Abstract
Pan-drug-resistant (PDR) Acinetobacter baumannii is an important nosocomial pathogen that poses therapeutic challenges. Tigecycline alone or in combination with agents such as colestimethate, imipenem, and/or amikacin is being used clinically to treat PDR A. baumannii infections. The purpose of this study was to compare in vitro susceptibility testing by epsilometric (Etest) methods and the checkerboard (CB) method with testing by time-kill analysis. PDR A. baumannii clinical strains representing eight unique pulsed-field gel electrophoresis clones selected from a total of 32 isolates were tested in vitro with tigecycline, colestimethate, imipenem, and amikacin in single- and two-drug combinations by using two different methods of Etest (with a fixed ratio method [method 1] and with the incorporation of the active drug in medium [method 2]) and by using CB. The three-drug combination of imipenem, tigecycline, and amikacin was also tested by CB. These results were compared to time-kill results. Synergy was consistently detected with the imipenem plus colestimethate and tigecycline plus imipenem combinations. The Etest method with active drug incorporated into the agar allowed us to detect synergy even in the presence of the active drug and was more comparable to CB and time-kill tests. Synergy was detected with the three-drug combination of imipenem, tigecycline, and amikacin by both CB and time-kill methods among several tested clones. These findings indicate the utility of synergy testing to predict activity of specific antibiotic combinations against PDR A. baumannii.
In recent years Acinetobacter baumannii, an aerobic, Gram-negative coccobacillus, has emerged as an important nosocomial pathogen due to multiple drug resistance mechanisms, and it can be an extremely difficult microorganism for the clinician to treat (3, 8, 20, 22). It causes a variety of infections that include pneumonia, wound, urinary tract, bloodstream, and intra-abdominal infections (3, 8). We had experienced an increased number of cases of A. baumannii with resistant or intermediate susceptibility patterns to carbapenems over a 2-year time frame at our medical center. Growing numbers of isolates locally, nationally, and internationally have shown resistance to antibiotics such as the carbapenems, which previously had excellent activity in vitro and clinically. Our annual medical center antibiograms and a review of the literature (1, 9, 10, 12, 20, 22, 23, 25, 26) support this. Nontraditional agents, such as colestimethate (polymyxin E) and polymyxin B, despite the associated high toxicities, are being used to treat patients infected with pan-drug-resistant (PDR) A. baumannii (with pan-drug resistance defined as resistance to all routinely tested antimicrobials including carbapenems). Antibiotic resistance has also developed among some strains during treatment with these agents (10, 20). Drug treatment with newer antimicrobials or antimicrobial combinations has become increasingly important to eradicate these infections.
Tigecycline was approved by the Food and Drug Administration in June 2005 for treatment of complicated skin and skin structure infections and complicated intra-abdominal infections caused by susceptible strains of bacteria. This glycylcycline antimicrobial has shown a broad spectrum of activity both in vitro and in vivo, and its spectrum includes A. baumannii (21).
Therapy with tigecycline alone or in combination with other antimicrobials has included colestimethate, imipenem, amikacin, and ampicillin-sulbactam. In vitro susceptibility data supporting or negating these antibiotics in combination are lacking; there are limited data on whether these combinations act synergistically, additively, or antagonistically. The purpose of this study was to determine combinations of agents that reveal in vitro antimicrobial synergy by two different Etest methods (fixed ratio method [method 1] and with the incorporation of the active drug in medium [method 2]) and the broth microdilution checkerboard (CB) method and to compare these results with time-kill analysis results.
MATERIALS AND METHODS
Study population, bacterial identification, and susceptibility testing.
We identified multidrug-resistant (MDR) A. baumannii-positive cultures (with multidrug resistance defined as resistance to all antimicrobials in each of three or more classes of antibiotics) from 199 patients during a 1-year time period (June 2005 to June 2006). Institutional Review Board approval was obtained, and chart reviews were performed. All personal identifiers were removed. Patients with a clinical history showing no association with an obvious infection were excluded from the present study. Further review was performed to identify those who were infected with carbapenem-resistant PDR A. baumannii.
