Abstract
Anthrax toxin (ATx) is composed of the binary exotoxins lethal toxin (LTx) and edema toxin (ETx). They have separate effector proteins (edema factor and lethal factor) but have the same binding protein, protective antigen (PA). PA is the primary immunogen in the current licensed vaccine anthrax vaccine adsorbed (AVA [BioThrax]). AVA confers protective immunity by stimulating production of ATx-neutralizing antibodies, which could block the intoxication process at several steps (binding of PA to the target cell surface, furin cleavage, toxin complex formation, and binding/translocation of ATx into the cell). To evaluate ATx neutralization by anti-AVA antibodies, we developed two low-temperature LTx neutralization activity (TNA) assays that distinguish antibody blocking before and after binding of PA to target cells (noncomplexed [NC] and receptor-bound [RB] TNA assays). These assays were used to investigate anti-PA antibody responses in AVA-vaccinated rhesus macaques (Macaca mulatta) that survived an aerosol challenge with Bacillus anthracis Ames spores. Results showed that macaque anti-AVA sera neutralized LTx in vitro, even when PA was prebound to cells. Neutralization titers in surviving versus nonsurviving animals and between prechallenge and postchallenge activities were highly correlated. These data demonstrate that AVA stimulates a myriad of antibodies that recognize multiple neutralizing epitopes and confirm that change, loss, or occlusion of epitopes after PA is processed from PA83 to PA63 at the cell surface does not significantly affect in vitro neutralizing efficacy. Furthermore, these data support the idea that the full-length PA83 monomer is an appropriate immunogen for inclusion in next-generation anthrax vaccines.
Anthrax is caused by infection with Bacillus anthracis, and its pathogenesis is associated with an antiphagocytic poly-d-glutamic acid capsule and a binary anthrax toxin (ATx). The ATx comprises two protein exotoxins: lethal toxin (LTx) and edema toxin (ETx). The two toxins both have a binding protein called protective antigen (PA) but have separate effector proteins, edema factor (EF) and lethal factor (LF) (3, 20, 56). LTx is composed of PA and LF, and ETx is composed of PA and EF. In the initial stages of infection by B. anthracis, full-length 83-kDa PA (PA83) secreted by the bacterium binds to either one or both of at least two host cell ATx receptors (ATRs): tumor endothelial marker 8 (TEM8) (7, 27, 57) or capillary morphogenesis protein 2 (CMG2) (51).
Vaccines containing PA as the major component confer protective efficacy in various animal models of multiple routes of infection (5, 14-16, 29, 42-44, 59). PA has four domains: an amino-terminal domain (domain 1, which is composed of subregions 1a and 1b) that contains two calcium ions and the S163RKKRS168 cleavage site for activating proteases, a heptamerization domain (domain 2) that contains a large flexible loop implicated in membrane insertion, a small domain (domain 3) hypothesized to aid in oligomerization, and a carboxy-terminal receptor-binding domain (domain 4) (26, 41). Upon binding to an ATR, PA is proteolytically cleaved by the cell surface protease furin to a 63-kDa polypeptide (PA63), releasing a 20-kDa amino-terminal fragment (domain 1a). Cleavage and release of domain 1a facilitate assembly of the PA prepore; a heptameric ring-shaped structure with a negatively charged lumen. Assembly of the prepore exposes a large hydrophobic surface for binding of LF and/or EF molecules to form ATx (9, 17, 19, 26, 33, 35, 37, 41). One PA63 heptamer is able to bind up to three LF and/or EF molecules. The ATx is then endocytosed by a lipid raft-mediated clathrin-dependent process (1, 34). The low-pH conditions (pH 5.5) in the endosome induce the prepore to undergo a conformational switch that translocates ATx to the target cell cytosol (6, 17, 18, 24, 32, 37, 41, 55).
The current licensed vaccine for use in humans is anthrax vaccine adsorbed (AVA [BioThrax]; Emergent BioSolutions, Lansing, MI). AVA is a cell-free filtrate from a toxigenic, nonencapsulated B. anthracis strain, V770-NP1-R (2, 10, 45). The primary immunogen is PA (59) adsorbed to aluminum hydroxide adjuvant (10, 29). The current AVA vaccination schedule consists of five 0.5-ml intramuscular (i.m.) injections at 0 and 4 weeks and 6, 12, and 18 months, with annual boosters (10, 30).
There are various potential molecular targets in which the host humoral antibody response to vaccination with AVA or PA can interfere with ATx-mediated cytotoxicity. These targets include, but are not limited to, (i) blocking of free PA83 binding to the host cell ATx receptor (TEM8 or CMG2); (ii) inhibition of PA83 proteolytic cleavage by the host cell surface furin-like enzyme or serum proteases, leaving the PA unprocessed and thus unable to form toxin complexes; (iii) interruption of PA63 heptamerization to form the prepore on the host cell surface; (iv) blocking the binding of LF and EF monomers to the PA heptamer prepore; and (v) disruption of internalization and translocation of the ATx. Consequently, PA has become a focal point in developing immunotherapies and next-generation vaccines for the prevention and treatment of anthrax (4, 13, 21, 22, 31, 36, 39, 40, 53, 58, 60).
Most of the anti-PA therapies under development specifically target PA domains 2 and 4, with domain 4 being the most frequent target (21, 53, 60). The therapeutic effects of antibodies targeted against domain 4 are considered to be based primarily on blocking the interaction of PA with its host cell receptor (26, 49). However, in active immunization, there will be multiple epitopes presented to the host immune system that are critical to mounting a protective immune response and, likewise, others that may make little or no contribution. Although PA20 is cleaved from PA83 and has no described role in the intoxication process, recent reports have proposed that in AVA-vaccinated humans, the PA20 fragment (domain 1a) contains immunodominant epitopes (48, 61). Therefore, it was postulated that vaccines containing full-length PA (PA83) may be suboptimal due to the dominance of PA20 and that perhaps PA63-based vaccines may be more advantageous (47, 48).
To address the question of suboptimal immune responses in PA83-based vaccine and therapeutic design, we developed two low-temperature anthrax lethal toxin (LTx) neutralization activity (TNA) assays, the noncomplexed TNA (NC-TNA) and receptor-bound TNA (RB-TNA) assays. These assays allow comparison of antibody-mediated neutralization of LTx both before and after receptor binding by PA. The goal of this work was to evaluate the ability of anti-PA antibody responses in AVA-vaccinated and inhalation anthrax-challenged rhesus macaques (Macaca mulatta) to neutralize anthrax LTx in vitro both before and after PA has bound to, and been processed at, the cell surface receptor.
MATERIALS AND METHODS
Materials.
Recombinant ATx PA (rPA), recombinant LF (rLF), and J774A.1 murine macrophages (TIB-67) were obtained from BEI Resources, Manassas, VA. Human reference standard AVR801 was provided by the Centers for Disease Control and Prevention (CDC), and its preparation and characterization are described elsewhere (52). AVR801 is available from the CDC and BEI Resources under the appropriate agreements.
Rabbit sera BMI001, BMI009, BMI023, and BMI025 were produced at the Battelle Memorial Institute as follows. BMI001 was made by pooling approximately equal volumes of serum from 50 New Zealand White (NZW; Oryctolagus cuniculus) rabbits vaccinated with 5 to 100 μg/ml proprietary investigational rPA83 vaccine adjuvanted with aluminum hydroxide (Alhydrogel). BMI009 was made by pooling approximately equal volumes of serum from nine NZW rabbits vaccinated with 82.5 μg/ml proprietary investigational rPA83 vaccine in Alhydrogel adjuvant. BMI023 was produced by pooling approximately equal volumes of serum from a nine NZW rabbits vaccinated with a 1:16 dilution of AVA. BMI025 was derived by pooling approximately equal volumes of serum from five NZW rabbits vaccinated with 50 μg/ml proprietary investigational rPA83 vaccine adjuvanted with Alhydrogel and then further diluting the serum approximately 1:5 in naïve rabbit serum.
Nonhuman primate (NHP) sera that were used for assay development, BMI280 and BMI281, were produced at the Battelle Memorial Institute as follows. BMI280 was made by pooling five different bleeds from a single rhesus macaque vaccinated with 50 μg/ml proprietary investigational rPA83 vaccine adjuvanted with Alhydrogel. BMI281 was made by pooling five different bleeds from a single rhesus macaque vaccinated with 50 μg/ml proprietary investigational rPA83 vaccine (different from the vaccine used to produce BMI280) adjuvanted with Alhydrogel.
