ab:H2 typical EPEC sample but not the adhesion of unrelated bacterium O127:H6 (Fig. 5).
FIG. 5.
Determination of the capacity of O111ac:H− antibodies to inhibit the adhesion of O111ab:H2 typical EPEC and O127:H6 typical EPEC to human epithelial cells. HEp-2 cells were incubated for 3 h with O111ab:H2 alone (A) or in the presence of rabbit antibodies against O111ac:H− EHEC (B) or with O127:H6 alone (C) or in the presence of rabbit antibodies against O111ac-H− EHEC (D). An Ocular 10 objective (×100) was used.
Antibodies against O111ac:H− EHEC were also able to inhibit the adhesion of O111ab:H12 (EAEC), O111ab:H2 (atypical EPEC), and the homologous sample O111ac:H− (EHEC) to human epithelial cells (data not shown).
Influence of antibodies against O111 E. coli on bacterial clearance by macrophages.
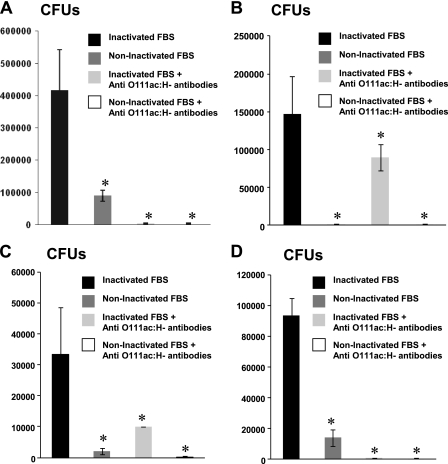
Macrophages were incubated with O111ab:H12 (EAEC), O111ab:H9 (atypical EPEC), O111ab:H2 (typical EPEC), and O127:H6 bacterial samples with or without antibodies against O111ac:H− EHEC to determine the ability of these antibodies to increase bacterial clearance. Noninactivated and inactivated fetal bovine sera were used as sources of noninactivated and inactivated complement, respectively. Both sera were received frozen and kept at −20°C until use.
The results obtained in this experiment show that, after 3 h of incubation, the macrophages are not able to completely eliminate the O127:H6 bacteria taken up by them when they are incubated in the presence of inactivated fetal bovine serum, either with or without antibodies against O111ac:H− EHEC. However, when the cells are incubated with O127:H6 bacteria in the presence of noninactivated fetal bovine serum alone, they are able to eliminate all bacteria ingested by them. It was also observed that the presence of noninactivated serum increases the clearance of O111ab:H2 (typical EPEC), O111ab:H9 (atypical EPEC), and O111ab:H12 (EAEC) by macrophages. However, macrophages are able to eliminate completely all O111 bacteria only in the presence of antibodies against O111ac:H− EHEC (Fig. 6).
FIG. 6.
Influence of complement and antibodies against O111 E. coli on bacterial clearance by macrophages. J774A.1 cells were incubated with the following bacterial samples: O111ab:H9 (atypical EPEC) (A), O111ab:H2 (typical EPEC) (B), O111ab:H12 (EAEC) (C), and O127:H6 (atypical EPEC) (D). The incubation was performed in the presence of inactivated fetal bovine serum (FBS), noninactivated fetal bovine serum, inactivated fetal bovine serum plus antibodies against O111ac:H− EHEC, or noninactivated fetal bovine serum plus antibodies against O111ac:H− EHEC. After incubation the cells were washed and lysed and the numbers of colony formation units (CFUs) were determined. *, statistically significant (P ≤ 0.05) difference between the group of macrophages incubated with bacteria in the presence of inactivated FBS and the other experimental groups.
Determination of O111 LPS core type by PCR analysis.
To determine which types of core are present in EHEC, EAEC, and EPEC belonging to the serogroup O111, the waa gene, which encodes the enzymes responsible for the biosynthesis of the LPS cores (R1, R2, R3, R4, and K12) from E. coli, was amplified in 73 samples of O111 E. coli by PCR using the respective sequence for each type of core. The results obtained in this experiment showed that all EAEC samples have core type R2 whereas all EHEC and typical EPEC samples have core type R3. Atypical EPEC samples, however, have core types R2 and R3 (Table 4). Core types R1 and K12 were not found in any of the samples tested.
