Abstract
Antigen-binding fragments (Fab fragments) and single-chain variable fragments (scFv) against staphylococcal enterotoxin B (SEB) were produced by phage display technology. SEB epitopes were first identified by phage display approach using the commercial anti-SEB monoclonal antibody ab53981 as the target. Heptamer and dodecamer mimotope peptides recognized by ab53981 were screened from Ph.D-7 or Ph.D-12 random peptide phage libraries expressed in Escherichia coli. The isolated 7-mer and 12-mer mimotopes were shown to share a sequence homologous to 8PDELHK14S in the amino acid sequence of SEB. The N-terminal 15-mer peptide of SEB was determined to be an epitope of ab53981. After immunization of mice with maltose-binding protein-tagged N-terminal 15-mer peptide, a phage display Fab library was constructed using cDNA prepared from the mRNAs of spleen cells. Three phage clones displaying the Fab molecule which recognized SEB were isolated through three rounds of panning. Only one of them produced a soluble Fab fragment from the transformed cells, and the fragment fused with a histidine tag sequence was produced in E. coli cells and converted into scFv. Surface plasmon resonance analysis showed that the dissociation constants of these proteins with SEB were (4.1 ± 1.1) × 10−9 M and (8.4 ± 2.3) × 10−10 M, respectively. The produced molecule was applied to the determination of SEB by enzyme-linked immunosorbent assay and Western blot analysis.
Staphylococcal enterotoxins (SEs) are extracellular toxic proteins with a molecular size range of 25 to 28 kDa that cause food poisoning (19). They are known as “superantigens,” releasing excessive amounts of cytokines by cross-linking with major histocompatibility complex II molecules and T-cell receptors (30, 35). Staphylococcal enterotoxin B (SEB) is one of six SEs antigenetically classified as antigen types A, B, C, D, E, and G (2). The SEB gene has been cloned from chromosomal DNA, and the crystal structure of SEB has been elucidated (15, 33). SEB is a thermostable protein that can withstand heating at 100°C for several minutes (14). Due to its structural stability and toxicity, SEB is listed as a potential biological warfare agent by the Centers for Disease Control and Prevention and the World Health Organization.
Because a small amount of SEs (∼0.1 mg) is sufficient to cause intoxication in humans, sensitive and rapid detection of SEs is therefore critical for successful medical treatment. Detection of SEs are commonly done by immunoassays, including enzyme-linked immunosorbent assay (ELISA) (5, 6), surface plasmon resonance (SPR) assay (27), and biomolecular interaction mass spectrometry (23). The antibodies used in these assays have been prepared by hybridoma technology or purified from antisera of animals immunized with enterotoxins, but there are several problems in the production of anti-toxic protein antibodies: (i) identical antisera cannot be prepared constantly, (ii) maintaining hybridoma involves high costs, and (iii) the preparation of antibodies against toxic proteins is dangerous.
As an alternative strategy, phage display technology has been widely used to generate the molecular recognition peptides and proteins (10). Compared to antibodies, smaller monovalent antibody fragments such as the fragment antigen-binding (Fab fragment) and single-chain variable fragment (scFv) may be favorable because of protein stability due to the small molecular size. An scFv is a fusion molecule of the variable regions of heavy and light antibody chains linked together with a short linker peptide (18). In phage display technology, the DNA regions encoding antibody fragments or short peptides are cloned into phagemid vectors and subsequently expressed as fusion proteins with phage coat proteins in Escherichia coli. A phage clone with antigen-binding activity can be isolated from a phage library by an in vitro selection process called “panning” (20, 21). Unlike conventional antibodies, recombinant antibodylike proteins can be permanently produced in large quantities at low cost. The affinity and specificity of recombinant antibodylike proteins can be improved by random or site-specific mutagenesis (4, 9).
Several investigators reported the production of recombinant antibodylike proteins that bound selectively to biological warfare agents such as Shiga toxin, ricin, and the spore forms of Bacillus anthracis (7, 11, 22). Hexamer and dodecamer peptide ligands that bind to SEB have been isolated from a combinatorial peptide library (8, 32, 34), but the affinity of these small-molecule ligands to SEB does not seem high. However, production of a SEB-specific recombinant antibodylike protein has not been reported.
The aim of the present study was to generate Fab fragments and scFv proteins binding to SEB using phage display technology. A unique method for preparing an anti-SEB Fab fragment library was developed. The SEB epitope was first elucidated by phage display screening from the Ph.D-7 and Ph.D-12 library against commercial anti-SEB antibody. Mice were immunized with carrier maltose-binding protein (MBP) fused directly with a SEB epitope peptide instead of toxic SEB directly. Positive clones were obtained from a phage display Fab library prepared from spleen cells, and soluble anti-SEB Fab fragment protein was isolated from one of the three clones. The obtained Fab fragment was converted into scFv, and both proteins were produced in E. coli and characterized for SEB binding affinity. The usefulness of these agents as molecular recognition tools was ascertained by successful application to the SEB determinants from serum by Western blotting. This is the first report associated with the anti-SEB Fab fragment and scFv.
MATERIALS AND METHODS
Materials.
SEB and other biochemicals were purchased from Sigma Chemical Company (St. Louis, MO). Anti-SEB monoclonal antibody (ab53981) was purchased form Abcam (Cambridge, United Kingdom). Anti-M13 monoclonal antibody, restriction endonucleases, Ph.D.-12 and Ph.D-7 phage display peptide libraries, and the pMal-PIII vector were purchased from New England Biolabs (Ipswich, MA). KOD-Plus DNA polymerase was purchased from Toyobo (Osaka, Japan). The pET3a vector was purchased from Novagen (Madison, WI). The pComb3 phagemid vector was provided from the Scripps Research Institute (La Jolla, CA).
DNA and protein manipulation.
Plasmids were isolated from E. coli with a Qiaprep spin miniprep kit (Qiagen, Valencia, CA). DNA fragments were separated by agarose gel electrophoresis (AGE) or polyacrylamide gel electrophoresis (PAGE) and extracted by using a QiaEX II gel extraction kit (Qiagen). Targeted DNA was amplified by PCR using 9700 GeneAmp PCR system (Applied Biosystems, Foster City, CA). DNA sequencing was done by the dideoxy chain termination method using ABI Prism BigDye terminator cycle sequencing ready reaction kits on an ABI Prism DNA sequencer (Applied Biosystems). PCR Primers were obtained from Hokkaido System Science (Sapporo, Japan).