The monoclonal antibodies (MAb) from clones 14B7 and 1G3 were generously provided by Stephen Little of USAMRIID, Fort Detrick, Frederick, MD. MAb 14B7 recognizes an epitope on PA domain 4 between residues Asp671 and Ile721 and inhibits binding of PA83 to the cell surface receptor but does not prevent binding of LF to the cell-bound PA63 heptamer (Fig. 1) (26, 49). The 14B7 antibody does not inhibit LTx activity in vitro after PA83 has bound to its cell surface receptor. In contrast, MAb 1G3 recognizes an epitope on a 17-kDa fragment located between residues Ser168 and Phe314 that partly overlaps PA domain 1b (residues 168 to 258). The region containing this epitope is involved in LF binding to PA (Fig. 1) (26). MAb 1G3 preferentially binds to the cleaved form PA63 and inhibits the binding of LF to PA bound to the cell surface. Removal of the 20-kDa PA domain 1a fragment exposes the epitope recognized by 1G3. 1G3 does not inhibit the binding of PA83 to the PA cell surface receptor (26). Both MAb 14B7 and 1G3 were an integral part of the development and routine testing of the NC- and RB-TNA assays. Throughout the study, these antibodies served as quality control (QC) samples tested on each plate and in both assay formats. As a QC reagent, 1G3 was required to neutralize the toxin in all of the types of TNA assays tested, whereas 14B7 only neutralized toxin when PA was not complexed to the cell.
FIG. 1.

Domains and critical residues of B. anthracis PA important for binding of 1G3 and 14B7. PA consists of domains 1 to 4. Domain 1 is composed of subdomains 1a (residues 1 to 167) and 1b (residues 168 to 258). Residues 1 to 258 contain a furin cleavage site, and upon cleavage by furin, a hydrophobic region of PA is exposed to allow binding by EF and/or LF (6, 32). Domain 2 is composed of residues 259 to 487 and is involved in oligomerization and formation of the channel for LF and EF entry into the host cell cytosol (6, 24, 41, 54). Domain 3 is made up of residues 488 to 595, and the only known function is thought to be oligomerization (32). Domain 4, which consists of residues 596 to 736, is essential for binding of host cell receptors (32). MAb 1G3 recognizes an epitope on a 17-kDa fragment located between residues Ser-168 and Phe-314 that partly overlaps domain 1 (specifically, residues 168 to 258). This epitope is involved in LF binding to PA. 1G3 preferentially binds to PA63 (specifically, between residues Ser-168 and Phe-314, a region which spans domains 1 and 2) and inhibits the binding of LF to PA bound to the cell surface. Removal of the 20-kDa fragment (domain 1a) is required to expose the epitope recognized by 1G3. 1G3 does not inhibit binding of PA83 to the PA cell surface receptor. MAb 14B7 recognizes an epitope on domain 4 between residues Asp-671 and Ile-721 and inhibits binding of PA83 to the cell surface receptor but does not prevent binding of LF to PA. 14B7 does not inhibit LTx activity after PA83 has bound to the PA cell surface receptor (32). Note that this image is for illustration and is not to actual scale.
Rhesus macaque (M. mulatta) sera.
Rhesus macaque sera were obtained from a cohort of animals from the CDC AVRP Immune Correlates and Duration of Protection study (30). Briefly, rhesus macaques received undiluted or diluted AVA (undiluted or diluted 1:5, 1:10, 1:20, or 1:40) under a three-dose i.m. schedule (0, 4, and 26 weeks) and challenged with aerosolized B. anthracis Ames spores at 12, 30, and 52 months postvaccination (Table 1). The full details of this study will be published elsewhere (50; C. P. Quinn et al., unpublished data). The antigen loads (AVA dilutions) differed for the purpose of modulating the magnitude of the rhesus macaque immune response. In the present study, we evaluated serum samples taken from 110 animals across all vaccine dilution groups at week 30, which corresponds to the peak response to the third i.m. vaccination with AVA. Additional sera from 20 of these animals (52-month time point only) were obtained at 14 days postexposure to B. anthracis Ames spores (28; Quinn et al., unpublished). Table 1 summarizes the animal groupings by challenge month, AVA dose, and percent survival for the samples used in these experiments. Rhesus macaque sera used for these analyses were collected at the Battelle Memorial Institute and the Emory Vaccine Center, Atlanta, GA, under study protocols approved by Institutional Animal Care and Use Committees at the CDC, the Battelle Memorial Institute, and the Emory Vaccine Center.
TABLE 1.
NHP challenge and dose groups
| Challenge time point (mo) and AVA dose | Survival frequencya [no. of survivors/total (%)] |
|---|---|
| 12 | |
| 1:10 | 6/8 (75) |
| 1:20 | 5/9 (56) |
| 1:20 | 6/10 (60) |
| 1:40 | 2/3 (66) |
| 1:40 | 7/8 (88) |
| 30 | |
| Undiluted | 9/9 (100) |
| 1:5 | 6/6 (100) |
| 1:10 | 6/9 (67) |
| 1:20 | 7/8 (88) |
| 52 | |
| Undiluted | 8/10 (80) |
| 1:5 | 9/9 (100) |
| 1:10 | 6/10 (60) |
The survival frequency represents the subset of rhesus macaque samples tested only in the NC- and RB-TNA assays.
TNA assays.
The low-temperature TNA assays (the NC- and RB-TNA assays described later) were performed similarly to the regular TNA assay (referred to here as the R-TNA assay) as described by Li et al. but with minor modifications (23, 38). Briefly, J774A.1 murine monocytes/macrophages (TIB-67) were cultured in a medium consisting of Dulbecco's modified Eagle's medium (Gibco BRL, Gaithersburg, MD) with 5% heat-inactivated fetal bovine serum (HyClone, Logan, UT), 1% penicillin-streptomycin (Gibco BRL), and 1% 1 M HEPES buffer (Gibco BRL). Cells were scraped from confluent cultures and used to seed 96-well plates (100 μl per well) at 3 × 105 to 5 × 105 cell/ml and incubated for 16 to 18 h at 37°C in 5% CO2 to fully attach. Similar to that described by Little et al. (25), two variants of the R-TNA assay were performed to study interactions among J774A.1 cells, PA, LF, MAb, and immune serum from NHPs. To distinguish the two types of low-temperature TNA assays conducted in part at 4°C from the R-TNA assay (run at 37°C) and each other, the low-temperature TNA assays will be referred to as the NC-TNA assay and the RB-TNA assay.
In the NC-TNA assay, the AVR801 reference standard, NHP antisera, and the MAb were serially diluted 1:2 in a 96-well plate (preparation plate) by adding the sera or MAb to row A and diluting the samples 1:2 through row G with cold (4°C) medium to produce a seven-point dilution curve. The PA and LF were diluted to final concentrations of 100 and 50 ng/ml, respectively, and preincubated with test and control antibodies at 4°C on ice for 2 h. Meanwhile, the 96-well plates seeded with the cell monolayers on the previous day were cooled by placement at 4°C on ice for 30 min. The growth medium was aspirated from the cell plates, and 100 μl of the serum-MAb-LF-PA cocktail was transferred from the preparation plate to the corresponding wells of the cell plate and incubated at 4°C on ice for 2 h. To complete the intoxication of the cells, the cell plates were gradually warmed by incubation first for 45 min at 4°C (no ice) and then at 37°C in 5% CO2 for 2 h 15 min.
In contrast to the NC-TNA assay, the RB-TNA assay cell plates were first cooled by placement at 4°C on ice for 30 min. After chilling, the growth medium was carefully aspirated and 100 μl of cold (4°C) medium containing PA was added to the cell plates to a final concentration of 100 ng/ml and incubated at 4°C on ice for an additional 60 min. During the PA incubation, the test and control antibodies were diluted 1:2 in a separate 96-well preparation plate to create a seven-point dilution curve. At the completion of the PA incubation with the cells, the medium and unbound PA were decanted and 100 μl of the antisera/MAb was transferred from the preparation plates to the cell plates and incubated for 1 h at 4°C on ice. The sera/MAb were then decanted from the wells, 100 μl of LF was added to a final concentration of 50 ng/ml, and the plates were incubated for 45 min at 4°C, followed by 2 h 15 min at 37°C in 5% CO2 to complete the cell intoxication.
For both assays, after the cell intoxication steps were completed and without removing the serum-MAb-toxin solutions, a 25-μl volume of tetrazolium salt, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT; Sigma Chemicals, St. Louis, MO) dissolved in 0.01 M phosphate-buffered saline (pH 7.4) was added to each well and incubated for 2 h at 37°C in 5% CO2. Without removing the MTT, 100 μl of solubilization buffer, an aqueous preparation of 20% (wt/vol) sodium dodecyl sulfate (Sigma Chemicals, St. Louis, MO) in 50% (vol/vol) N,N,-dimethylformamide (Fisher Scientific, Pittsburgh, PA) was added to each well, and the plates were incubated for 16 to 20 h at 37°C in 5% CO2. LTx cell lysis was measured by determining cell viability colorimetrically using MTT as the reporter system. Serum- or MAb-mediated neutralization of LTx was manifested as suppression of cytotoxicity and hence preservation of cell viability. Cell viability was measured by the reduction of MTT by viable cells that produced a color change.