TABLE 4.
Types of LPS core found in different categories of O111 E. coli
| O111 category | Serotype | No. of samples with core type: |
Total | ||||
|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | K1 | |||
| EAEC | O111ab:H12 | 17 | 17 | ||||
| EAEC | O111ab:H10 | 2 | 2 | ||||
| EAEC | O111ac:H4 | 1 | 1 | ||||
| EAEC | O111ab:H21 | 2 | 2 | ||||
| Typical EPEC | O111ab:H2 | 17 | 17 | ||||
| Typical EPEC | O111ab:H− | 15 | 15 | ||||
| EHEC | O111ac:H8 | 3 | 3 | ||||
| EHEC | O111ac:H− | 7 | 7 | ||||
| Atypical EPEC | O111ac:H9 | 1 | 2 | 3 | |||
| Atypical EPEC | O111ab:H9 | 1 | 1 | ||||
| Atypical EPEC | O111ab:H− | 1 | 2 | 3 | |||
| Atypical EPEC | O111ab:H25 | 2 | 2 | ||||
DISCUSSION
It is well documented that antibodies against O111 polysaccharides are able to recognize homologous E. coli (20, 40). However, the ability of O111 polysaccharides when used as vaccine antigens to induce antibodies able to prevent infection by O111 E. coli strains with different clinical, epidemiological, and pathogenic profiles has not been evaluated. This study investigated whether O111 polysaccharides could be used as an antigen target for the development of a universal vaccine against all categories (EPEC, EHEC, and EAEC) of O111 pathogenic E. coli. The results obtained in this work demonstrated that some strains of O111 E. coli have twice as many colitose units as others, making these polysaccharides especially suitable for use as antigens in vaccine formulations, since colitose is the major antigenic determinant of the molecule (17).
It was also observed in this study that, despite all antigenic variations found in the O111 polysaccharide, antibodies raised against a particular category of O111 E. coli strain (O111ac:H− EHEC) can recognize and inhibit the adhesion of different bacterial strains regardless of their category, their mechanism of virulence, the nature of their flagellar antigen, or their O111 polysaccharide subtype. In contrast, they were not able to recognize or inhibit the adhesion of a bacterial sample from a heterologous serogroup. The same results were obtained with the serum absorbed with E2348/69 (data not shown).
These results are in accord with previous studies which demonstrated that the adherence to human intestinal epithelial cells of Shiga toxin-producing E. coli of O111 and O157 serogroups was 95% inhibited by homologous, but not heterologous, antibodies (41).
It was also demonstrated in this work that normal fetal bovine serum, as a source of complement, increased bacterial clearance by macrophages, although complete elimination of O111 bacteria was achieved only in the presence of homologous antibodies. This result indicates that the presence of specific antibodies at the initial phase of infection is essential to prevent diarrheal diseases, since Gram-negative bacteria, including O111 E. coli strains, are in general resistant to the direct action of the complement system (18, 27, 43, 47).
It is reasonable to suppose that the LPS core can also be used as an antigen target for immunotherapy or vaccination (2, 10, 11, 51, 57). In the case of E. coli, this hypothesis is supported by studies which demonstrated that human antibodies against the LPS core of the Escherichia coli J5 strain, whose core is devoid of O antigen, were able to protect human patients against death from bacteremia due to Gram-negative bacteria (58) and sepsis when given passively as treatment for neutropenic rats (9). However, in order to construct a universal vaccine against all categories of O111 E. coli using the LPS core as an antigen, it is necessary to know which subtypes of core are the most appropriate to target all O111 E. coli strains. The results obtained in this study suggest that core type R2 can be used as an antigen target against all O111 EAEC strains whereas core type R3 is the best choice to target EHEC and typical EPEC but that antibodies against core types R2 and R3 would be required to recognize all atypical EPEC strains. It is interesting to note that core type R1, which is the most prevalent in clinical isolates (1, 15, 16), was not detected in any of the O111 samples tested. On the other hand, core type R3, which in this work was detected in all O111 EHEC samples, is also the most prevalent type of core in Shiga toxin-producing E. coli strains regardless of their serogroup (1). It is interesting to note that core type R4, which in healthy humans and cattle induces the highest level of circulating antibodies, was not detected in any of the O111 samples tested. Core type K12 was not detected in any sample either, which is in accord with other reports in the scientific literature indicating that, along with core type R4, it is the least prevalent in E. coli (1).
