Abstract
Tetracycline, clindamycin, and other protein synthesis inhibitors at subinhibitory concentrations significantly increased the expression of the pivotal virulence regulator agr and production of the agr-regulated cytolytic phenol-soluble modulins in the community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strain USA300. Our results suggest that such protein synthesis inhibitors may exacerbate the progression of CA-MRSA disease when applied at concentrations that are too low or when treating infections caused by strains resistant to those antibiotics.
Phenol-soluble modulins (PSMs) are key virulence factors of Staphylococcus aureus that lyse neutrophils and other cell types and elicit a series of proinflammatory responses (14). PSMs are produced by most S. aureus strains and contribute significantly to the progression of skin infection caused by epidemic community-associated methicillin-resistant S. aureus (CA-MRSA) (5, 14).
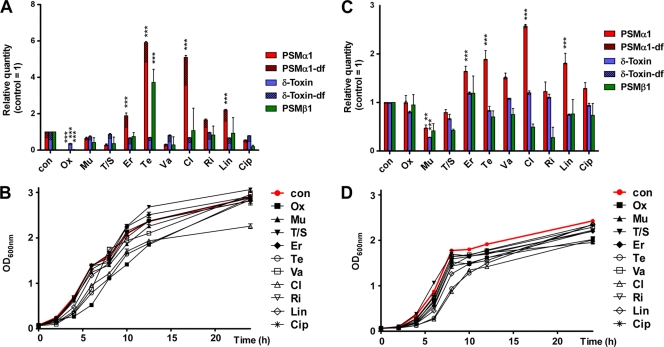
Subinhibitory concentrations of antibiotics may trigger differential expression of bacterial virulence determinants (4), which may lead to exacerbated disease, for example, when antibiotic regimes are not strictly followed by the patient. The effect of subinhibitory antibiotic concentrations on the expression of PSMs has not been investigated. Here, we analyzed PSM production under the influence of subinhibitory concentrations of several antibiotics in clinical use, focusing on antibiotics used for the treatment of hospital-associated MRSA (HA-MRSA) and CA-MRSA, including multidrug-resistant CA-MRSA (6) (Fig. 1 A and C). Of note, “subinhibitory” is often defined solely as having no effect on overnight growth. However, growth patterns may be severely affected even if strains reach comparable optical density values overnight; in that case, observed effects may simply be due to growth inhibition. Therefore, we ascertained that the used antibiotic concentrations did not cause considerable growth defects over the entire growth curve (Fig. 1B and D). As test strains, we chose the CA-MRSA strain USA300 (clone LAC), which is by far the most frequent cause of CA-MRSA infections in the United States (7, 11), and strain Sanger 252, representing a prevalent hospital-associated MRSA clone (9). To select optimal subinhibitory antibiotic concentrations (SICs) (Table 1), we first applied 1/10 of the published MICs. If under those conditions we found severe growth defects, SICs were gradually adjusted until only minimal effects on growth were observed. If no effect on PSM production was observed, we increased the SIC as much as possible without seeing a considerable growth defect. PSM concentrations were determined in triplicate by reversed-phase high-pressure chromatography/electrospray ionization mass spectrometry (RP-HPLC/ESI-MS) as described previously (14). One-way analysis of variances (ANOVAs) with Dunnett's multiple comparison tests were used to evaluate differences in PSM expression. Increased PSM levels were seen with tetracycline, clindamycin, erythromycin, and linezolid, and decreased levels were seen especially with oxacillin (Fig. 1A). In strain Sanger 252, changes in PSM levels were similar to those in strain LAC but in general less pronounced (Fig. 1C). However, in contrast to strain LAC, no significant change in PSM levels by oxacillin were observed, although oxacillin was applied at much higher concentrations for strain Sanger 252 (Table 1). Furthermore, there was decreased PSM production with mupirocin, which was not observed in strain LAC.
FIG. 1.
Effects of antibiotics at subinhibitory concentrations on the production of PSMs. Different antibiotics, at the concentrations shown in Table 1, were added to cultures of S. aureus strains at the time of inoculation (1:100 from precultures) into 5-ml tubes filled with 1 ml of tryptic soy broth. Tubes were shaken for 24 h at 200 rpm, and PSM concentrations were determined in the culture filtrates using RP-HPLC/ESI-MS (14). Growth under the same conditions was monitored by determining the optical density at 600 nm (OD600). PSM concentrations in S. aureus LAC (USA300) (A), growth curves for S. aureus LAC (USA300) (B), PSM concentrations in S. aureus Sanger 252 (C), growth curves for S. aureus Sanger 252 (D). The parts of bars that are striped show the amount of N-deformylated (df) PSM among total PSM peptide concentration. Concentrations of PSMα1 are shown as an example of PSMα peptides. Statistical analysis is shown for the effects and antibiotics described in the text (Er, Te, Cl, Lin for both strains, Ox for LAC, and Mu for Sanger 252); **, P < 0.01; ***, P < 0.001; con, control (without antibiotic). Error bars represent means ± standard deviations.