The plates were read on a BioTek ELx800 microplate reader using the software KC4 Signature at a wavelength of 570 nm and at 690 nm as a reference filter (BioTek Instruments, Inc., Winooski, VT). The raw data were exported from KC4 and analyzed using SAS Version 9.1 (SAS Institute, Cary, NC) running the customized assay endpoint calculation algorithm developed by the CDC (version ED50.61CORE03.sas and ED50.61BETA03_printing plots.sas) (23). A four-parameter logistic curve fit was applied to the standard curve using the equation y = (A − B)/[1 + (x/C)D̂] + B, where A represents the y value which corresponds to the lower asymptote (i.e., zero neutralization response), B represents the y value which corresponds to the upper asymptote (i.e., maximum neutralization response), D represents the slope factor, and C represents the inflection point which is the x value associated with the y value at the midpoint between A and B (11). The reciprocal of C is the dilution at which there is 50% neutralization, or the 50% effective dilution (ED50). The code used a four-parameter logistic model to fit a dose-response curve to the data. For evaluation of sera of low reactivity, the code contains a pattern recognition algorithm which distinguishes among fully formed sigmoid neutralization curves and partially formed curves and is further described by Li et al. (23). In addition to the ED50, the 50% effective concentration (EC50) was also calculated during assay development to help compare the neutralizing activities of MAb to those of sera. The EC50 was calculated as the quotient of the ED50 and the mean anti-PA IgG concentration titer as determined by enzyme-linked immunosorbent assay (data not shown) and is presented in μg/ml. The EC50s are only presented in conjunction with the ED50 data in Table 2, and thereafter, all data are reported as ED50s only.
TABLE 2.
NC- and RB-TNA assay development
| Sample, vaccine, and reportable value | Geometric mean ED50 or EC50 titerf (n) |
Ratio compared to R-TNA assay (95% CI) |
Ratio of RB-TNA assay to NC-TNA assay (95% CI) | Test of significance (P value) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| R-TNA assay | NC-TNA assay | RB-TNA assay | NC-TNA assay | RB-TNA assay | NC-TNA assay vs R-TNA assay | RB-TNA assay vs R-TNA assay | RB-TNA assay vs NC-TNA assay | ||
| AVR801 (human) AVA | |||||||||
| ED50 | 529 (15) | 254 (15) | 122 (30) | 2.1 (1.7, 2.6) | 4.3 (3.6, 5.2) | 2.1 (1.7, 2.5) | <0.0001a | <0.0001a | <0.0001a |
| EC50 | 0.207 (15) | 0.431 (15) | 0.898 (30) | 0.480 (0.391, 0.590) | 0.230 (0.193, 0.276) | 0.48 (0.402, 0.574) | <0.0001a | <0.0001a | <0.0001a |
| BMI001 (rabbit) rPA | |||||||||
| ED50 | 1,160 (20) | 520 (16) | 232 (37) | 2.2 (1.9, 2.6) | 5 (4.4, 5.7) | 2.2 (1.9, 2.6) | <0.0001a | <0.0001a | <0.0001a |
| EC50 | 0.069 (20) | 0.154 (16) | 0.344 (37) | 0.449 (0.382, 0.527) | 0.200 (0.175, 0.229) | 0.446 (0.386, 0.515) | <0.0001a | <0.0001a | <0.0001a |
| BMI009 (rabbit) rPA | |||||||||
| ED50 | 4,741 (9) | 2,362 (13) | 1,233 (38) | 2 (1.6, 2.5) | 3.8 (3.2, 4.7) | 1.9 (1.6, 2.3) | <0.0001a | <0.0001a | <0.0001a |
| EC50 | 0.090 (9) | 0.180 (13) | 0.344 (38) | 0.498 (0.396, 0.627) | 0.260 (0.214, 0.317) | 0.522 (0.441, 0.619) | <0.0001a | <0.0001a | <0.0001a |
| BMI023 (rabbit) AVA | |||||||||
| ED50 | 298 (30) | 155 (12) | 76 (46) | 1.9 (1.6, 2.3) | 3.9 (3.5, 4.4) | 2 (1.7, 2.4) | <0.0001a | <0.0001a | <0.0001a |
| EC50 | 0.117 (30) | 0.226 (12) | 0.461 (46) | 0.520 (0.437, 0.619) | 0.255 (0.226, 0.287) | 0.489 (0.415, 0.577) | <0.0001a | <0.0001a | <0.0001a |
| BMI025 (rabbit) rPA | |||||||||
| ED50 | 542 (12) | 292 (15) | 167 (24) | 1.9 (1.5, 2.2) | 3.3 (2.7, 3.9) | 1.8 (1.5, 2.1) | <0.0001a | <0.0001a | <0.0001a |
| EC50 | 0.218 (12) | 0.404 (15) | 0.709 (24) | 0.540 (0.445, 0.654) | 0.307 (0.258, 0.366) | 0.569 (0.484, 0.67) | <0.0001a | <0.0001a | <0.0001a |
| BMI280 rPA | |||||||||
| ED50 | 7,419d (9) | 3,291 (14) | 1,560 (16) | 4,079e (4,175, 3,983) | 5,815e (5,910, 5,721) | 2.1 (1.8, 2.5) | <0.0001a | <0.0001a | <0.0001a |
| EC50 | 5.763d (9) | 0.129 (14) | 0.272 (16) | 5.632e (3.052, 8.212) | 5.484e (2.896, 8.071) | 0.474 (0.404, 0.556) | <0.0001a | <0.0001a | <0.0001a |
| BMI281 rPA | |||||||||
| ED50 | 2,669d (15) | 861 (15) | 390 (16) | 1,803e (1,856, 1,751) | 2,258e (2,311, 2,206) | 2.2 (1.8, 2.7) | <0.0001a | <0.0001a | <0.0001a |
| EC50 | 9.288d (15) | 0.493 (15) | 1.088 (16) | 8.793e (5.739, 11.847) | 8.135e (5.02, 11.25) | 0.453 (0.375, 0.546) | <0.0001a | <0.0001a | <0.0001a |
| MAb 1G3 | |||||||||
| ED50 | 98,659 (30) | 75,296 (16) | 9,560 (44) | 1.3 (1, 1.7) | 10.3 (8.4, 12.8) | 7.9 (6.1, 10.2) | 0.0555 | <0.0001a | <0.0001a |
| EC50 | 0.052 (30) | 0.068 (16) | 0.533 (44) | 0.763 (0.579, 1.007) | 0.097 (0.078, 0.120) | 0.127 (0.098, 0.165) | 0.0555 | <0.0001a | <0.0001a |
| MAb 14B7 | |||||||||
| ED50 | 7,349 (12) | 11,616 (16) | <LODb (18) | 0.6 (0.5, 0.7) | 734.9 (636.1, 849.0) | 11,615.6 (10,168.1, 13,269.2) | <0.0001a | <0.0001a | <0.0001a |
| EC50 | 0.354 (12) | 0.224 (16) | <LODc (18) | 1.581 (1.363, 1.833) | 0.354 (0.306, 0.409) | 0.224 (0.197, 0.255) | <0.0001a | <0.0001a | <0.0001a |
The ratio is statistically significantly different from 1 at a 0.05 level of significance. Thus, there is a statistically significant difference in potency between the assays.
The RB-TNA assay ED50 for 14B7 was below the LOD of 12, but to perform these statistical computations, a value of 1 was substituted for “<LOD.”
The RB-TNA assay EC50 for 14B7 was below the LOD of 12, but to perform these statistical computations, a value of 1 was substituted for “<LOD.”
The R-TNA assay ED50 and EC50 values are taken from historical data from assays run prior to this study.
Because only historical R-TNA assay data were available for BMI280 and BMI281, the difference of the NC- and RB-TNA assays from the R-TNA assay using arithmetic means is reported instead of a ratio.
EC50s are in micrograms per milliliter.
Data handling and statistical analyses.
All ED50 data were included in the analyses, with the exception of the samples that presented ED50 values that were below the limit of detection (LOD) in the NC-TNA and/or RB-TNA assays. Of the 110 NHP serum samples from the 30-week postvaccination time point, there were nine samples in this category. Of these nine samples, seven had ED50s below the LOD in the RB-TNA assay and two had ED50s below the LOD in both the RB- and NC-TNA assays (23).
An analysis of variance model was fitted to the log-transformed assay development ED50 and EC50 data separately for each sample to determine if there was a significant difference between (i) the R-TNA and NC-TNA assays and (ii) the R-TNA and RB-TNA assays using Tukey's test at a 0.05 level of significance. Next, only the ED50 data from the 30-week postvaccination time point (the remaining 99 samples) were log10 transformed and plotted and a linear regression model was fitted to determine the relationship between the NC-TNA (PA83) and RB-TNA (PA63) assays with respect to the ED50 endpoint. Linear regression was also used to determine if the relationship between the NC- and RB-TNA assays remained consistent among animals that survived the challenge and animals that did not survive the challenge. Additionally, logistic regression models on survival were fitted to the log10-transformed ED50 data in order to assess whether the NC- and RB-TNA assays were predictive of survival across challenge times. If challenge time was not a significant factor in the relationship, then a logistic regression model based on survival was fitted to the data regardless of the challenge time to estimate the antibody levels and the Fieller approximate 95% confidence intervals (CIs) associated with 80%, 90%, and 95% survival probabilities. Finally, linear regression analysis was used to determine whether the same relationship between the NC- and RB-TNA assays held for postchallenge sera as for prechallenge sera (samples from 20 macaques from the 52-month challenge time point only).