In summary, the data presented here suggest that the O111 polysaccharide of LPS and its core are potential antigen candidates for the construction of a universal vaccine against O111 pathogens capable of preventing adherence and the establishment of infection in humans by food-borne pathogens (56) and of preventing the development of diarrhea and acute renal failure, which in children can lead to the need for chronic renal replacement therapy (49).
Acknowledgments
This work was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), Brazil, grant 4796506-06. L.M.-P. thanks the National Institute for Science and Technology-Vaccine (INCT-V) for support.
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Amor, K., D. E. Heinrichs, E. Fridrich, K. Ziebell, R. P. Johnson, and R. C. Whitfield. 2000. Distribution of core oligosaccharide types in lipopolysaccharide from Escherichia coli. Infect. Immun. 68:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, S. R., T. Guthrie, G. R. Guile, J. Kolberg, S. Hou, L. Hyland, and S. Y. C. Wong. 2002. Cross-reactive polyclonal antibodies to the inner core of lipopolysaccharide from Neisseria meningitidis. Infect. Immun. 70:1293-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bestebroer, J., C. J. C. de Hass, and J. A. G. van Strip. 2010. How microorganisms avoid phagocyte attraction. FEMS Microbiol. Rev. 34:395-414. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, J. T., D. Bergmire-Sweat, M. Kennedy, K. Hendricks, M. Garcia, L. Marengo, J. Wells, M. Ying, W. Bidd, P. M. Griffin, R. M. Hoekstra, and C. R. Friedman. 2004. Outbreak of Shiga toxin-producing Escherichia coli O111:H8 infections among attendees of a high school cheerleading camp. Clin. Infect. Dis. 38:190-198. [DOI] [PubMed] [Google Scholar]
- 5.Calderon, V. E., Q. Chang, M. McDermott, M. B. Lythe, G. Mckee, K. Rodriguez, D. A. Rasko, V. Sperandio, and A. G. Torres. 2010. Outbreak caused by cad-negative Shiga toxin-producing Escherichia coli O111, Oklahoma. Foodborne Pathog. Dis. 7:107-109. [DOI] [PubMed] [Google Scholar]
- 6.Campos, L. C., T. S. Whittam, T. A. Gomes, J. R. Andrade, and L. R. Trabulsi. 1994. Escherichia coli serogroup O111 includes several clones of diarrheagenic strains with different virulence properties. Infect. Immun. 62:3282-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos, L. C., M. R. Franzolin, and L. R. Trabulsi. 2004. Diarrheagenic Escherichia coli categories among the traditional enteropathogenic E. coli O serogroups—a review. Mem. Inst. Oswaldo Cruz 99:545-552. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2000. Escherichia coli O111:H8 outbreak among teenage campers, Texas, 1999. MMWR Morbid. Mortal. Wkly. Rep. 49:321-324. [PubMed] [Google Scholar]
- 9.Cross, A. S., S. Opal, P. Cook, J. Drabick, and A. Bhattacharjee. 2004. Development of an anti-core lipopolysaccharide vaccine for the prevention and treatment of sepsis. Vaccine 22:812-817. [DOI] [PubMed] [Google Scholar]
- 10.Di Padova, F. E., H. Brade, G. R. Barclay, I. R. Poxton, E. Liehl, E. Schuetze, P. H. Kocher, G. Ramsay, M. H. Schreier, R. L. McClelland, and E. T. Rietschel. 1993. A broadly cross-protective monoclonal antibody binding to Escherichia coli and Salmonella lipopolysaccharides. Infect. Immun. 61:3863-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dissanayake, D. R., T. G. Wijewardana, G. A. Gunawardena, and I. R. Poxton. 2010. Potential use of a liposome-encapsulated mixture of lipopolysaccharide core types (R1, R2, R3 and R4) of Escherichia coli in controlling colisepticaemia in chickens. J. Med. Microbiol. 59:100-107. [DOI] [PubMed] [Google Scholar]
- 12.Elliott, E. J., R. M. Robbins-Broune, L. V. Loughlin, V. Bennet-Wood, J. Bourke, P. Henning, G. G. Hogg, J. Knight, H. Powell, D. Redmond, and Contributors to the Australian Paedriatric Surveillance Unit. 2001. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological and epidemiological features. Arch. Dis. Child. 85:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewing, W. H. 1986. Edwards and Ewing's identification of Enterobacteriaceae, 4th ed. Elsevier Science Publishing Co., New York, NY.