Isolation and susceptibility testing of A. baumannii were done as routinely performed in the clinical microbiology laboratory. Specimens were plated on 5% sheep blood agar and MacConkey agar plates and incubated overnight at 35°C for growth and identification of A. baumannii by using the MicroScan Walkaway system (Siemens Diagnostics Inc., CA). When exhibiting positive growth, the bacterial Gram-positive and Gram-negative organisms recovered were identified to species level, and susceptibility testing was performed using the MicroScan system. The MicroScan panel NUC45 was utilized for identification and susceptibility testing of A. baumannii. The antimicrobials routinely tested included ampicillin-sulbactam, cefepime, imipenem, ciprofloxacin, gentamicin, tobramycin, amikacin, and trimethoprim-sulfamethoxazole. Piperacillin-tazobactam was tested by the epsilometric test (Etest; bioMérieux, Marcy l'Etoile, France). If the organism displayed PDR, additional antibiotics such as tigecycline and colestimethate were tested by Etest following the manufacturer's instructions. Tigecycline testing implemented breakpoints for Enterobacteriaceae. Isolates were stored at −70°C in Trypticase soy broth (Difco Laboratories, Detroit, MI) supplemented with 15% glycerol.
Genotyping of isolates.
Strain typing was determined by pulsed-field gel electrophoresis (PFGE). For this, slight modifications were made to the Pulse Net protocol for molecular subtyping of Yersinia pestis (www.cdc.gov/pulsenet). DNA was digested by ApaI, and the fragments were separated in a clamped homogenous electric field (CHEF) mapper unit (Bio-Rad Laboratories, Hercules, CA) for 18.5 h using the following running parameters: 6 V/cm; initial switch, 7 s; final switch, 20 s. PFGE profiles were interpreted as described previously by Tenover et al. (24). Fingerprint images were analyzed by using Bionumerics software v. 4.61 (Applied Maths NV, Belgium) using a DICE similarity index for cluster analysis and the unweighted pair group average (UPGMA) method for tree building. All isolates with PFGE banding patterns with a similarity index of >75% were grouped within the same cluster. Banding patterns were compared with 1.5% optimization and 1.0% band position tolerance.
Synergy testing.
Antibiotics for synergy testing were selected after reviewing the medical center's antibiograms and a review of the literature on the potentially potent antibiotic combinations for PDR A. baumannii. The antibiotics included in the study were tigecycline, colestimethate, imipenem, and amikacin. Tigecycline was combined in two-drug combinations. Colestimethate was combined with each of the study drugs except amikacin in two-drug combinations. Colestimethate and amikacin were not used in combination clinically due to overlapping nephrotoxicity concerns. Synergy testing was first performed on these isolates by Etest methods 1 and 2 and by CB. These results were then compared to time-kill analysis results. Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853) strains were used as quality control strains for all MIC determinations. The MICs for imipenem, amikacin, tigecycline, and colestimethate were determined by broth microdilution and Etest methods. The cutoffs for resistance for the antibiotics tested were as follows: tigecycline, ≥8 μg/ml (intermediate, 4 μg/ml); imipenem, ≥16 μg/ml; amikacin, ≥64 μg/ml; colestimethate, ≥4 μg/ml. Determination of the MICs by Etest was performed according to the manufacturer's recommendations (Etest application sheet EAS 023 and personal communications). The Etest strip antibiotic concentration range was 0.002 to 32 μg/ml for imipenem, 0.016 to 256 μg/ml for amikacin, 0.016 to 256 μg/ml for tigecycline, and 0.016 to 256 μg/ml for colestimethate.
MIC determinations. (i) Etest method 1 (fixed ratio method).