As part of the linear regression analysis, the regression coefficients (r2 values) and slopes were calculated for the ED50 value groupings. When testing whether there was a statistically significant difference between slopes, a t test at a 0.05 level of significance was used. The data sets used in each analysis are further described in Results.
RESULTS
The rationale for developing the NC-TNA assay was to provide a comparative functional control for the effects of running the assay at reduced temperatures and to bridge the data from the RB-TNA assay performed at 4°C with those from the traditional R-TNA assay performed at 37°C. Figure 2 compares the three types of TNA assays. In the NC-TNA assay, antisera or MAb were preincubated with both the PA and LF toxin components in solution. The NC-TNA assay was similar to the R-TNA assay in that it was designed to quantify the antibody response that neutralized at any of several steps in the intoxication pathway: (i) the PA in solution prior to binding to the ATR, (ii) activation of the PA to form the heptamer, (iii) and/or binding of LF to form the lethal toxin complex (so neutralization could also take place with the RB 63-kDa form of the PA in this assay). In contrast to the R-TNA assay, which is conducted entirely at 37°C, in the NC-TNA assay, the PA and LF neutralization incubation was conducted at 4°C on ice. Additionally, in the NC-TNA assay, the cell monolayer was first cooled before the intoxication step by incubating the cells at 4°C on ice and the cold PA-LF-serum cocktail was then transferred to the chilled cell monolayers. The cell monolayers were warmed to 37°C, at which temperature cell intoxication progressed (Fig. 2).
FIG. 2.
Schematic of the R-, NC-, and RB-TNA assays (Ab represents either antiserum or MAb). The traditional R-TNA assay (A) performed as described by Li et al. (23) and the NC-TNA assay (B) both quantify neutralization from antibodies that recognize unbound PA83 in solution, although some recognition of bound PA63 can occur after the cocktail of PA, LF, and Ab is added to the cells. The NC-TNA assay (B) was developed solely to include cell cooling steps so that the results of the RB-TNA assay may be compared to those of the traditional R-TNA assay. The cooling step performed in the NC-TNA assay slightly affected the potency of Ab neutralization (as evident in Table 2; comparison of R- and NC-TNA assays), although the level of neutralization by the Ab is proportional between the assays. The RB-TNA assay (C) was developed to delineate the level of neutralization from antibodies that complexed with only the RB form of PA (PA63), as unbound PA is removed from the cells prior to the addition of Ab and LF. The cooling steps for this assay served to cool the lipid bilayer cell membrane to slow PA internalization so that after unbound PA was removed, the subsequently added anti-ATx Ab could effectively bind surface-bound PA without the presence of LF. The LF was added after unbound Ab was removed so that the cells could be warmed and intoxication could proceed.
The purpose of the RB-TNA assay was to delineate the level of neutralization from antibodies that complexed with only the RB form of PA (PA63). The RB-TNA assay approach to cell intoxication by LTx proceeded in a two-stage fashion. In this assay, the cell monolayers were first cooled to 4°C to minimize receptor internalization and facilitate measurement of receptor-binding events (46). In the first stage, only PA83 (no LF) was incubated directly with the cooled cell monolayers. This first cooled incubation step allowed for binding and cleavage of PA83 to the 63-kDa form (PA63) on the cell surface before the addition of the LF. This differs from the R-TNA and NC-TNA assays, where both PA and LF were preincubated together with antisera in solution at 37°C and 4°C, respectively. Unbound PA83 and cleaved 20-kDa PA fragments were removed, leaving only the PA63 form bound to the lipid bilayer. Antiserum containing anti-PA antibody was then added. The cell plates and all reagents were kept cold at all times; therefore, the cooled membranes slowed the otherwise rapid degradation or internalization of the PA63 complexes so as to enable the binding of antibodies that were specific to the 63-kDa form of the RB PA. In the second stage of the RB-TNA assay, the antiserum was removed and LF was added, the cell monolayers were warmed to 37°C, and toxicity progressed so that the cells lysed, with the exception of where the antibodies neutralized the toxin in a dose-dependent fashion (Fig. 2).
Development of low-temperature TNA assays.
Using MAb 14B7 and 1G3 as controls, we first demonstrated that the NC- and RB-TNA assays could distinguish between the NC and RB PAs. In addition to the MAb, antisera from different genera (humans, NHPs, and rabbits) that received different PA-based vaccines (AVA or rPA) were tested to determine if the assays were comparable (Table 2). The approximately 10-fold lower potency of 1G3 in the RB-TNA assay than in both the R- and NC-TNA assays is an intriguing observation that may merit further investigation (Table 2). An explanation for this phenomenon may be that receptor binding by PA affects the affinity or accessibility of the epitope for 1G3 such that PA63-bound MAb is more readily displaced by LF in the later stages of the assay and toxin internalization process. The ratio of reportable values for 1G3 in the R-TNA assay and NC-TNA assay, 1.3, indicates that the potency of this antibody in these in vitro assays was not significantly affected by temperature (P = 0.0555, Table 2). Despite the decrease in relative activity, 1G3 still provided substantial neutralization in the RB-TNA assay, with an ED50 of 9,560 (EC50 of 0.533 μg/ml), whereas there was no detectable neutralization by 14B7 (Table 2). These data confirmed that neutralization by 14B7 is a pre-receptor-binding event, whereas neutralization by 1G3 is a post-receptor-binding event, and demonstrated that the assays were performing as intended. Thus, domain 4 of PA, to which 14B7 binds, was blocked in the RB-TNA assay since the PA was prebound to the ATR. Moreover, 1G3 was effective in neutralizing in all three assays because it binds to exposed domains 1b to 3 of PA63. Domains 1b, 2, and 3 are the only sites on PA that are accessible for antibody binding in the RB-TNA assay and are accessible in later steps of the R- and NC-TNA assays. The results demonstrated that the assays distinguish effectively between bound and unbound PA because only 1G3 neutralized PA in all three (R-, NC-, and RB-TNA) assay formats whereas 14B7 did not neutralize in the RB-TNA assay.
The ED50 ratio of the NC-TNA assay to the R-TNA assay was consistently lower than the ED50 ratio of the RB-TNA assay to the R-TNA assay for all of the sera tested (Table 2). The results revealed a statistically significant shift in LTx neutralization potency among the three (R-, NC-, and RB-TNA) assays (Table 2). For polyclonal serum, the magnitude in the shift in ED50 values of the R-TNA assay compared to those of the NC-TNA assay is approximately 2-fold lower, and the ED50 values of the R-TNA assay are approximately 4-fold lower than those of the RB-TNA assay. Likewise, for the EC50 data, the NC-TNA assay potency is approximately half that of the R-TNA assay and the RB-TNA assay potency is approximately one-quarter that of the R-TNA assay. When they were compared to each other using either ED50 or EC50 assay readouts, there was an approximately 2-fold difference between the NC- and RB-TNA assays (Table 2). These data indicate that the reduction in assay operating temperature effected an approximately 2-fold reduction in polyclonal anti-PA antiserum potency and the combined effect of temperature reduction and receptor binding effected a 4-fold reduction in potency. The ratios of the potencies of the polyclonal serum tested between the NC- and RB-TNA assays compared to the R-TNA assay were remarkably consistent among the three species tested (humans, NHPs, and rabbits) and the PA-based vaccines (AVA and rPA) (Table 2). Collectively, these data demonstrate that the NC-TNA assay and RB-TNA assay are able to distinguish the in vitro neutralization of RB and nonbound PA.
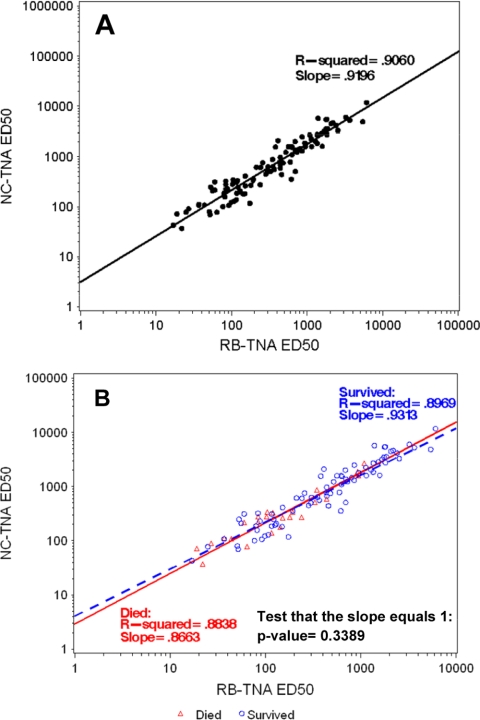
When the week 30 postvaccination macaque samples were tested, linear regression analyses of the ED50 endpoint data were performed comparing the NC-TNA assay and RB-TNA assay ED50 outputs, which demonstrated an r2 value of 0.9060 and a slope of 0.9196 (Fig. 3A). At the midpoint of the regression line, there was a 1.9-fold change using the slope of 0.9196 and the intercept of 0.4413. These data indicated a direct relationship between serum in vitro neutralizing efficacies in the two assay systems, manifest as a consistent 2-fold shift in ED50 between the NC-TNA and RB-TNA assays. This is consistent with the ED50 and EC50 data presented in Table 2 from assay development.