ELISA was carried out using a polystyrene 96-well microtiter plate (Maxisorp; Nunc, Naperville, IL). Wells were coated with 100 μl of 10 μg of SEB/ml per well in 50 mM carbonate buffer (pH 9.6) by overnight incubation. Wells were blocked with the same buffer containing 0.5% (wt/vol) bovine serum albumin (BSA; fraction V) for 1 h. After the wells were washed with TBST buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% [vol/vol] Tween 20; pH 7.5), binding materials, such as mouse antisera, Fab-displaying phage solution, or serially diluted recombinant antibodylike proteins, were sequentially added, followed by incubation at 37°C for 1 h. After the wells were washed with TBST buffer, the different horseradish peroxidase (HRP)-labeled secondary antibodies were added to detect binding materials as follows. HRP-labeled anti-mouse IgG (1:5,000 dilution), HRP-labeled anti-M13 polyclonal antibody (1:10,000 dilution), and HRP-labeled protein L (1:5,000 dilution) (Pierce, Rockford, IL) were used to detect anti-mouse antibody and Fab fragment, phage clones, and recombinant scFv proteins, respectively. After the wells were washed with TBST buffer, 100 μl of One-Step Ultra TMB ELISA solution (Pierce) was added to the wells. Color development was stopped by the addition of 2 M sulfuric acid. The optical density at 450 nm (OD450) was measured with a microplate reader using SpectraMax 250 (Molecular Devices, Sunnyvale, CA). Protein content was measured by a modified Bradford assay (Coomassie protein assay kit; Thermo Scientific) using BSA as a standard.
Screening of mimotope peptides from random peptide phage libraries.
Mouse anti-SEB monoclonal antibody ab53981 was used as a target of epitope mapping. The cross-reactivities of ab53981 against enterotoxin type C2 and type D were 54 and 40.9%, respectively, according to the manufacturer's data sheet. The mimotope peptides reactive with ab53981 were screened from Ph.D.-12 and Ph.D-7 phage display peptide libraries. The wells of the microtiter plate were coated with 100 μl of 10 μg of ab53981/ml per well in 50 mM carbonate buffer (pH 9.6) by overnight incubation and then blocked with the same buffer containing 0.5% (wt/vol) BSA for 1 h. After the wells were washed six times with TBST buffer, phage solution (2 × 1011 PFU/well) was sequentially poured, followed by incubation at 37°C for 1 h. Unbound phages in the supernatant were discarded. Wells were washed six times with TBST buffer. Bound phages were detached from the wells by pouring 0.2 M glycine-HCl buffer (pH 2.2) containing 0.1% (wt/vol) BSA. The eluate was immediately neutralized with 1 M Tris-HCl buffer (pH 9.1). It was transferred to 20 ml of freshly prepared E. coli ER2738 culture and further incubated at 37°C overnight. The cells were removed by centrifugation at 10,000 × g for 10 min. The resulting supernatant was added to 1/6 volume of polyethylene glycol (PEG)-NaCl solution (20% [wt/vol] PEG 8000, 2.5 M NaCl). Precipitated phages were collected by centrifugation at 10,000 × g for 15 min. They were then suspended in 1 ml of TBS buffer (50 mM Tris-HCl, 150 mM NaCl; pH 7.5) and used for the next round of panning.
Construction of an expression vector and the expression and purification of a MBP-tagged SEB epitope.
A DNA fragment of 108 nucleotides coding N-terminal 15-mer peptide of SEB was prepared by PCR using four primers: N1-15F, N1-15R, Acc65I Forward, and EagI Reverse (Table 1). The PCR mixture contained 1× PCR buffer, 0.2 μM N1-15F, 0.2 μM N1-15R, 2 μM Acc65I Forward, 2 μM EagI Reverse, 0.25 mM deoxynucleoside triphosphates (dNTPs), 1 mM MgSO4, and 1 U of KOD-Plus DNA polymerase in a final volume of 50 μl. The reaction conditions were denaturation at 94°C for 2 min, followed by 25 cycles of 98°C for 10 s, 50°C for 30 s, and 68°C for 1 min. The amplified 108-bp DNA fragment was separated by PAGE. It was then extracted, double digested with Acc65I and EagI, and inserted into the AccI-EagI site of the pMal-pIII vector. The vector was transformed into E. coli XL2-Blue (Stratagene, La Jolla, CA). The resultant plasmid was named pMal-N15-MBP.
TABLE 1.
PCR primers used in this study
| Primer | DNA sequence (5′-3′)a |
|---|---|
| N1-15F | TTCTATTCTCACTCTGAGAGTCAACCAGATCCTAAACCAGAT |
| N1-15R | GGCCGAACCTCCACCACTCGATTTGTGCAACTCATCTGGTTTAGGATC |
| Acc65I Forward | ATTCCTTTAGTGGTACCTTTCTATTCTCACTCT |
| EagI Reverse | TTCAACAGTTTCGGCCGAACCTCCACC |
| His-tag F | GAGTCATTCTGCGGCCGCACATCATCATCATCATCACGCCGCATAGACTGTTGAAAGTTGTTT |
| His-tag R | GGCCAGTGAATTCTTATTAAGACTCCTTATTAC |
| pET Forward | GGAGATTTTCCATATGAAAAAATTATTATTCGC |
| pET Reverse | AAACAACTTTCGGATCCCTATGCGGCGTGATGATG |
| MBP-scFv Forward | CCTTTCTATGCGGCCCAGGGATCCATGGCCCAGGT |
| IA+IB | SAGGTGCAGCTKCTCGAGTCAGGACCTRGC |
| IIA | SAGGTYCAGCTGCTCGAGTCTGGASCTGAG |
| IIB | CAGGTCCARCTGCTCGAGYCTGGGGCTGAG |
| IIC | GAGGTTCAGCTGCTCGAGTCTGKGGCWGAG |
| 3A | GARGTGAAGGTGCTCGAGTCTGGRGGAGGC |
| 3B+3C | GARGTGAAGCTTCTCGAGTCTGGAGGWGGC |
| 3D | GARGTGCAGCTGCTCGAGGGKGGGGGAGGA |
| IgG1 | CGCGCGACTAGTACCACAATCCCTGGGCACAATTTT |
| κI | GCGCGCGAGCTCGACRTTGTGATGWCACAGTCTCCATCCTYC |
| κII | GCGCGCGAGCTCGATRTTKTGATGACCCARACTCCACTCTCC |
| κIII | GCGCGCGAGCTCGACATTGTGCTGACMCARTCTCCWGCTTCC |
| κIV | GCGCGCGAGCTCSAAAWTGTKCTCACCCAGTCTCCAGCAATC |
| κV | GCGCGCGAGCTCGAYATYCAGATGACMCAGWCTMCATCCTCC |
| κVI | GCGCGCGAGCTCCAAATTGTKCTCWCCCAGTCTCCAGCAATC |
| κ | GCGCGCTATCTAGAATTAACACTCATTCCTGTTGAAGCTCTT |
Restriction sites are indicated by underlining.
E. coli transformant harboring pMal-N15-MBP was cultivated in LB medium (28) containing 100 μg of ampicillin/ml at 37°C. When the OD600 of the culture solution reached 0.6, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.3 mM, and the culture continued at 15°C for 15 h. The cells were harvested by centrifugation at 10,000 × g for 15 min, suspended in 12 ml of TBST buffer, disrupted on ice by sonication (output 5, duty cycle 50, ultrasonic disruptor; TOMY, Tokyo, Japan), and centrifuged at 15,000 × g for 10 min. The resulting supernatant was added to 48 ml of buffer A (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA; pH 7.5). The mixture was applied to a 4-ml column of amylose resin (New England Biolabs) equilibrated with buffer A. After the column was washed with buffer A, the proteins were eluted with buffer A containing 10 mM maltose and dialyzed against 50 mM Tris-HCl (pH 8.0) at 4°C overnight. The dialysate was applied to a 5-ml column of HiTrap Q equilibrated with the same 50 mM Tris-HCl buffer using the fast-protein liquid chromatography (FPLC) system (GE Healthcare Biosciences, Little Chalfont, United Kingdom). After being washed with the same buffer, proteins were eluted with a linear gradient of 0 to 1,000 mM NaCl in the same buffer. Fractions containing recombinant proteins were collected and dialyzed against 20 mM sodium phosphate buffer (pH 7.2) containing 150 mM NaCl at 4°C overnight. The dialysate was concentrated by centrifugation with a Centricon YM-30 machine (Millipore, Billerica, MA). As a control, MBP was purified from E. coli transformant harboring empty pMal-pIII vector.