- 14.Gerber, A., H. Karch, F. Allerberger, H. M. Verweyen, and L. B. Zimmerhacki. 2002. Clinical course and the role of Shiga toxin-producing Escherichia coli infection in the hemolytic-uremic syndrome in pediatric patients, 1997-2000, in Germany and Austria: a prospective study. J. Infect. Dis. 186:493-500. [DOI] [PubMed] [Google Scholar]
- 15.Gibb, A. P., G. R. Barclay, I. R. Poxton, and F. Di Padova. 1992. Frequencies of lipopolysaccharide core types among clinical isolates of Escherichia coli defined with monoclonal antibodies. J. Infect. Dis. 166:1051-1057. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs, R. J., J. Stwart, and I. R. Poxton. 2004. The distribution of, and antibody response to, the core lipopolysaccharide region of Escherichia coli isolated from the faeces of health humans and cattle. J. Med. Microbiol. 53:959-996. [DOI] [PubMed] [Google Scholar]
- 17.Goldman, R. C., D. White, F. Orskov, I. Orskov, P. D. Kick, M. S. Lewis, A. K. Bhattacharjee, and L. Leive. 1982. A surface polysaccharide of Escherichia coli O111 contains O-antigens and inhibits agglutination of cells by O-antiserum. J. Bacteriol. 151:1210-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldman, R. C., K. Joiner, and L. Leive. 1984. Serum-resistant mutants of Escherichia coli O111 contain increased lipopolysaccharide, lack an O antigen-containing capsule, and cover more of their lipid A core with O antigen. J. Bacteriol. 159:877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory, S. H., W. H. Chen, S. Mott, J. E. Palardy, N. A. Parejo, S. Heninger, C. A. Anderson, A. W. Artenstein, S. M. Opal, and A. S. Cross. 2010. Detoxified endotoxin vaccine (J5dLPS/OMP) protects mice against lethal respiratory challenge with Francisella tularensis Schu S4. Vaccine 28:2908-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta, R. K., W. Egan, D. A. Bryla, J. B. Robbins, and S. C. Szu. 1995. Comparative immunogenicity of conjugates composed of Escherichia coli O111 O-specific polysaccharide, prepared by treatment with acetic acid or hydrazine, bound to tetanus toxoid by two synthetic schemes. Infect. Immun. 63:2805-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyles, C. L. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85(Suppl. 13):E45-E62. [DOI] [PubMed] [Google Scholar]
- 22.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes and silver-staining polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iguchi, A., N. R. Thomson, Y. Ogura, D. Saunders, T. Ooka, I. R. Henderson, D. Harris, M. Asadulghani, K. Kurokawa, P. Dean, B. Kenny, M. A. Quail, S. Thurston, G. Dougan, T. Hayashi, J. Parkhill, and G. Frankel. 2009. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191:347-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacques, M. 1996. Role of lipo-oligosaccharides and lipopolysaccharides in bacterial adherence. Trends Microbiol. 4:408-410. [DOI] [PubMed] [Google Scholar]
- 25.Janeway, C. A., Jr. 2001. How the immune system protects the host from infection. Microbes Infect. 3:1167-1171. [DOI] [PubMed] [Google Scholar]
- 26.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 27.Joiner, K. A., R. C. Goldman, C. H. Hammer, L. Leive, and M. M. Frank. 1983. Studies on the mechanism of bacterial resistance to complement-mediated killing VI-IgG increases the bactericidal efficiency of C5b-9 for E. coli O111B4 by acting at a step before C5 cleavage. J. Immunol. 131:2570-2575. [PubMed] [Google Scholar]