FIG. 3.
Correlation of log10 ED50 titers for week 30 postvaccination samples combined between the NC- and RB-TNA assays and separately based on survivors versus nonsurvivors. Linear regression analysis was performed on the log10 ED50 data from both the NC- and RB-TNA assays to determine if the assays correlated with each other using rhesus macaque sera taken from the AVRP project. Panel A represents the week 30 postvaccination samples combined (n = 101), whereas panel B represents the week 30 postvaccination samples analyzed separately based on survivors versus nonsurvivors. As part of the analysis, the r2 and slope were calculated. For the combined analysis, there was a strong positive correlation (r2 = 0.9060) between the RB- and NC-TNA assay ED50 titers. In addition, separate regression lines were fitted to the 30-week postvaccination samples from surviving (n = 76) and nonsurviving (n = 25) animals to determine if there was a difference in assay correlation for either cohort of animals. As part of the analysis, the r2 and slope were calculated for the ED50 titers. To test whether there was a statistically significant difference between the slopes of the surviving animals and the nonsurviving animals, a t test at a 0.05 level of significance was used. The slopes were not statistically significantly different (P = 0.3389), and there was a strong positive correlation (survivor r2 = 0.8969, nonsurvivor r2 = 0.8838) between RB- and NC-TNA assay ED50 titers.
Evaluation of rhesus macaque vaccination responses and association with protection.
The comparability of the NC- and RB-TNA assays was established during development using the MAb and polyclonal sera. Consequently, peak response sera (4 weeks after the third vaccination) from AVA-vaccinated rhesus macaques were analyzed in the NC- and RB-TNA assays to determine whether a differentiation between RB and unbound PA neutralization in response to vaccination correlated with survival of inhalation anthrax.
Linear regression analyses were used to determine whether the RB- and NC-TNA assays were able to distinguish vaccine-induced neutralization responses in macaques that subsequently survived the challenge from those that succumbed to infection (Fig. 3B). These data demonstrated a strong positive correlation between week 30 ED50 values in both assays for survivors (r2 = 0.8969) and nonsurvivors (r2 = 0.8838). The regression slopes for survivors and nonsurvivors were 0.9313 and 0.8663, respectively, and these were not statistically significantly different (P = 0.3389). These data indicated that the ability of week 30 antibody responses to neutralize RB PA was predictive of protection but not in itself a correlate of protection for AVA-vaccinated rhesus macaques.
NC- and RB-TNA assay titers are predictive of survival.
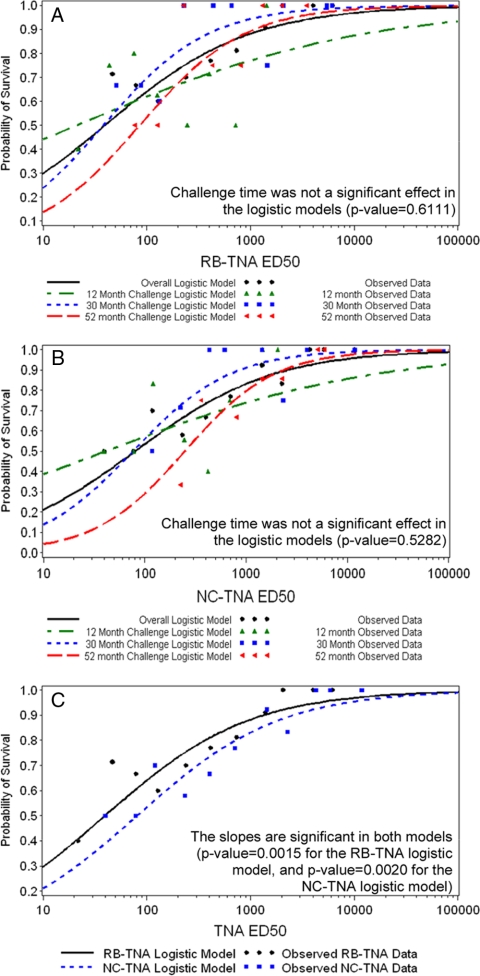
We determined that NC- and RB-TNA assay responses were predictive of survival across challenge times, whether the challenge was months or years after vaccination. The animal groups consisted of animals challenged at 12, 30, and 52 months (Table 1). Logistic regression curves were fitted to the log10 ED50 data using the 30-week postvaccination samples from both the NC- and RB-TNA assays to determine if the assays were predictive of survival (Fig. 4 ). The models were fitted to each challenge time (12, 30, or 52 months) and each assay separately (RB- or NC-TNA assay), where it was determined that challenge time was not significant (Fig. 4A and B; RB-TNA assay, P value of 0.6111; NC-TNA assay, P value of 0.5282). This allowed for a combined logistic model to be fitted so that data from each assay could be directly compared (Fig. 4C). The antibody levels associated with 80%, 90%, and 95% survival probabilities were assessed (Table 3) using the model from Fig. 4C. An LTx-neutralizing ED50 titer of 721 in the NC-TNA assay or a titer of 366 in the RB-TNA assay at week 30 (using a 0-, 4-, and 26-week i.m. regimen) was predictive of at least 80% protection against an aerosol challenge with virulent B. anthracis Ames spores up to 52 months after the first vaccination (Table 3). According to the model, higher ED50 values provided additional protection with an improved probability of survival (Table 3).
FIG. 4.
Logistic regression curves of log10 ED50 titers from week 30 postvaccination samples. Logistic regression curves were fitted to the log10 ED50 data using the 30-week postvaccination samples (n = 101) from both the NC- and RB-TNA assays to determine if the assays were predictive of survival. (A) First, models were fitted to the log10 ED50 RB-TNA assay data by challenge month (12 months, green line, n = 39; 30 months, blue line, n = 33; 52 months, red line, n = 29). It was determined that challenge time was not a significant effect in the RB-TNA assay logistic models (P value of 0.6111), which permitted analysis of the RB-TNA assay data regardless of the challenge month (overall model, black line). (B) Next, the models were fitted to the log10 ED50 NC-TNA assay data by challenge month (12 months, green line, n = 39; 30 months, blue line, n = 33; 52 months, red line, n = 29). It was determined that challenge time was not a significant effect in the NC-TNA assay logistic models (P value of 0.5282), which permitted analysis of the NC-TNA assay data regardless of the challenge month (overall model, black line). (C) Finally, since the challenge times did not significantly affect the RB- and NC-TNA assay models, the overall models from the assays were combined and assessed together regardless of the challenge month (black line, RB-TNA assay; blue line, NC-TNA assay). From this analysis, it was found that the slopes are significant in both models (P value of 0.0015 for the RB-TNA assay logistic model and P value of 0.0020 for the NC-TNA assay logistic model); thus, both the NC- and RB-TNA assays were predictive of survival, regardless of the challenge month.
TABLE 3.
ED50 levels in the NC- and RB-TNA assays associated with NHP survival of inhalation challenge with B. anthracis spores
| Survival probability (%) | ED50 |
|
|---|---|---|
| RB-TNA assay | NC-TNA assay | |
| 80 | 366 (169, 1,838)a | 721 (338, 3,831) |
| 90 | 1,340 (500, 41,061) | 2,594 (975, 97,031) |
| 95 | 4,431 (1,136, 858,337) | 8,441 (2,169, 2,275,952) |
The values in parentheses are 95% CIs.
Comparison of immune responses to AVA vaccination and native PA from a B. anthracis aerosol challenge.