Immunization of mice and construction of a phage display Fab library.
Female 6-week-old BALB/c mice (Japan SLC, Hamamatsu, Japan) were inoculated via the intraperitoneal route with 60 μg of N15-MBP emulsified in 690 μl of complete Freund adjuvant (Difco, Franklin Lake, NJ) as a priming injection. Mice were boosted on day 15 and day 30 using the same protocol except that incomplete Freund adjuvant was used instead of complete Freund adjuvant. On day 43, 10 μg of N15-MBP diluted with saline solution was injected intravenously as a final booster. On day 46, mice were killed to obtain spleens. Serum was taken to monitor SEB-specific antibody titer by ELISA using HRP-labeled anti-mouse immunoglobulin (Dako, Copenhagen, Denmark) as a secondary detection. Nonimmune mice and mice immunized with MBP were used as controls.
A Fab-displaying phage library was constructed according to the method of Ito et al. (12, 13). The mRNA was extracted from the mouse spleen cells by using a QuickPrep Micro mRNA purification kit (Qiagen). First-strand cDNA was synthesized by using an avian myeloblastosis virus (AMV) reverse transcriptase first-strand cDNA synthesis kit (Roche Diagnostics, Mannheim, Germany). The reverse transcription reaction mixture [which contained 1× reaction buffer, 5 mM MgSO4, 1 mM dNTPs, 50 U of RNase inhibitor, 20 U of AMV reverse transcriptase, 1 μg of mRNA, and 1.6 μg of oligo(dT) primer in a final volume of 50 μl] was incubated at 25°C for 10 min, at 42°C for 60 min, and then at 4°C. DNA coding the “fragment different” (Fd) region of the heavy chain and DNA coding the entire κ light chain were amplified by PCR using the family-specific variable region and the isotype-specific constant region primers shown in Table 1. DNA coding Fd region of the heavy chain was amplified using seven sense primers (IA+IB, IIA, IIB, IIC, 3A, 3B+3C, and 3D) and one antisense primer (IgG1). DNA coding the light chain was amplified by using six sense primers (κI, κII, κIII, κIV, κV, and κVI) and one antisense primer (κ). PCR was carried out in 100 μl. The mixture contained 1× reaction buffer, 5 mM MgSO4, 1 mM dNTPs, 3 μl of cDNA, and 5 U of Taq DNA polymerase, and the process consisted of denaturation at 94°C for 1 min, followed by 25 cycles of 94°C for 60 s, 57°C for 1 min, and 72°C for 1 min. Amplified DNA was separated by AGE and double-digested with SpeI/XhoI and SacI/XbaI to obtain the DNAs coding Fd fragment and light chain, respectively. Both DNA digests were ligated sequentially into phagemid vector pComb3. These heavy- and light-chain combinatorial DNA repertoires were electroporated into E. coli XL1-Blue (Stratagene). The transformed E. coli cells were cultivated in 100 ml of Superbroth (28) containing 100 μg of carbenicillin/ml for 1 h at 37°C. Fab-displayed phage clones were rescued by infecting VCSM13 helper phage (Stratagene) and further cultivated overnight at 37°C. Phages were collected from the supernatant by precipitation with a PEG-NaCl solution, centrifugation at 10,000 × g for 15 min, and suspension in 1 ml of phosphate-buffered saline containing 1% BSA. The phages were used for affinity selection.
Selection of phage clones displaying Fab fragment binding to SEB and preparation of the fragment.
The wells of the microtiter plate were coated with 100 μl of 10 μg of SEB/ml per well in 50 mM carbonate buffer (pH 9.6) by overnight incubation. Wells were blocked with the same buffer containing 0.5% (wt/vol) BSA for 1 h. After the wells were washed with TBST buffer, 100 μl of Fab-displayed phages (2 × 1011 PFU/well) was sequentially poured, followed by incubation at 37°C for 1 h. Unbound phages in the supernatant were discarded, and the wells were washed six times with TBST buffer. Bound phages were detached from the wells by adding glycine-HCl solution (pH 2.2) containing 1 mg of BSA/ml and neutralized with 1 M Tris-HCl buffer (pH 9.1). Eluted phages were added to 10 ml of freshly prepared E. coli XL1-Blue culture, and the mixture was incubated at 37°C for 1 h. After cultivation, 15 ml of Superbroth and VCSM13 helper phage containing 1011 PFU were supplemented, and the culture continued for a further 2 h. Then, 35 ml of Superbroth and kanamycin (70 μg/ml) were added, and the culture continued at 37°C overnight. After cultivation, cells were removed by centrifugation at 10,000 × g for 10 min. The resulting supernatant was added to a 1/6 volume of PEG-NaCl solution, and the precipitated phages were collected by centrifugation at 10,000 × g for 15 min. Phage clones were suspended into 1 ml of TBS buffer and used for the next round of panning. After the final round of panning, phages displaying Fab fragment were produced according to the method of Scheffer et al. (29), and the phages binding to SEB were screened by ELISA.
Phagemid DNA from phage clones displaying Fab fragment binding to SEB was processed to remove the gIII coat gene by the following procedure. Isolated phage clones were added to 2 ml of freshly prepared E. coli XL1-Blue culture, followed by incubation at 37°C for 1 h. Ampicillin was added to a final concentration of 50 μg/ml and further incubated overnight. Phagemid DNA was extracted and double digested with NheI and SpeI. Large DNA fragments were collected and recircularized by self-ligation. Constructed plasmids were transformed into E. coli XL2-Blue.
E. coli transformants were cultivated in 2YT medium (28) containing 100 μg of ampicillin/ml at 37°C. When the OD600 of the culture solution reached 0.6, IPTG was added to a final concentration of 1 mM, followed by further cultivation at 37°C for 15 h. Cells were harvested by centrifugation at 10,000 × g for 15 min and suspended in buffer B (50 mM Tris-HCl, 150 mM NaCl; pH 8.0). After disruption of the cells by freezing and thawing, the solution was centrifuged at 27,000 × g for 20 min, and the resulting supernatant was loaded onto a 2-ml column of nickel-chelating resin (Novagen) equilibrated with buffer B. After being washed with buffer B containing 20 mM imidazole, the proteins were eluted with buffer B containing 300 mM imidazole. The proteins were concentrated by centrifugation by a Centricon YM-10 unit. They were then subjected to a gel filtration on Sephacryl S-200 column and eluted with buffer B using an FPLC system. Fractions containing Fab fragment were collected and concentrated by centrifugation.