- 28.Kato, K., R. Shimoura, K. Yoshifuzi, N. Sakuri, H. Kodama, and S. Kuramoto. 2005. Outbreak of enterohemorrhagic Escherichia coli O111 among high school participants on excursion to Korea. Jpn. J. Infect. Dis. 58:332-333. [PubMed] [Google Scholar]
- 29.Kenne, L., B. Lindberg, E. Soderholm, D. R. Bundle, and D. W. Griffith. 1983. Structural studies of the O-antigens from Salmonella greenside and Salmonella adelaide. Carbohydr. Res. 111:289-296. [DOI] [PubMed] [Google Scholar]
- 30.Laarman, A., F. Milder, J. van-Strip, and S. Rooyakkers. 2010. Complement inhibition by gram-positive pathogens: molecular mechanisms and therapeutic implications. J. Mol. Med. 88:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lee, G. Y., H. I. Jang, I. G. Hwang, and M. S. Rhee. 2009. Prevalence and classification of pathogenic Escherichia coli isolated from fresh beef, poultry and pork in Korea. Int. J. Food Microbiol. 134:196-200. [DOI] [PubMed] [Google Scholar]
- 33.Lerouge, I., and J. Vanderleyden. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26:17-47. [DOI] [PubMed] [Google Scholar]
- 34.Lux, A., S. Aschermann, G. Bilburger, and F. Nimmerjahn. 2010. The pro and anti-inflammatory activities of immunoglobulin G. Ann. Rheum. Dis. 69(Suppl. 1):i92-i96. [DOI] [PubMed] [Google Scholar]
- 35.Morabito, S., H. Karch, P. Mariain-Kurkdjan, H. Schmidt, F. Minelli, E. Bingen, and A. Caprioli. 1998. Enteroaggregative, Shiga toxin-producing Escherichia coli O111:H2 associated with an outbreak of hemolytic-uremic syndrome. J. Clin. Microbiol. 36:840-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moura, A. R., M. P. Sircili, L. Leomil, M. H. Matté, L. R. Trabulsi, W. P. Elias, K. Irino, and A. F. Pestana de Castro. 2009. Clonal relationship among atypical enteropathogenic Escherichia coli strains isolated from different animal species and humans. Appl. Environ. Microbiol. 75:7399-7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller-Loennies, S., L. Brade, and H. Brade. 2007. Neutralizing and cross-reactive antibodies against enterobacterial lipopolysaccharide. Int. J. Med. Microbiol. 297:321-340. [DOI] [PubMed] [Google Scholar]
- 38.Nagy, G., and T. Pál. 2008. Lipopolysaccharide: a tool and target in enterobacterial vaccine development. Biol. Chem. 389:513-520. [DOI] [PubMed] [Google Scholar]
- 39.Opal, S. M., J. E. Palardy, W. H. Chen, N. A. Parejo, A. K. Bhattacharjee, and A. S. Cross. 2005. Active immunization with a detoxified endotoxin vaccine protects against lethal polymicrobial sepsis: its use with CpG adjuvant and potential mechanisms. J. Infect. Dis. 192:2074-2080. [DOI] [PubMed] [Google Scholar]
- 40.Palmeira, P., S. B. Carbonare, B. E. Guth, C. B. Carbonare, G. N. Pontes, M. Tino-De-Franco, L. B. Zapata-Quintanilla, and M. Carneiro-Sampaio. 2009. Acquisition of serum antibodies reactive with enterohemorrhagic Escherichia coli virulence-associated factors by healthy Brazilian children and adults. Pediatr. Infect. Dis. J. 28:1089-1094. [DOI] [PubMed] [Google Scholar]
- 41.Paton, A. W., E. Voss, P. A. Manning, and J. C. Paton. 1998. Antibodies to lipopolysaccharide block adherence of Shiga toxin-producing Escherichia coli to human intestinal epithelial cells. Microb. Pathog. 24:57-63. [DOI] [PubMed] [Google Scholar]
- 42.Pollard, A. J., K. P. Perrett, and P. C. Beverly. 2009. Maintaining protection against invasive bacteria with protein-polysaccharide conjugate vaccines. Nat. Rev. Immunol. 9:213-220. [DOI] [PubMed] [Google Scholar]