For Etest method 1 (Etest 1), Mueller-Hinton agar plates (150-mm diameter) were inoculated with suspensions of study isolates grown to an optic density of 0.5 McFarland units. Determinations of standard MICs of drug A and drug B were performed along with the combination setup (Etest customer information sheet [CIS] 007). For combination setups, Etest strips containing the study antibiotics were added to the bacterial lawn sequentially; the first Etest strip (strip A) was incubated for 1 h at room temperature, removed, cleaned with alcohol, and saved as an MIC reading scale. The second Etest strip (strip B) was added immediately over the imprint of the first Etest strip, strip A. The plates were incubated for 18 h at 35°C. Respective MIC strips/scales were used to read MICs by placing them in each gradient's position. Four Etest strips were placed onto each Mueller-Hinton agar plate. For bactericidal drugs, i.e., imipenem, amikacin, and colestimethate, the MICs of single drugs and combinations of the drugs were read at the point of complete inhibition of all growth. The MIC was interpreted as the value at which the inhibition zone intersected the scale on the Etest strip. For tigecycline, which is bacteriostatic, the MIC was read at the point of significant inhibition of growth (80% inhibition according to Etest CIS 007) when used either singly or in combination. Etest strip results were rounded up to the nearest 2-fold dilution value for the purpose of comparison with broth microdilution MIC results.
(ii) Etest 2.
Plates containing colestimethate and those containing tigecycline with one-half the MIC and one-fourth the MIC were manufactured by Trek Diagnostics, Cleveland, OH. Imipenem and tigecycline Etest strips were tested on plates containing colestimethate. Imipenem and amikacin Etest strips were tested on plates containing tigecycline. The MICs were interpreted as described above for Etest 1.
Broth microdilution CB MICs.
Customized panels of 96-well microtiter plates containing lyophilized concentrations of the above antimicrobial drugs alone and in CB combination were manufactured by Trek Diagnostics. The MICs of the individual drugs imipenem (0.5 to 32 μg/ml), amikacin (0.5 to 32 μg/ml), tigecycline (0.015 to 16 μg/ml), and colestimethate (0.25 to 16 μg/ml) and of the combinations were determined using the broth microdilution technique as recommended by the CLSI and described in the literature (6, 7, 13, 18). Briefly, the broth microdilution plates were inoculated with each test organism to yield the appropriate density (105 CFU/ml) in 100 μl Mueller-Hinton broth (MHB) and incubated for 24 h at 35°C in ambient air. One well with no antibiotic was used as a positive growth control on each plate. Plates were read for visual turbidity, and results were recorded after 24 h of incubation at 35°C in ambient air by using a magnifying mirror reader, as turbidity in wells indicated growth of the microorganism. The MIC was determined as the well in the microtiter plate with the lowest drug concentration at which there was no visible growth. The MICs of single drugs A and B (MICA and MICB) and in combination (MICAB and MICBA) were determined after 24 h of incubation at 35°C in ambient air. MICAB was defined as the MIC of drug A in the presence of drug B; MICBA was defined as the MIC of drug B in the presence of drug A.
Synergy interpretations for Etest 1 and the broth microdilution CB method.
The fractional inhibitory concentration index (FICI) was calculated for each antibiotic in each combination (Etest application sheet EAS 021) by using the following formula: FICA + FICB = FICI, where FICA equals the MIC of drug A in combination divided by the MIC of drug A alone and FICB equals the MIC of drug B in combination divided by the MIC of drug B alone. The FICIs were interpreted as follows: synergy, FICI of ≤0.5; additivity, FICI of >0.5 to ≤1; no interaction (indifference), FICI of >1 to ≤4; antagonism, FICI of >4. Additionally, for the CB method the FICIm (minimum value of FICI in the microtiter plate) and FICIM (maximum value of FICI in the microtiter plate) were calculated.
Synergy interpretations for Etest 2.