TABLE 1.
Subinhibitory concentrations of antibiotics used in this studya
| Antibiotic | Abbreviation | LAC (USA300) |
Sanger 252 |
||
|---|---|---|---|---|---|
| MIC (μg/ml) | SIC (μg/ml) | MIC (μg/ml) | SIC (μg/ml) | ||
| Oxacillin | Ox | >16 | 0.4 | >16 | 32 |
| Mupirocin | Mu | 0.12-0.25 (0.5) | 0.03 | 0.25 (0.5) | 0.06 |
| Trimethoprim | T/S | 0.25 | 0.1 | 0.25 | 0.2 |
| Sulfamethoxazole | 4.75 | 2 | 4.75 | 4 | |
| Erythromycin | Er | >8 | 0.8 | >8 | 16 |
| Tetracycline | Te | 1 (1) | 0.08 | 1 (1) | 0.05 |
| Vancomycin | Va | 1 | 2 | 0.5 | 1 |
| Clindamycin | CL | 0.12 | 0.08 | >4 | 0.3 |
| Rifampin | Ri | 0.004-0.016 (0.0625) | 0.006 | 0.004-256 (0.03125) | 0.006 |
| Linezolid | Lin | 2 | 0.2 | 1 | 0.2 |
| Ciprofloxacin | Cip | 2 | 3.2 | >16 | 3.2 |
All MICs are from the NARSA website (www.narsa.net), except those for mupirocin, tetracycline, and rifampin, for which general MICs for S. aureus were obtained from the literature (1-3) and MICs were determined for the Sanger 252 and LAC strains (shown in parentheses). SIC, subinhibitory concentration used.
PSM peptides of S. aureus include the PSMα peptides (PSMα1, PSMα2, PSMα3, PSMα4), which are encoded in the psmα operon, the PSMβ peptides (PSMβ1, PSMβ2), which are encoded in the psmβ operon, and the δ-toxin, which is encoded within the region coding for RNAIII, the intracellular effector molecule of agr-mediated quorum sensing (12-14). PSMα peptides and δ-toxin are cytolytic, while PSMβ peptides lack that activity at physiological concentrations (14). In general, changes seen with antibiotics were most pronounced for PSMα1 and PSMα2. PSMα3 and PSMα4 were less affected, but the direction of changes was consistent with all PSMα peptides. Concentrations of the δ-toxin were not considerably affected, except for decreased production under the influence of oxacillin in strain LAC and mupirocin in strain Sanger 252. Concentrations of PSMβ peptides were very low and only PSMβ1 could be measured in most samples. Changes observed with PSMβ1 were overall consistent with those observed for PSMα peptides.
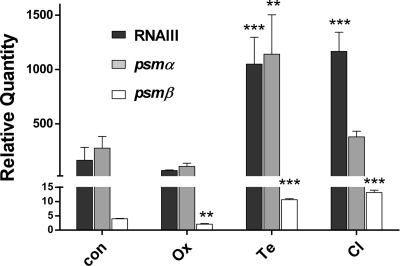
Expression of the psmα and psmβ operons is under strict and direct control of the agr quorum-sensing system (13-14). In addition, expression of the δ-toxin gene hld is directly linked to agr, as it is encoded within RNAIII (10). Overall, changes in PSM production under antibiotic influence were consistent among the different PSM peptides, suggesting that the various antibiotics may affect PSM production by an impact on agr activity. Thus, to evaluate whether agr contributes to the observed effects, we measured transcript levels of RNAIII, using those antibiotics for which the most pronounced changes in PSM levels were observed. In addition, we determined whether amounts of psmα and psmβ transcripts were affected, as can be expected in the case of an agr effect given the strong impact of agr on PSM expression (13-14). This experiment demonstrated that agr activity was significantly affected by clindamycin and tetracycline, and psmα and psmβ transcript levels changed accordingly. This suggests that an effect of agr is likely involved in the increased PSM production with those antibiotics, while the decreased production of PSMs under the influence of oxacillin did not appear to be mediated through agr (Fig. 2). In support of the former, agr activity is greater in strain LAC than in strain Sanger 252 (14), which is in accordance with the more pronounced changes in PSM production in LAC than in Sanger 252.
FIG. 2.