- 43.Porat, R., M. A. Johns, and R. W. McCabe. 1987. Selective pressures and lipopolysaccharide subunits as determinants of resistance of clinical isolates of gram-negative bacilli to human serum. Infect. Immun. 55:320-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qadri, F., A. M. Svennerholm, A. S. G. Faruque, and R. B. Sack. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeves, P. 1995. Role of O antigen variation in the immune response. Trends Microbiol. 3:381-386. [DOI] [PubMed] [Google Scholar]
- 46.Rivero, M. A., J. A. Passucci, E. M. Rodriguez, and A. E. Parma. 2010. Role and clinical course of verotoxogenic Escherichia coli infections in childhood acute diarrhoea in Argentina. J. Med. Microbiol. 59:345-352. [DOI] [PubMed] [Google Scholar]
- 47.Rizvi, M., and S. Kumar. 2003. Serum resistance of Escherichia coli strains causing urinary tract infection and diarrhoea in relation to alpha haemolysin production and O type. Ind. J. Pathol. Microbiol. 46:504-506. [PubMed] [Google Scholar]
- 48.Scaletsky, I. C. A., T. B. Souza, K. R. S. Orand, and I. N. Okeke. 2010. Genetic elements associated with antimicrobial resistance in enteropathogenic Escherichia coli (PEC) from Brazil. BMC Microbiol. 10:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheiring, J., S. P. Andreoli, and L. B. Zimmerhackl. 2008. Treatment and outcome of Shiga-toxin-associated hemolytic uremic syndrome (HUS). Pediatr. Nephrol. 23:1749-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sperandeo, P., G. Deho, and A. Polissi. 2009. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta 1791:594-602. [DOI] [PubMed] [Google Scholar]
- 51.Stanislavsky, E. S., T. A. Makarenko, E. V. Kholodkova, and C. Lugowski. 1997. R-form lipopolysaccharides (LPS) of Gram-negative bacteria as possible vaccine antigens. FEMS Immunol. Med. Microbiol. 18:139-145. [DOI] [PubMed] [Google Scholar]
- 52.Sweeley, C. C., R. Bentley, M. Makita, and W. W. Wells. 1963. Gas-liquid chromatography of trimethylsilyl derivatives of sugars and related substances. J. Am. Chem. Soc. 85:2497-2507. [Google Scholar]
- 53.Whitfield, C., and S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 54.Whitfield, C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39-68. [DOI] [PubMed] [Google Scholar]
- 55.Wight, J. P., P. Rhodes, P. A. Chapman, S. M. Lee, and P. Finner. 1997. Outbreaks of food poisoning adults due to Escherichia coli O111 and campylobacter associated with coach trips to northern France. Epidemiol. Infect. 119:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xicohtencatl-Cortes, J., V. Monteiro-Neto, M. A. Ledesma, D. M. Jordan, O. Francetic, J. B. Kaper, J. L. Puente, and J. A. Giron. 2007. Intestinal adherence associated with type IV pili of enterohemorrhagic Escherichia coli O157:H7. J. Clin. Invest. 117:3519-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamasaki, S. 2002. Development of vaccine for enterohemorrhagic Escherichia coli infection. Nippon Rinsho 60:1083-1088. [PubMed] [Google Scholar]
- 58.Ziegler, E. J., J. A. McCutchan, J. Fierer, M. P. Glauser, J. C. Sadoff, H. Douglas, and A. Braude. 1982. Treatment of gram-negative bacteremia and shock with human antiserum to a mutant Escherichia coli. N. Engl. J. Med. 307:1225-1230. [DOI] [PubMed] [Google Scholar]
- 59.Zuliani, M. E., and L. R. Trabulsi. 1969. Estudos sobre a E. coli O111:B4. Tipos sorológicos e comportamento bioquímico. Rev. Inst. Med. Trop. São Paulo 11:250-257. [PubMed] [Google Scholar]