Construction of an expression vector for histidine-tagged scFv and preparation of scFv.
The E-tag sequence of pCANTAB5E (GE Healthcare Biosciences) was changed to a histidine-tagged (His-tag) sequence as follows. The 1.3-kb gIII gene in the pCANTAB5E vector was amplified by PCR using two primers of His-tag F and His-tag R (Table 1). After digestion with NotI and EcoRI, the fragment was cloned back into the NotI/EcoRI site of pCANTAB5E. The resultant plasmid was named pCANTAB5H. The scFv gene was constructed using the cloned Fab gene as a template DNA according to the method of Pope et al. (26). The constructed scFv gene was digested with SfiI and NotI and inserted into the pCANTAB5H vector. The scFv gene, including the gIII signal sequence and His-tag codon, was amplified by using the primers pET Forward and pET Reverse (Table 1). After digestion with NdeI and BamHI, the DNA fragment was inserted into the NdeI-BamHI site of the pET3a vector. The resultant plasmid was transformed into E. coli Origami 2 (DE3)-competent cells (Novagen). Expression and purification of His-tagged scFv was done by the same procedure as for the Fab fragment.
Construction of an expression vector and expression and purification of an MBP-tagged scFv protein.
The 1.4-kb malE gene coding MBP of the pMal-pIII vector was excised by double digestion with NdeI and BamHI and subcloned into the NdeI-BamHI site of the pET-3a vector. The resultant plasmid was named pET-malE. The 0.75-kb scFv gene was then amplified by PCR with the pCANTAB5H harboring scFv gene as a template DNA using the primers MBP-scFv Forward and pET Reverse (Table 1). It was digested with BamHI and inserted into the BamHI site of the pET-malE vector. The resultant plasmid was transformed into E. coli Origami 2 (DE3)-competent cells. E. coli transformants were cultivated in 2YT medium (28) containing 100 μg of ampicillin/ml at 37°C. When the OD600 of the culture solution reached 0.6, IPTG was added to a final concentration of 1 mM and further cultivated at 30°C for 15 h. The MBP-tagged scFv protein was purified as described for the MBP-tagged SEB epitope protein.
SPR assay.
Binding kinetics were determined by SPR assay using BIAcore T100 (GE Healthcare Biosciences). Experiments were done at 25°C using HBS-T buffer (10 mM HEPES, 150 mM NaCl, 0.005% Tween 20; pH 7.4). SEB (30 μg/ml in 10 mM sodium acetate [pH 4.5]) was covalently immobilized on a CM3 sensor chip activated with a solution consisting of 100 mM N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide hydrochloride and 400 mM hydroxysuccinimide (EDC/NHS). Analyte proteins serially diluted with HBS-T buffer were injected into the apparatus at a flow rate of 30 μl/min. The injection periods for association and dissociation were 120 s. After each measurement, the chip surface was regenerated with 10 μl of glycine 2.0 (GE Healthcare Biosciences). The obtained sensorgrams were analyzed with a 1:1 binding mass transfer model using BIAcore T100 evaluation software (version 1.1.1).
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate (SDS)-PAGE analysis of proteins was done according to the method of Laemmli (17). After electrophoresis, the gel was stained with Coomassie brilliant blue R-250. The molecular weights of proteins were estimated by comparison with protein markers (Bio-Rad, Hercules, CA). Proteins in the polyacrylamide gel were transferred onto a polyvinylidene difluoride membrane (ATTO, Tokyo, Japan). The membrane was blocked for 1 h in TBS buffer containing 3% BSA and washed with TBST buffer. It was incubated for 1 h with TBST buffer containing 1 ng/ml of molecular recognition protein against SEB. After being washed with TBST buffer, the membrane was soaked in HRP-labeled protein L solution (1:2,000 dilution) or HRP-labeled anti-MBP monoclonal antibody (1:10,000 dilution) for 1 h. Proteins on the membrane were visualized by using EzWest Blue (ATTO).
Gene recombination, animal experiments, and safety considerations.
The microbial gene recombination experiments were approved by the ethical committee of National Research Institute of Police Science. Animal care and experiments were performed in accordance with the guidelines for the care and use of laboratory animals of the University of Shizuoka. Ricin is toxic, so this compound should be handled carefully and destroyed by autoclaving or with sodium hypochlorite after use. The usage of ricin here was approved by the Minister of Economy, Trade, and Industry of Japan.
RESULTS
Strategy for generation of molecular recognition protein against SEB by epitope mapping and phage display technology.
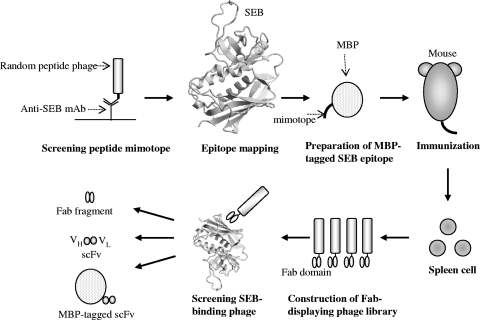
Figure 1 shows the scheme for generating a molecular recognition protein against SEB by phage display technology. In the present study, the SEB epitope was mapped by comparison of the amino acid sequence of a peptide mimotope that was recognized by the commercially available anti-SEB monoclonal antibody ab53981, and an MBP-tagged SEB epitope was used for immunization. This is the most strategic point for generating an antibodylike SEB recognition molecule because of the induction of an SEB-immune response without using native or toxic SEB in the immunization step. A Fab-displayed phage library was constructed by using cDNA from the spleens of immunized mice. SEB-reactive Fab-displaying phages were then screened. The Fab fragment was converted into scFv as a small protein, and its potential for detection of SEB was investigated.
FIG. 1.
Schematic diagram showing generation of the anti-SEB Fab fragment and scFv. The three-dimensional structure of SEB (PDB ID; 3SEB) was produced with CueMol software (http://cuemol.sourceforge.jp/en/).
Production of a SEB mimotope from a random peptide phage library.
By screening from the Ph.D-7 random peptide phage library using monoclonal anti-SEB antibody ab53981 as the target, seven phage clones were obtained after four rounds of panning. Phage DNA was sequenced, and the amino acid sequences of the peptide regions were determined: YLCKFGC (1/7), SPDELHK (2/7), REPLVYW (1/7), AEPIIYW (2/7) and KQPIVFW (1/7). The amino acid sequence SPDELHK was almost identical to that of SEB 7DPDELH13K. By screening from the Ph.D-12 random peptide phage library, 20 phage clones were obtained, and the amino acid sequences were determined: MGGTLIASDQYQ (17/20), QSLPASMSYQTA (1/20), SLSASMDFMMYA (1/20), and NALRASNSFMDE (1/20). These 12-mer mimotopes showed a conserved motif, LXXS, that is homologous to SEB 11LHK14S. The common sequence HLK was observed from 7-mer and 12-mer mimotopes, suggesting that the epitope of antibody ab53981 was located around the site of 8PDELHK14S within the N-terminal residues of SEB. We selected the N-terminal 15-residue peptide as the antigen for the immunization because the N-terminal portion might be necessary for the stable expression in E. coli cells.