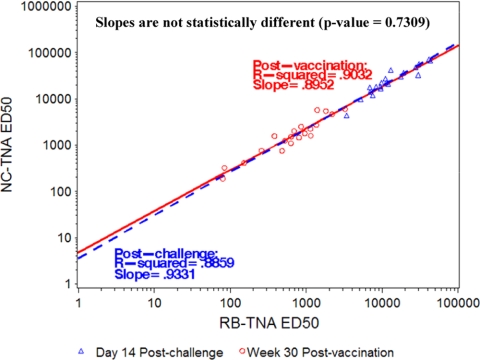
Due to the high positive correlation between the NC- and RB-TNA assay results, it appeared that prebinding of lethal toxin to the receptor did not have a measurable effect on the ability of anti-AVA antiserum to neutralize LTx, and therefore, there was not a bias in antibody response to AVA to a specific PA domain. However, it was not clear if the same conclusion applied to anti-PA antibodies that were generated postinfection in response to a B. anthracis aerosol challenge. Therefore, we sought to determine whether the same relationship held for postchallenge sera, as the native PA antigen presented during infection may be different from the PA presented to the host from vaccination. We hypothesized that perhaps specific PA domain dominance might be elicited postinfection. Twenty samples from the 52-month challenge group that survived an aerosol challenge with virulent B. anthracis spores (collected 14 days postchallenge, irrespective of the challenge month) were tested in both the NC- and RB-TNA assays, and the resulting data were compared to the corresponding week 30 postvaccination time point data for each animal using linear regression analysis. The ED50 response postchallenge was at least 100-fold higher than the response after vaccination, but it did not appear to be biased toward the NC or RB mechanism of neutralization. This is demonstrated by the nearly identical r2 values of 0.8952 and 0.9331 (P value of 0.7309) for the 30-week postvaccination and 14-day postchallenge samples, respectively (Fig. 5).
FIG. 5.
Relationship between NC- and RB-TNA assay ED50 titers in surviving animals (n = 20) at week 30 postvaccination and at day 14 postchallenge (log10 ED50). Linear regression analysis was performed on the log10 ED50 data using the 30-week postvaccination samples and day 14 postchallenge samples from both the NC- and RB-TNA assays to determine if the assays correlated with each other. Separate regression lines were fitted to the 30-week postvaccination titers and the day 14 postchallenge titers to determine if there was a difference in assay correlation for either cohort of animals. As part of the analysis, the r2 and slope were calculated for the ED50 values. To test whether there was a statistically significant difference between the slopes of the 30-week postvaccination samples and day 14 postchallenge samples, a t test at a 0.05 level of significance was used. The slopes were found to be not statistically significantly different (P value of 0.7309).
DISCUSSION
It has been reported previously that sera from rhesus macaques immunized with AVA, rPA, or the United Kingdom licensed anthrax vaccine precipitated (AVP) contain toxin-neutralizing antibodies that recognize all four of the domains of PA (47, 48, 60). While antibody recognition of the N terminus of PA was predominant in macaques immunized with the existing vaccines (AVP and AVA), macaques immunized with rPA recognized both the N- and C-terminal domains of PA (60). Nonetheless, other studies of the anti-PA responses to AVA have proposed that the PA20 fragment (domain 1a) may carry the immunodominant epitopes in AVA-vaccinated donors. This suggests that removing the immunodominant epitopes found in PA20 from the vaccine would improve the vaccine by targeting RB PA epitopes (47, 48, 61). Therefore, the PA63 conformer has received some attention as a vaccine component, whether as the recombinant protein, as a truncated gene transcript expressed in Salmonella spp., or as a component of purified DNA vaccines (8, 12). The rationale for this approach resides in the concepts that PA63 may present different, more relevant, epitopes to the immunized host than does PA83 and that direct presentation of PA63 eliminates the need for proteolytic processing of the full-length molecule. However, despite some success in animal models, the development of PA63 as a vaccine component is technically challenging because it is difficult to produce and purify (e.g., problems with aggregates and mixture of conformers). Therefore, the focus of recombinant PA vaccine research and development still remains on full-length PA83.
In this study, we demonstrated that LTx-neutralizing responses elicited by AVA vaccination in rhesus macaques can neutralize LTx in vitro even after it has bound to the cell surface receptor. Receptor binding was confirmed by loss of neutralization by MAb 14B7 while retaining neutralization by MAb 1G3 (26). The consistent correlations between the NC- and RB-TNA assays indicate that vaccination with AVA generates a repertoire of anti-PA immune responses capable of neutralizing free and RB PA. The results of this study indicate that vaccination by AVA provides protection against multiple targets in LTx intoxication of host cells, perhaps by blocking the PA-LF interaction required for LTx formation, thereby preventing LTx internalization/translocation or by successfully displacing PA from the cell surface receptor. We interpret the high correlation between vaccine-induced and postinfection serum activities in the NC- and RB-TNA assays to mean that the anti-PA response elicited by AVA is qualitatively not different from that following exposure of vaccinated animals to PA during inhalation anthrax.
The neutralizing response in the RB-TNA assay was very consistent across low and high ED50 values, survivors and nonsurvivors, challenge times, and postvaccination and postchallenge, indicating that the anti-PA neutralizing response occurs irrespective of blocking of the interaction between domain 4 of PA and the host cell receptor. It was also determined during these studies that there was a strong positive correlation in both the NC- and RB-TNA assays between the magnitude of the ED50 response and survival. These findings are consistent with results of other studies that employed the traditional R-TNA assay (23, 38).
It is important to note that one must be careful when assigning a value to a protective ED50 titer because it depends on the vaccine used and its formulation, the assay from which the ED50 was obtained, the timing of sample collection, and the treatment of the donor prior to sampling (e.g., the number of vaccinations, etc.). For example, the results of this study demonstrated that an ED50 of 2,941 in the RB-TNA assay from the 30-week postvaccination samples was predictive of up to 95% protection. However, this ED50 value may not apply to other vaccines or regimens. Interestingly, observations from the NC- and RB-TNA assays align with data from the R-TNA assay such that the nature of the peak response immediately following the third vaccination with AVA (which, using the original AVA vaccination schedule, was at 4 weeks) gives the best correlation with 95% protection (28).
The data presented here demonstrate that there was not a functional bias in the immune response to PA following AVA vaccination, as indicated by the analysis of the pre- and postchallenge samples. In fact, the only clear difference was an increase in the magnitude of the peak anti-PA IgG response, as judged by higher ED50s, to toxin exposure postchallenge. Thus, although the magnitude of the ED50 response was greater after aerosol infection, the relationship between neutralization in the RB assay and in the NC assay did not change compared to the vaccine-induced response alone. Because the animals were primed by PA from the AVA vaccination, the higher neutralizing response after challenge was an anamnestic response due to common epitopes in AVA PA and infection PA.
Taken together, these data demonstrate that the PA component of AVA is highly immunogenic and the humoral immune response generated by vaccination with AVA is potent in neutralizing multiple mechanistic steps of intoxication by anthrax LTx. Importantly, this study showed that the regions of PA occluded during receptor binding via domain 4 are not the only protective epitopes in a PA-based vaccine. Given that the PA domain 4 epitopes can be modified without affecting toxin activity (49), we conclude that domain-based recombinant vaccines must be evaluated with great care. The results of this study, specifically, the RB-TNA assay results, found that credence should be given to using vaccines that include the 83-kDa form of PA, as it includes not only domain 4 but also domains 2 and 3 and part of domain 1, which provide epitopes necessary for a comprehensive antigenic repertoire and an effective vaccine. In conclusion, these data provide additional evidence that functionally competent full-length PA83 is an excellent vaccine antigen, and the relative ease of its production and purification, compared to properly folded, conformation-competent PA domain-based vaccines, is justification for continuing to pursue PA83 as the primary vaccine candidate.
Acknowledgments
This study was funded by the CDC Anthrax Vaccine Research Program, DHHS CDC contract 200-2000-10065. We are grateful for the following reagents provided by the NIH Biodefense and Emerging Infections Research Resources Repository (BEI Resources), NIAID, NIH: anthrax PA, recombinant from B. anthracis, NR-140, and anthrax LF, recombinant from B. anthracis, NR-142.
We acknowledge the contributions of Nina Marano and Jennifer Wright, former and current CDC principal investigators for the CDC Anthrax Vaccine Research Program (AVRP), respectively, and Walter Holt, Jr., AVRP coordinator. We acknowledge Robert E. Hunt, Pamela Olson, and Michelle Vassar at the Battelle Memorial Institute and Robert Mittler at the Emory Vaccine Center, Emory University School of Medicine, Atlanta, GA, for supplying the NHP serum samples and the human clinical study principal investigators for supplying the human serum samples.
The findings and conclusions in this report are ours and do not necessarily represent the official position of the CDC.