Activity of agr in comparison to psmα and psmβ transcript levels under the influence of subinhibitory concentrations of tetracycline, clindamycin, and oxacillin. The activity of agr was determined by real-time quantitative reverse transcription-PCR (qRT-PCR) of RNAIII in strain LAC (USA300). The expression of psm operons was determined by qRT-PCR of psmα and psmβ transcripts under the same conditions (after 8 h of growth in shaken 125-ml flasks). Primers and conditions for qRT-PCR were as described, using the gyrB transcript as a control (13-14). Experiments were performed in triplicate. Error bars represent means ± standard deviations. The antibiotic concentrations used are shown in Table 1. **, P < 0.01; ***, P < 0.001.
To our knowledge, these results demonstrate for the first time that subinhibitory concentrations of tetracycline and clindamycin have a strong stimulatory effect on the activity of agr. This represents an important finding, as agr controls a series of key virulence determinants of S. aureus, and tetracycline and clindamycin are frequently used for the treatment of MRSA infections. These infections may thus be severely exacerbated when using tetracycline or clindamycin at subinhibitory concentrations, such as when not following regimes or when dealing with strains that are resistant to those antibiotics. We are aware that our results contradict those achieved by Herbert et al., who reported that subinhibitory concentrations of clindamycin have no or a negative effect on agr expression (8). However, those authors used laboratory strains of S. aureus that may differ in many aspects from the clinical strains investigated herein.
Intriguingly, the antibiotics that caused increased activity of agr and production of PSMs work through the inhibition of protein synthesis by binding to the ribosome complex. In addition, although the treatment of a MRSA infection with oxacillin is certainly clinically wrong, the mechanism by which subinhibitory concentrations of oxacillin reduce PSM production in strain LAC is of great interest.
In conclusion, our study shows that the use of important antibiotics leads to increased expression of agr, PSMs, and possibly further virulence determinants and may thus exacerbate disease progression. This calls for rigid surveillance of regimes using these antibiotics and frequent susceptibility testing to avoid promoting virulence in bacterial strains resistant to those antibiotics.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), U.S. National Institutes of Health (NIH).
Footnotes
Published ahead of print on 16 August 2010.
REFERENCES
- 1.Bismuth, R., R. Zilhao, H. Sakamoto, J. L. Guesdon, and P. Courvalin. 1990. Gene heterogeneity for tetracycline resistance in Staphylococcus spp. Antimicrob. Agents Chemother. 34:1611-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Credito, K., G. Lin, and P. C. Appelbaum. 2007. Activity of daptomycin alone and in combination with rifampin and gentamicin against Staphylococcus aureus assessed by time-kill methodology. Antimicrob. Agents Chemother. 51:1504-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Critchley, I. A., C. L. Young, K. C. Stone, U. A. Ochsner, J. Guiles, T. Tarasow, and N. Janjic. 2005. Antibacterial activity of REP8839, a new antibiotic for topical use. Antimicrob. Agents Chemother. 49:4247-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies, J., G. B. Spiegelman, and G. Yim. 2006. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 9:445-453. [DOI] [PubMed] [Google Scholar]
- 5.DeLeo, F. R., M. Otto, B. N. Kreiswirth, and H. F. Chambers. 2010. Community-associated methicillin-resistant Staphylococcus aureus. Lancet 375:1557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diep, B. A., H. F. Chambers, C. J. Graber, J. D. Szumowski, L. G. Miller, L. L. Han, J. H. Chen, F. Lin, J. Lin, T. Haivan Phan, H. A. Carleton, L. K. McDougal, F. C. Tenover, D. E. Cohen, K. H. Mayer, G. F. Sensabaugh, and F. Perdreau-Remington. 2008. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann. Intern. Med. 148:249-257. [DOI] [PubMed] [Google Scholar]
- 7.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 8.Herbert, S., P. Barry, and R. P. Novick. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 69:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janzon, L., S. Lofdahl, and S. Arvidson. 1989. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Genet. 219:480-485. [DOI] [PubMed] [Google Scholar]
- 11.Moran, G. J., A. Krishnadasan, R. J. Gorwitz, G. E. Fosheim, L. K. McDougal, R. B. Carey, and D. A. Talan. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666-674. [DOI] [PubMed] [Google Scholar]
- 12.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Queck, S. Y., M. Jameson-Lee, A. E. Villaruz, T. H. Bach, B. A. Khan, D. E. Sturdevant, S. M. Ricklefs, M. Li, and M. Otto. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, R., K. R. Braughton, D. Kretschmer, T. H. Bach, S. Y. Queck, M. Li, A. D. Kennedy, D. W. Dorward, S. J. Klebanoff, A. Peschel, F. R. DeLeo, and M. Otto. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13:1510-1514. [DOI] [PubMed] [Google Scholar]