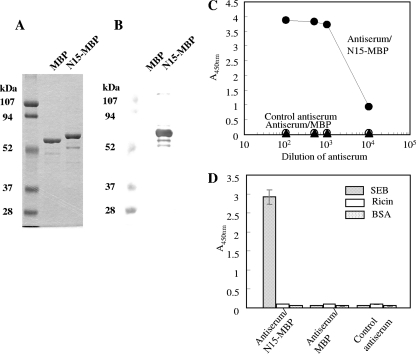
Next, the DNA fragment coding the SEB N-terminal 15-residues peptide 1ESQPDPKPDELHKS15S was synthesized by PCR. Two primers, N1-15F and N1-15R, annealed to yield a fragment 73-bp template, and then two primers, Acc65I Forward and EagI Reverse, introduced the restriction site (Table 1). The produced 108-bp DNA fragment was inserted into the pMal-pIII vector, and the resultant plasmid was used for the expression of an MBP-tagged 15-mer peptide named N15-MBP. MBP fusion protein could be successfully expressed in E. coli and purified by affinity chromatography and ion-exchange chromatography. As shown in Fig. 2A, the bands of MBP and N15-MBP were observed at locations corresponding to the expected molecular masses of 53 and 55 kDa, respectively. Western blot analyses showed that ab53981 was bound to N15-MBP but did not bind to MBP (Fig. 2B). It was concluded that the region within the 15-mer N-terminal peptide of SEB was an epitope of ab53981. Approximately 420 μg of N15-MBP was purified from a 1.2-liter culture.
FIG. 2.
Binding of ab53981 antibody to the MBP-tagged 15-mer peptide of SEB and characterization of antisera obtained from N15-MBP immunized mice. (A) SDS-PAGE of the purified MBP and N15-MBP protein. Each protein (1 μg) was subjected to SDS-PAGE and stained by Coomassie blue staining. Left lane: molecular mass marker proteins. (B) Western blot. MBP and N15-MBP (1 μg) were separated by SDA-PAGE and subjected to Western blot analysis with ab53981 for primary detection. (C) Serially diluted antisera from nonimmune mice (▴), sacrificed mice immunized with N15-MBP (•), and sacrificed mice immunized with MBP (○) were subjected to ELISA. HRP-labeled anti-mouse IgG was used for secondary detection. (D) SEB, BSA, and ricin were coated on a 96-well microtiter plate, and mouse antisera at 1:1,000 dilution was added. HRP-labeled anti-mouse IgG was used for secondary detection.
Purified N15-MBP protein was used to immunize mice. No titer was detected in antisera from nonimmunized mice or mice immunized with MBP, but antisera from sacrificed mice immunized with N15-MBP showed high titers (<10,000) (Fig. 2C). ELISA analysis (Fig. 2D) showed that the antisera selectively bound to SEB but did not bind to BSA or ricin. These results indicate that the N-terminal 15-residue peptide possessed immunogenicity and the capacity to produce antibodies recognizing SEB.
Production of Fab fragment binding to SEB.
From mRNAs prepared from the spleen cells of immunized mice, a heavy- and light-chain combinatorial library was constructed using a PCR-amplified Fd fragment gene and the entire κ light-chain gene. The resultant library contained 3.8 × 107 transformants. After three rounds of panning, 60 phage clones were obtained. They were subjected to ELISA to select a SEB-reactive phage clone using an SEB-coated or an SEB-uncoated plate (data not shown). Three phages—clones 12, 14, and 49—were found to bind to SEB. In the absence of SEB, the ELISA OD450s of phage clones 12, 14, and 49 were 0.17, 0.1, and <0.01, respectively.
These three positive phage clones were treated to remove the phage gIII coat-protein coding gene, and phagemid DNA was double digested with SpeI and NheI and recircularized by self-ligation. The processed phagemid DNAs, named pComb-12, pComb-14, and pComb-49, were transformed into E. coli XL2-Blue, and soluble Fab fragments were expressed using IPTG induction. Although no titer was detected in the cell lysate from transformants harboring pComb-12 and pComb-14, the transformant harboring pComb-49 showed a sufficient titer (data not shown). These results indicated that the soluble Fab fragment derived from only pComb-49 possessed SEB binding ability. The variable part of the heavy-chain (VH) and the variable part of the light-chain (VL) region genes of pComb-49 were sequenced. The deduced amino acid sequences are shown in Fig. 3. The amino acid sequences of both VH and VL region gene segments were 100% identical to mouse germ line clone VHSM7.a3.93 (heavy chain) and mouse germ line clone 8-30 (light chain).
FIG. 3.
Amino acid sequence of the variable region of heavy- and light-chain-selected Fab fragments binding to SEB.
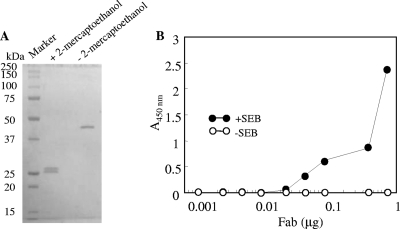
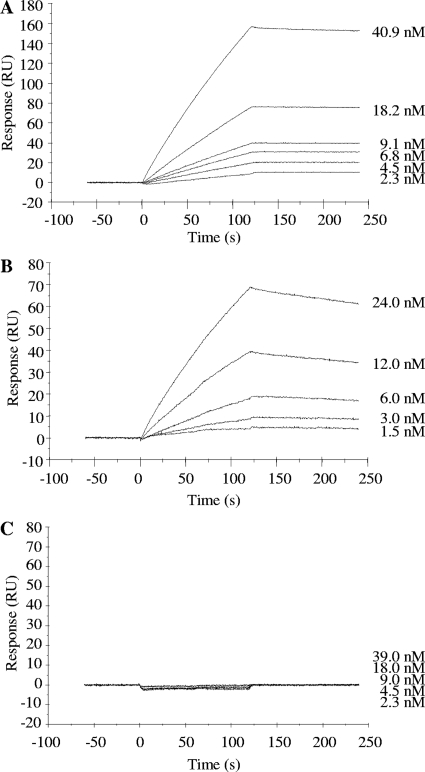
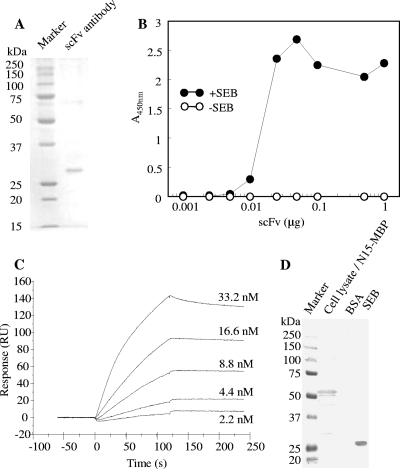
The soluble Fab fragment was purified from the cell lysate of transformant harboring pComb-49 by nickel affinity column chromatography and gel filtration. SDS-PAGE of the purified Fab fragment (Fig. 4A) showed that one and two bands corresponding to heavy and light chains were observed under nonreducing and reducing conditions, respectively. Approximately 50 μg of Fab fragment protein was purified from a 1-liter culture. ELISA analysis showed that the Fab fragment reacted with SEB and that the staining level was increased in a dose-dependent manner (Fig. 4B). SPR assay (Fig. 5) showed that interaction between immobilized SEB and BSA was not detected but that ab53981 and Fab fragment could interact with immobilized SEB. The association rate constant (Ka), dissociation rate constant (Kd), and dissociation constant (KD) of ab53981 were calculated to be (1.5 ± 0.5) × 105 M−1 s−1, (1.8 ± 0.4) × 10−4 M−1, and (1.4 ± 0.3) × 10−9 M, respectively. The Ka, Kd, and KD values for the Fab fragment were determined to be (2.5 ± 0.6) × 105 M−1 s−1, (9.9 ± 0.5), × 10−4 M−1, and (4.1 ± 1.1) × 10−9 M, respectively.