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Abrami, L., S. Liu, P. Cosson, S. H. Leppla, and F. G. van der Goot. 2003. Anthrax toxin triggers endocytosis of its receptor via a lipid raft-mediated clathrin-dependent process. J. Cell Biol. 160:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach, S., and G. G. Wright. 1955. Studies on immunity in anthrax. VI. Immunizing activity of protective antigen against various strains of Bacillus anthracis. J. Immunol. 75:129-133. [PubMed] [Google Scholar]
- 3.Beall, F. A., M. J. Taylor, and C. B. Thorne. 1962. Rapid lethal effect in rats of a third component found upon fractionating the toxin of Bacillus anthracis. J. Bacteriol. 83:1274-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belova, E. V., S. A. Dubilei, T. B. Kravchenko, A. V. Kolesnikov, M. I. Zakharova, and I. G. Shemiakin. 2004. Monoclonal antibodies to B. anthracis protective antigen are capable to neutralize and to enhance the anthrax lethal toxin action in vitro. Mol. Gen. Mikrobiol. Virusol. 2004:21-26. (In Russian.) [PubMed] [Google Scholar]
- 5.Belton, F. C., H. M. Darlow, and D. W. Henderson. 1956. The use of anthrax antigen to immunise man and monkey. Lancet 271:476-479. [DOI] [PubMed] [Google Scholar]
- 6.Blaustein, R. O., T. M. Koehler, R. J. Collier, and A. Finkelstein. 1989. Anthrax toxin: channel-forming activity of protective antigen in planar phospholipid bilayers. Proc. Natl. Acad. Sci. U. S. A. 86:2209-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley, K. A., J. Mogridge, M. Mourez, R. J. Collier, and J. A. Young. 2001. Identification of the cellular receptor for anthrax toxin. Nature 414:225-229. [DOI] [PubMed] [Google Scholar]
- 8.Coulson, N. M., M. Fulop, and R. W. Titball. 1994. Bacillus anthracis protective antigen, expressed in Salmonella typhimurium SL 3261, affords protection against anthrax spore challenge. Vaccine 12:1395-1401. [DOI] [PubMed] [Google Scholar]
- 9.Elliott, J. L., J. Mogridge, and R. J. Collier. 2000. A quantitative study of the interactions of Bacillus anthracis edema factor and lethal factor with activated protective antigen. Biochemistry 39:6706-6713. [DOI] [PubMed] [Google Scholar]
- 10.Emergent BioSolutions. 2008. BioThrax (anthrax vaccine adsorbed) product prescribing information. http://www.emergentbiosolutions.com/pdf/emergent_biothrax_us.pdf.
- 11.Findlay, J. W., and R. F. Dillard. 2007. Appropriate calibration curve fitting in ligand binding assays. AAPS J. 9:E260-E267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garmory, H. S., R. W. Titball, K. F. Griffin, U. Hahn, R. Bohm, and W. Beyer. 2003. Salmonella enterica serovar Typhimurium expressing a chromosomally integrated copy of the Bacillus anthracis protective antigen gene protects mice against an anthrax spore challenge. Infect. Immun. 71:3831-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubbins, M. J., L. Schmidt, R. S. Tsang, J. D. Berry, A. Kabani, and D. I. Stewart. 2007. Development of a competitive enzyme linked immunosorbent assay to identify epitope specific antibodies in recipients of the U.S. licensed anthrax vaccine. J. Immunoassay Immunochem. 28:213-225. [DOI] [PubMed] [Google Scholar]
- 14.Ivins, B. E., J. W. Ezzell, Jr., J. Jemski, K. W. Hedlund, J. D. Ristroph, and S. H. Leppla. 1986. Immunization studies with attenuated strains of Bacillus anthracis. Infect. Immun. 52:454-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivins, B. E., P. F. Fellows, and G. O. Nelson. 1994. Efficacy of a standard human anthrax vaccine against Bacillus anthracis spore challenge in guinea-pigs. Vaccine 12:872-874. [DOI] [PubMed] [Google Scholar]
- 16.Ivins, B. E., M. L. Pitt, P. F. Fellows, J. W. Farchaus, G. E. Benner, D. M. Waag, S. F. Little, G. W. Anderson, Jr., P. H. Gibbs, and A. M. Friedlander. 1998. Comparative efficacy of experimental anthrax vaccine candidates against inhalation anthrax in rhesus macaques. Vaccine 16:1141-1148. [DOI] [PubMed] [Google Scholar]
- 17.Klimpel, K. R., S. S. Molloy, G. Thomas, and S. H. Leppla. 1992. Anthrax toxin protective antigen is activated by a cell surface protease with the sequence specificity and catalytic properties of furin. Proc. Natl. Acad. Sci. U. S. A. 89:10277-10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koehler, T. M., and R. J. Collier. 1991. Anthrax toxin protective antigen: low-pH-induced hydrophobicity and channel formation in liposomes. Mol. Microbiol. 5:1501-1506. [DOI] [PubMed] [Google Scholar]
- 19.Leppla, S. H. 1988. Production and purification of anthrax toxin. Methods Enzymol. 165:103-116. [DOI] [PubMed] [Google Scholar]
- 20.Leppla, S. H. 2000. Anthrax toxin, p. 445-472. In K. Aktories and I. Just (ed.), Bacterial protein toxins. Springer, Berlin, Germany.
- 21.Leysath, C. E., A. F. Monzingo, J. A. Maynard, J. Barnett, G. Georgiou, B. L. Iverson, and J. D. Robertus. 2009. Crystal structure of the engineered neutralizing antibody M18 complexed to domain 4 of the anthrax protective antigen. J. Mol. Biol. 387:680-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, G., Y. Qu, C. Cai, Y. Kong, S. Liu, J. Zhang, J. Zhao, L. Fu, J. Xu, and W. Chen. 2009. The inhibition of the interaction between the anthrax toxin and its cellular receptor by an anti-receptor monoclonal antibody. Biochem. Biophys. Res. Commun. 385:591-595. [DOI] [PubMed] [Google Scholar]
- 23.Li, H., S. D. Soroka, T. H. Taylor, Jr., K. L. Stamey, K. W. Stinson, A. E. Freeman, D. R. Abramson, R. Desai, L. X. Cronin, J. W. Oxford, J. Caba, C. Pleatman, S. Pathak, D. S. Schmidt, V. A. Semenova, S. K. Martin, P. P. Wilkins, and C. P. Quinn. 2008. Standardized, mathematical model-based and validated in vitro analysis of anthrax lethal toxin neutralization. J. Immunol. Methods 333:89-106. [DOI] [PubMed] [Google Scholar]
- 24.Little, S. F., B. E. Ivins, P. F. Fellows, and A. M. Friedlander. 1997. Passive protection by polyclonal antibodies against Bacillus anthracis infection in guinea pigs. Infect. Immun. 65:5171-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little, S. F., S. H. Leppla, and A. M. Friedlander. 1990. Production and characterization of monoclonal antibodies against the lethal factor component of Bacillus anthracis lethal toxin. Infect. Immun. 58:1606-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little, S. F., J. M. Novak, J. R. Lowe, S. H. Leppla, Y. Singh, K. R. Klimpel, B. C. Lidgerding, and A. M. Friedlander. 1996. Characterization of lethal factor binding and cell receptor binding domains of protective antigen of Bacillus anthracis using monoclonal antibodies. Microbiology 142(Pt. 3):707-715. [DOI] [PubMed] [Google Scholar]
- 27.Liu, S., and S. H. Leppla. 2003. Cell surface tumor endothelium marker 8 cytoplasmic tail-independent anthrax toxin binding, proteolytic processing, oligomer formation, and internalization. J. Biol. Chem. 278:5227-5234. [DOI] [PubMed] [Google Scholar]
- 28.Madigan, D. 8 November 2007. Anthrax vaccines: bridging correlates of protection in animals to immunogenicity in humans. Center for Biologics Evaluation and Research, Food and Drug Administration, U.S. Department of Health and Human Services, Washington, DC.