FIG. 4.
Characterization of purified Fab fragment binding to SEB. (A) SDS-PAGE of the purified Fab fragment. Purified Fab fragment (1 μg) was subjected to SDS-PAGE under nonreducing or reducing conditions and stained by Coomassie blue staining. Left lane: molecular mass marker proteins. (B) Serially diluted Fab fragment was poured onto SEB-coated (•) or uncoated (○) polystylene plates. After washing, the plate Fab fragment was detected by using HRP-labeled protein L.
FIG. 5.
SPR analysis of SEB binding Fab fragment. SEB was covalently immobilized by amine coupling to the dextran matrix of a sensor chip CM3 to yield 1,200 resonance units. Followed by blocking with ethanol amine, serially diluted ab53981 (A), Fab fragment (B), and BSA (C) were introduced to the chip at a flow rate of 30 μg/min, and the responses were recorded.
Production of scFv binding to SEB.
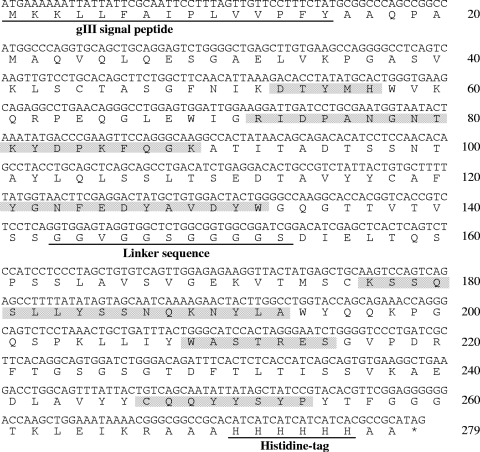
The variable-region gene of the heavy and light chains within the pComb-49 was connected together with short-linker DNA coding (GlyGlyGlyGlySer)3. The resultant scFv gene was inserted into the phagemid vector pCANTAB-5H in which the E-tag sequence had been replaced with a His6 tag. The resultant plasmid was transformed into E. coli XL2-Blue and then induced with IPTG. One transformant from randomly selected six transformants showed a sufficient titer in ELISA analysis for SEB-binding activity (data not shown). The nucleotide sequence and deduced amino acid sequence of this positive scFv gene is shown in Fig. 6. The amino acid sequences of the complementarity-determining regions (CDRs) were identical to that of the Fab fragment, but deletion of four glycine residues and mutation of serine to valine were found in the linker peptide region.
FIG. 6.
Nucleotide and deduced amino acid sequences of scFv binding to SEB. The gene III signal sequence (amino acids 1 to 15), linker peptide (amino acids 143 to 153), and histidine tag (amino acids 271 to 277) are underlined. The CDRs are shown in shaded gray boxes.
The scFv gene containing the gIII signal sequence and His6 codon was cloned into a pET-3a vector and transformed into E. coli Origami 2 (DE3) cells. scFv was purified to homogeneity by nickel affinity chromatography and gel filtration. SDS-PAGE showed that one band with a molecular mass of 29 kDa was observed, a finding consistent with the value calculated from the amino acid sequence (Fig. 7A). Approximately of 70 μg of scFv protein was purified from a 1-liter culture. ELISA analysis (Fig. 7B) showed that scFv was bound to immobilized SEB, and the staining level increased in a dose-dependent manner. Compared to the Fab fragment, even a small amount of scFv (0.01 μg) was sufficient to detect SEB. An SPR assay yielded the following the kinetic parameters: Ka, (7.2 ± 1.6) × 105 M−1 s−1; Kd, (6.0 ± 1.4) × 10−4 M−1; and KD, (8.4 ± 2.3) × 10−10 M (Fig. 7C).
FIG. 7.
Characterization of scFv binding to SEB. (A) Purified scFv (1 μg) was subjected to SDS-PAGE and Coomassie blue stained. (B) Serially diluted scFv was poured into an SEB-coated (•) or an uncoated (○) polystyrene plate. After the plate was washed with buffer, scFv was detected by using HRP-labeled protein L. (C) Serially diluted scFv were introduced to sensorchip-immobilized SEB. (D) E. coli cell lysates containing N15-MBP, 1 μg of BSA, and 1 μg of SEB were separated by SDS-PAGE and subjected to Western blot analysis with scFv antibody for primary detection.
E. coli cell lysate containing N15-MBP protein was separated by SDS-PAGE and subjected to Western blotting with scFv for primary detection. The bands corresponding to N15-MBP and SEB were detected, indicating that the recognition site of scFv was identical to that of ab53981 (Fig. 7D).
Application of the generated SEB-binding scFv protein to detection of SEB in serum samples by Western blotting.
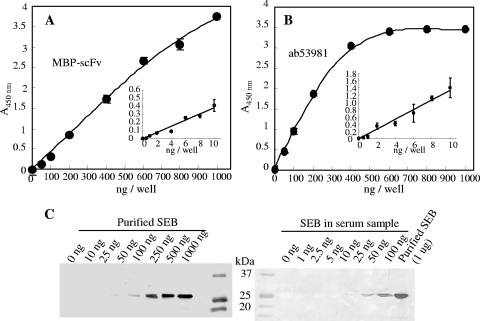
Although the scFv binding to SEB was successfully produced in E. coli cells, its productivity was not high. To improve the productivity of scFv, the DNA coding scFv gene was connected downstream of the malE gene to express scFv as a MBP-fused protein (MBP-scFv). Soluble MBP-scFv was effectively produced in E. coli cells and could be purified by affinity column and ion-exchange chromatography. The E. coli transformant produced ∼700 μg of MBP-scFv from a 1-liter culture. SPR analysis demonstrated interaction between immobilized SEB and MBP-scFv (data not shown).
Purified MBP-scFv was applied to the determination of SEB by ELISA. The lower detection limit of MBP-scFv against SEB was 20 ng/ml (Fig. 8A). The sensitivity of MBP-scFv was almost equal to that of monoclonal antibody ab53981. Western blotting was done to detect purified SEB and SEB in serum samples (Japanese Red Cross Society, Tokyo, Japan) (Fig. 8B). We detected 50 and 25 ng of SEB in the control and serum samples, respectively.
FIG. 8.
Application of MBP-scFv by ELISA and Western blotting. (A and B) Detection of SEB by ELISA. MBP-scFv (A) or ab53981 (B) was poured onto an SEB-coated polystyrene plate. After the plate was washed with buffer, SEB was detected by using HRP-labeled anti-MBP monoclonal antibody (A) or HRP-labeled anti-mouse IgG polyclonal antibody (B). Color development was done for 5 and 30 min using 50 to 1,000 ng of SEB and 0.5 to 10 ng of SEB, respectively. (C) Western blot detection of SEB by MBP-scFv. SEB solutions diluted with TBS (left) or serum (right) were separated by SDS-PAGE and subjected to Western blot analysis with MBP-scFv antibody for primary detection.
DISCUSSION
Wang et al. (34) also produced SEB-binding hexamer peptides by screening the solid-phase combinatorial peptide library. Goldman et al. (8) selected the SEB-binding phage screened from the random 12-mer library. Soykut et al. (32) produced SEB-binding 12-mer peptides selected from the same library, and the binding constant was not high (4.2 × 105 M−1). We first carried out screening of SEB-binding peptide from the Ph.D-7 and Ph.D-12 random peptide phage libraries. Phage clones displaying SEB-binding peptide were not concentrated through several rounds of panning (data not shown). The short peptides obtained by Wang et al. (34), Goldman et al. (8), and Soykut et al. (32) do not seem adequate as a high-affinity ligand to SEB. Small peptides such the 7-mer and 12-mer phage library peptides may not be sufficient to bind SEB with high affinity. We therefore decided to generate an antibodylike molecular recognition protein by phage display techniques.
Several high-affinity recombinant antibodies have been generated by phage display (1, 3), and immune libraries are indispensable for obtaining high-affinity recombinant antibodies. An immunization procedure using native antigen was used to construct phage libraries. However, the use of toxins in standard laboratories is restricted or illegal. In the present study, epitope peptide fused to MBP was adopted for immunization instead of native toxic antigen. As a result, induction of a high immune response against native SEB was achieved. The combination of epitope mapping by phage display and epitope immunization appears to be effective for constructing a high-quality immune library for toxic proteins.
Based on the amino acid sequence alignment between mimotope peptides and SEB, the region within the N-terminal 15-residue sequence of SEB was identified as an epitope for ab53981. The selected heptamer and dodecamer mimotopes that bound to ab53981 shared common amino acid sequences, i.e., 11LHK14S. The amino acid sequence alignment of SEs showed that the LHK motif was conserved among type B, C1 to C3, and D toxins (Fig. 9A). Considering ab53981 cross-reactivity to type C2 and D toxins, it is reasonable that this LHK motif is essential for ab53981 binding to antigen toxins. Although the N-terminal 20-residue amino acid sequences of enterotoxin types C1 to C3 were almost identical to that of SEB, the binding power of ab53981 against type C2 toxin was 54% that of SEB. According to the crystal structure, 12H-17F of SEB forms a short α-helix structure (Fig. 9B, left) (14, 24). The N-terminal 20-residue amino acid sequence of type C2 toxin contains two short α-helix structures at 7TPDE11L and 12KSSE17F (Fig. 9B, right) (24). The difference in reactivity of ab53981 against SEB and type C2 toxin may be due to the structural difference at the N-terminal region.
FIG. 9.
N-terminal region of SEB recognized by Fab fragment and scFv. (A) The amino acid sequences of SEs were aligned by using CLUSTAL W software. The conserved LHK motif is underlined. (B) Comparison of the three-dimensional structure between SEB and SEC2 (PDB ID; 1STE) at the N-terminal 20-residue region. The short α-helix structures of SEB and SEC2 are indicated by a bold black line.
After three rounds of panning, three positive phage clones displaying an SEB-binding Fab fragment were isolated from the newly created phage display Fab library. A soluble Fab fragment was successfully expressed from one of the three clones in E. coli (Fig. 4A). We succeeded in producing a smaller molecular recognition tool, scFv, without affecting recognition properties (Fig. 7D). It is reported that that apparent affinity of Fab or the scFv molecule is 10- to 1,000-fold lower than that of the corresponding immunoglobulins (25), but the dissociation constant of our Fab fragment was almost equal to that of ab53981. The dissociation constant of scFv was reduced compared to the Fab fragment and ab53981. The increase in affinity of scFv was also supported by ELISA data, in which a small amount of scFv was sufficient to detect SEB (Fig. 4B and 7B). A mutation was not observed in the CDR in the VH and VL domains of the scFv gene (Fig. 6), so the dodecamer linker peptide GGSGGVGGGGS may be suitable in the protein folding and association of the VH and VL domain for scFv to raise binding affinity. Further study is needed to clarify the reason for the increased affinity for SEB.
E. coli cells have been used as a host strain for the production of recombinant antibodies (7, 8, 29, 35). The high yield of recombinant antibodies from E. coli cells is important. As an alternative, Pichia pastoris (31) and Bacillus megaterium (16) have been used as the host strain to improve the production yield of recombinant antibodies. We succeeded in improving the productivity of SEB-reactive scFv by expressing it as an MBP-fused protein. The purified MBP-fused scFv could be applied to detecting SEB in ELISA and Western blotting (Fig. 8A and B). The detection limits of SEB in aqueous samples by ELISA and in serum samples by Western blotting were 0.5 and 25 ng, respectively; both values are lower than the amount of SEB that produces symptoms of enterotoxin intoxication. The detection limits of SEB by ELISA, SPR assay, and biomolecular interaction mass spectrometry were reported to be nearly 1 ng (5, 6, 23, 27). Therefore, the scFv constructed in the present study can detect SEB and can be used as a low-cost SEB recognition tool instead of anti-SEB immunoglobulins.
Acknowledgments
This study was undertaken under the “Research and Development Program for Resolving Critical Issues” research program sponsored by the Special Coordination Funds for Promoting Science and Technology, which is supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Arakawa, M., T. Yamashiro, G. Uechi, M. Tadano, and A. Nishizono. 2007. Construction of human Fab (γ1/κ) library and identification of human monoclonal Fab possessing neutralizing potency against Japanese encephalitis virus. Microbiol. Immunol. 51:617-625. [DOI] [PubMed] [Google Scholar]
- 2.Bergdol, M. S. 1983. Staphylococci and staphylococcal infections. Academic Press, London, England.