- 29.Mahlandt, B. G., F. Klein, R. E. Lincoln, B. W. Haines, W. I. Jones, Jr., and R. H. Friedman. 1966. Immunologic studies of anthrax. IV. Evaluation of the immunogenicity of three components of anthrax toxin. J. Immunol. 96:727-733. [DOI] [PubMed] [Google Scholar]
- 30.Marano, N., B. D. Plikaytis, S. W. Martin, C. Rose, V. A. Semenova, S. K. Martin, A. E. Freeman, H. Li, M. J. Mulligan, S. D. Parker, J. Babcock, W. Keitel, S. H. El, G. A. Poland, R. M. Jacobson, H. L. Keyserling, S. D. Soroka, S. P. Fox, J. L. Stamper, M. M. McNeil, B. A. Perkins, N. Messonnier, and C. P. Quinn. 2008. Effects of a reduced dose schedule and intramuscular administration of anthrax vaccine adsorbed on immunogenicity and safety at 7 months: a randomized trial. JAMA 300:1532-1543. [DOI] [PubMed] [Google Scholar]
- 31.McConnell, M. J., P. C. Hanna, and M. J. Imperiale. 2007. Adenovirus-based prime-boost immunization for rapid vaccination against anthrax. Mol. Ther. 15:203-210. [DOI] [PubMed] [Google Scholar]
- 32.Milne, J. C., and R. J. Collier. 1993. pH-dependent permeabilization of the plasma membrane of mammalian cells by anthrax protective antigen. Mol. Microbiol. 10:647-653. [DOI] [PubMed] [Google Scholar]
- 33.Milne, J. C., D. Furlong, P. C. Hanna, J. S. Wall, and R. J. Collier. 1994. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J. Biol. Chem. 269:20607-20612. [PubMed] [Google Scholar]
- 34.Mogridge, J., K. Cunningham, and R. J. Collier. 2002. Stoichiometry of anthrax toxin complexes. Biochemistry 41:1079-1082. [DOI] [PubMed] [Google Scholar]
- 35.Mogridge, J., K. Cunningham, D. B. Lacy, M. Mourez, and R. J. Collier. 2002. The lethal and edema factors of anthrax toxin bind only to oligomeric forms of the protective antigen. Proc. Natl. Acad. Sci. U. S. A. 99:7045-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohamed, N., M. Clagett, J. Li, S. Jones, S. Pincus, G. D'Alia, L. Nardone, M. Babin, G. Spitalny, and L. Casey. 2005. A high-affinity monoclonal antibody to anthrax protective antigen passively protects rabbits before and after aerosolized Bacillus anthracis spore challenge. Infect. Immun. 73:795-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molloy, S. S., P. A. Bresnahan, S. H. Leppla, K. R. Klimpel, and G. Thomas. 1992. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 267:16396-16402. [PubMed] [Google Scholar]
- 38.Omland, K. S., A. Brys, D. Lansky, K. Clement, and F. Lynn. 2008. Interlaboratory comparison of results of an anthrax lethal toxin neutralization assay for assessment of functional antibodies in multiple species. Clin. Vaccine Immunol. 15:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson, J. W., J. E. Comer, W. B. Baze, D. M. Noffsinger, A. Wenglikowski, K. G. Walberg, J. Hardcastle, J. Pawlik, K. Bush, J. Taormina, S. Moen, J. Thomas, B. M. Chatuev, L. Sower, A. K. Chopra, L. R. Stanberry, R. Sawada, W. W. Scholz, and J. Sircar. 2007. Human monoclonal antibody AVP-21D9 to protective antigen reduces dissemination of the Bacillus anthracis Ames strain from the lungs in a rabbit model. Infect. Immun. 75:3414-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson, J. W., J. E. Comer, D. M. Noffsinger, A. Wenglikowski, K. G. Walberg, B. M. Chatuev, A. K. Chopra, L. R. Stanberry, A. S. Kang, W. W. Scholz, and J. Sircar. 2006. Human monoclonal anti-protective antigen antibody completely protects rabbits and is synergistic with ciprofloxacin in protecting mice and guinea pigs against inhalation anthrax. Infect. Immun. 74:1016-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petosa, C., R. J. Collier, K. R. Klimpel, S. H. Leppla, and R. C. Liddington. 1997. Crystal structure of the anthrax toxin protective antigen. Nature 385:833-838. [DOI] [PubMed] [Google Scholar]
- 42.Pitt, M. L., S. Little, B. E. Ivins, P. Fellows, J. Boles, J. Barth, J. Hewetson, and A. M. Friedlander. 1999. In vitro correlate of immunity in an animal model of inhalational anthrax. J. Appl. Microbiol. 87:304. [DOI] [PubMed] [Google Scholar]
- 43.Pitt, M. L., S. F. Little, B. E. Ivins, P. Fellows, J. Barth, J. Hewetson, P. Gibbs, M. Dertzbaugh, and A. M. Friedlander. 2001. In vitro correlate of immunity in a rabbit model of inhalational anthrax. Vaccine 19:4768-4773. [DOI] [PubMed] [Google Scholar]
- 44.Pitt, M. L., B. E. Ivins, J. E. Estep, J. Farchaus, and A. M. Friedlander. 1995. Comparison of the efficacy of purified protective antigen and MDPH [AVA] to protect non-human primates from inhalation anthrax. Salisbury Med. Bull. 87(Suppl.):130. [Google Scholar]
- 45.Puziss, M., L. C. Manning, J. W. Lynch, E. Barclaye, I. Abelow, and G. G. Wright. 1963. Large-scale production of protective antigen of Bacillus anthracis in anaerobic cultures. Appl. Microbiol. 11:330-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinn, C. P., Y. Singh, K. R. Klimpel, and S. H. Leppla. 1991. Functional mapping of anthrax toxin lethal factor by in-frame insertion mutagenesis. J. Biol. Chem. 266:20124-20130. [PubMed] [Google Scholar]
- 47.Reason, D., J. Liberato, J. Sun, W. Keitel, and J. Zhou. 2009. Frequency and domain specificity of toxin-neutralizing paratopes in the human antibody response to anthrax vaccine adsorbed. Infect. Immun. 77:2030-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reason, D. C., A. Ullal, J. Liberato, J. Sun, W. Keitel, and J. Zhou. 2008. Domain specificity of the human antibody response to Bacillus anthracis protective antigen. Vaccine 26:4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosovitz, M. J., P. Schuck, M. Varughese, A. P. Chopra, V. Mehra, Y. Singh, L. M. McGinnis, and S. H. Leppla. 2003. Alanine-scanning mutations in domain 4 of anthrax toxin protective antigen reveal residues important for binding to the cellular receptor and to a neutralizing monoclonal antibody. J. Biol. Chem. 278:30936-30944. [DOI] [PubMed] [Google Scholar]
- 50.Sabourin, C. L., T. L. Rudge, R. L. Warren, H. J. Mayfield, N. A. Niemuth, R. E. Hunt, J. E. Estep, D. M. Robinson, R. Mittler, D. Madigan, N. Marano, and C. P. Quinn. 2009. Assessment of anti-PA IgG as a correlate of protection following anthrax vaccine adsorbed immunization in rhesus macaques, abstr. S5:1, p. 25. In Bacillus-ACT 2009: the International Bacillus anthracis, B. cereus, and B. thuringiensis Conference, 30 August to 3 September 2009, Santa Fe, New Mexico. American Society for Microbiology, Washington, DC.
- 51.Scobie, H. M., and J. A. Young. 2005. Interactions between anthrax toxin receptors and protective antigen. Curr. Opin. Microbiol. 8:106-112. [DOI] [PubMed] [Google Scholar]
- 52.Semenova, V. A., E. Steward-Clark, K. L. Stamey, T. H. Taylor, Jr., D. S. Schmidt, S. K. Martin, N. Marano, and C. P. Quinn. 2004. Mass value assignment of total and subclass immunoglobulin G in a human standard anthrax reference serum. Clin. Diagn. Lab. Immunol. 11:919-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sivasubramanian, A., J. A. Maynard, and J. J. Gray. 2008. Modeling the structure of mAb 14B7 bound to the anthrax protective antigen. Proteins 70:218-230. [DOI] [PubMed] [Google Scholar]
- 54.Smith, H., and J. Keppie. 1954. Observations on experimental anthrax; demonstration of a specific lethal factor produced in vivo by Bacillus anthracis. Nature 173:869-870. [DOI] [PubMed] [Google Scholar]
- 55.Smith, H., J. Keppie, and J. L. Stanley. 1954. Observations on the cause of death in experimental anthrax. Lancet 267:474-476. [DOI] [PubMed] [Google Scholar]
- 56.Stanley, J. L., and H. Smith. 1961. Purification of factor I and recognition of a third factor of the anthrax toxin. J. Gen. Microbiol. 26:49-63. [DOI] [PubMed] [Google Scholar]
- 57.St Croix, B., C. Rago, V. Velculescu, G. Traverso, K. E. Romans, E. Montgomery, A. Lal, G. J. Riggins, C. Lengauer, B. Vogelstein, and K. W. Kinzler. 2000. Genes expressed in human tumor endothelium. Science 289:1197-1202. [DOI] [PubMed] [Google Scholar]
- 58.Subramanian, G. M., P. W. Cronin, G. Poley, A. Weinstein, S. M. Stoughton, J. Zhong, Y. Ou, J. F. Zmuda, B. L. Osborn, and W. W. Freimuth. 2005. A phase 1 study of PAmAb, a fully human monoclonal antibody against Bacillus anthracis protective antigen, in healthy volunteers. Clin. Infect. Dis. 41:12-20. [DOI] [PubMed] [Google Scholar]
- 59.Turnbull, P. C., M. G. Broster, J. A. Carman, R. J. Manchee, and J. Melling. 1986. Development of antibodies to protective antigen and lethal factor components of anthrax toxin in humans and guinea pigs and their relevance to protective immunity. Infect. Immun. 52:356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williamson, E. D., I. Hodgson, N. J. Walker, A. W. Topping, M. G. Duchars, J. M. Mott, J. Estep, C. Lebutt, H. C. Flick-Smith, H. E. Jones, H. Li, and C. P. Quinn. 2005. Immunogenicity of recombinant protective antigen and efficacy against aerosol challenge with anthrax. Infect. Immun. 73:5978-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, J., A. Ullal, J. Liberato, J. Sun, W. Keitel, and D. C. Reason. 2008. Paratope diversity in the human antibody response to Bacillus anthracis protective antigen. Mol. Immunol. 45:338-347. [DOI] [PMC free article] [PubMed] [Google Scholar]