- 3.Bugli, F., S. Manzara, R. Torelli, R. Graffeo, R. Santangelo, P. Cattani, and G. Fadda. 2004. Human monoclonal antibody fragment specific for glycoprotein G in herpes simplex virus type 2 with applications for serotype-specific diagnosis. J. Clin. Microbiol. 42:1250-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Pascalis, R., R. N. Gonzales, E. A. Padlan, P. Schuck, S. K. Batra, J. Schlom, and S. V. S. Kashmiri. 2003. In vitro affinity maturation of a specificity-determining region-grafted humanized anticarcinoma antibody: isolation and characterization of minimally immunogenic high-affinity variants. Clin. Cancer Res. 9:5521-5531. [PubMed] [Google Scholar]
- 5.Ewald, S., and S. Christensen. 1987. Detection of enterotoxin production by Staphylococcus aureus from aviation catering meals by the ELISA and the microslide immunodiffusion test. Int. J. Food Microbiol. 5:87-91. [Google Scholar]
- 6.Ewald, S. 1988. Evaluation of enzyme-linked immunosorbent assay (ELISA) for detection of staphylococcal enterotoxin in foods. Int. J. Food Microbiol. 6:141-153. [DOI] [PubMed] [Google Scholar]
- 7.Gao, C., S. Mao, C. Kaufmann, P. Wirsching, R. A. Lerner, and K. D. Janda. 2002. A method for the generation of combinatorial antibody libraries using pIX phage display. Proc. Natl. Acad. Sci. U. S. A. 99:12612-12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman, E. R., M. P. Pazirandeh, J. M. Mauro, K. D. King, J. C. Frey, and G. P. Anderson. 2000. Phage-displayed peptides as biosensor reagents. J. Mol. Recognit. 13:382-387. [DOI] [PubMed] [Google Scholar]
- 9.Griep, R. A., C. van Twisk, J. M. van der Wolf, and A. Schots. 1999. Fluobodies: green fluorescent single-chain Fv fusion proteins. J. Immunol. Methods 230:121-130. [DOI] [PubMed] [Google Scholar]
- 10.Hoess, R. H. 2001. Protein design and phage display. Chem. Rev. 101:3205-3218. [DOI] [PubMed] [Google Scholar]
- 11.Inoue, K., K. Itoh, H. Nakao, T. Takeda, and T. Suzuki. 2004. Characterization of a Shiga toxin 1-neutralizing recombinant Fab fragment isolated by phage display system. Tohoku J. Exp. Med. 203:295-303. [DOI] [PubMed] [Google Scholar]
- 12.Itoh, K., K. Inoue, K. Hirooka, K. Maruyama, M. Ohkawa, K. Matsui, H. Tada, T. Enomoto, Y. Hashimoto, T. Suzuki, and T. Masuko. 2001. Phage display cloning and characterization of monoclonal antibody genes and recombinant Fab fragment against the CD98 oncoprotein. Jpn. J. Cancer Res. 92:1313-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh, K., M. Ohnishi, M. Sonobe, M. Saito, A. Yoshida, H. Hayashi, K. Inoue, and T. Masuko. 2009. Antibody epitope peptides as potential inducers of IgG antibodies against CD98 oncoprotein. Cancer Sci. 100:126-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamlang, E. M., M. L. Bartlett, and H. E. Snyder. 1971. Effect of pH, protein concentration, and ionic strength on heat inactivation of staphylococcal enterotoxin B1. Appl. Environ. Microbiol. 22:1034-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, C. L., and S. A. Khan. 1986. Nucleotide sequence of the enterotoxin B gene from Staphylococcus aureus, J. Bacteriol. 166:29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan, E., L. A. Halabi, T. Schirmann, M. Hust, and S. Dübel. 2007. Production of single chain Fab (scFab) fragments in Bacillus megaterium. Microb. Cell Fact. 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. R. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lerner, R. A., A. S. Kang, J. D. Bain, D. R. Burton, and C. F. Barbas III. 1992. Antibodies without immunization. Science 258:1313-1314. [DOI] [PubMed] [Google Scholar]
- 19.Marrack, P., and J. Kappler. 1990. The staphylococcal enterotoxins and their relatives. Science 248:705-711. [DOI] [PubMed] [Google Scholar]
- 20.McCafferty, J., A. D. Griffiths, G. Winter, and D. J. Chiswell. 1990. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348:552-554. [DOI] [PubMed] [Google Scholar]
- 21.McConnell, S. J., T. Dinh, M.-H. Le, and D. G. Spinella. 1999. Biopanning phage display libraries using magnetic beads vs. polystyrene plates. Biotechniques 26:208-214. [DOI] [PubMed] [Google Scholar]
- 22.Mechaly, A., E. Zahavy, and M. Fisher. 2008. Development and implementation of a single-chain Fv antibody for specific detection of Bacillus anthracis spores. Appl. Environ. Microbiol. 74:818-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nedelkov, D., and R. W. Nelson. 2003. Detection of staphylococcal enterotoxin B via biomolecular interaction analysis mass spectrometry. Appl. Environ. Microbiol. 69:5212-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papageorgiou, A. C., K. R. Acharya, R. Shapiro, E. F. Passalacqua, R. D. Brehm, and H. S. Tranter. 1995. Crystal structure of the superantigen enterotoxin C2 from Staphylococcus aureus revels a zinc-binding site. Structure 3:769-779. [DOI] [PubMed] [Google Scholar]
- 25.Pini, A., A. Giuliani, C. Ricci, Y. Runci, and L. Bracci. 2004. Strategies for the construction and use of peptide and antibody libraries displayed on phages. Curr. Protein Peptide Sci. 5:487-496. [DOI] [PubMed] [Google Scholar]
- 26.Pope, A. R., M. J. Embleton, and R. Mernaugh. 1996. Construction and use of antibody gene repertoires, p. 1-40. In J. McCafferty, H. Hoogenboom, and D. Chiswell (ed.), Antibody engineering: a practical approach. IRL Press, Oxford, England.
- 27.Rassoly, A. 2001. Surface plasmon resonance analysis of staphylococcal enterotoxin B in food. J. Food Prot. 64:37-43. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Scheffer, G. L., A. W. Reurs, B. Jutten, S. H. W. Beiboer, R. van Amerongen, M. Schoester, E. A. S. Wiemer, H. R. Hoogenboom, and R. J. Scheper. 2002. Selection and characterization of a phage-displayed human antibody (Fab) reactive to the lung resistance-related major vault protein. Br. J. Cancer 86:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherer, M. T., L. Ignatowicz, G. Winslow, J. W. Kappler, and P. Marrack. 1993. Superantigens: bacterial and viral proteins that manipulate the immune system. Annu. Rev. Cell Biol. 9:101-128. [DOI] [PubMed] [Google Scholar]
- 31.Schoonooghe, S., V. Kaigorodov, M. Zawisza, C. Dumolyn, J. Haustraete, J. Grooten, and N. Mertens. 2009. Efficient production of human bivalent and trivalent anti-MUCI Fab-scFv antibodies in Pichia pastoris. BMC Biotechnol. 9:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soykut, E. A., F. C. Dudak, and I. H. Boyaci. 2008. Selection of staphylococcal enterotoxin B (SEB)-binding peptide using phage display technology. Biochem. Biophys. Res. Commun. 370:104-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swaminathan, S., W. Furey, J. Pletcher, and M. Sax. 1992. Crystal structure of staphylococcal enterotoxin B, a superantigen. Nature 359:801-806. [DOI] [PubMed] [Google Scholar]
- 34.Wang, G., J. De, J. S. Schoeniger, D. C. Roe, and R. G. Carbonell. 2004. A hexamer peptide ligand that binds selectively to staphylococcal enterotoxin B: isolation from a solid phase combinatorial library. J. Peptide Res. 64:51-64. [DOI] [PubMed] [Google Scholar]
- 35.Webb, S. R., and N. R. J. Gascoigne. 1994. T-cell activation by superantigens. Curr. Opin. Immunol. 6:467-475. [DOI] [PubMed] [Google Scholar]