The results of combination testing were interpreted by noting the MIC of drug A (imipenem, tigecycline, or amikacin) by Etest with and without one-fourth and one-half the MIC of drug B (tigecycline or colestimethate) in the medium. If the MIC of imipenem, tigecycline, or amikacin changed within a 1-fold dilution in the respective plates, the result was interpreted as indifference; if it was reduced by 2-fold dilutions, the result was considered additive; if it was reduced by >3-fold dilutions, the result was considered synergistic (Etest application sheet EAS 021).
E. coli (ATCC 25922) and P. aeruginosa (ATCC 27853) strains were used for quality control and were set up in conjunction with the synergy screen for both the Etest and CB methods. All tests were performed in duplicate.
Time-kill analysis.
Time-kill assays were performed only on the antibiotic combinations found to be “synergistic” or “additive” by CB and/or Etest methods. Time-kill analysis was performed according to previously published techniques (14, 17). Flasks containing MHB with one-, two-, or three-drug combinations were inoculated with a mid-log-phase aliquot of the test organism to a density of approximately 106 CFU/ml in a final volume of 100 ml and incubated in a shaking incubator at 35°C in ambient air. Aliquots were removed at times 0, 6, and 24 h postinoculation and serially diluted in sterile 0.85% sodium chloride solution for determination of viable counts. Diluted samples, in 0.05-ml aliquots, were plated in duplicate on Trypticase soy agar plates using a spiral plater. The total bacterial log10 CFU/ml was determined after 18 h of incubation at 35°C. Synergy was defined as a ≥2-log10 decrease in colony count at 6 or 24 h with the antimicrobial combination compared to the most active single agent. Indifference was defined as a <2-log10 increase or decrease in colony count at 6 or 24 h with the combination compared with the most active drug alone. Antagonism was defined as a ≥2-log10 increase in colony count at 6 or 24 h with the combination compared with that by the most active drug alone (17).
RESULTS
Patient clinical characteristics.
From June 2005 to June 2006, 199 patients with MDR A. baumannii were identified. From this group, 32 patients with bacteremia and/or symptoms and signs of respiratory illness with radiologic confirmation of pneumonia with PDR A. baumannii were selected. Clinical characteristics such as age, gender, comorbidities, residence in an extended care facility, presence of malignancy, transplant recipient status, and death within the same hospitalization or within 1 year were identified. A total of 32 isolates (20 respiratory and 12 from blood) from 32 patients were identified and included for synergy studies.
PFGE genotyping and antimicrobial susceptibility.
PFGE testing was performed to determine strain relatedness among the 32 PDR A. baumannii isolates. The PFGE patterns classified these isolates into eight distinct clonal types. Of the 32 isolates, 17 were classified as clonal type 1, 3 as clonal type 2, 4 as clonal type 3, 2 as clonal type 4, 3 as clonal type 5, and 1 each as clonal type 6, 7, or 8. A representative clone from each of the eight clonal types was included for antibiotic synergy testing. All of the eight clonal types of PDR A. baumannii exhibited resistance to amikacin (>32 μg/ml), ampicillin-sulbactam (>16/8 μg/ml), ciprofloxacin (>2 μg/ml), cefepime (>16 μg/ml), imipenem (>8 μg/ml), and piperacillin-tazobactam (>64/4 μg/ml). All clones were susceptible to colestimethate (≤2 μg/ml). Antimicrobial susceptibility testing results for tigecycline ranged from <2 to 8 μg/ml.
Synergy testing.
Synergy testing was first performed on the eight clones by Etest 1, Etest 2, and CB. The synergy results were then compared to time-kill analysis results. Etest 1 yielded indifference for all antibiotic combinations tested among all of the clones regardless of the order in which the antibiotics were placed (data not shown). When the tigecycline and colestimethate combinations were tested by Etest 2, all the PDR A. baumannii clones showed indifference at both one-fourth and one-half the MIC of colestimethate. Four clones (numbers 3, 6, 7, and 8) showed synergy at one-fourth and one-half the MIC of tigecycline when the imipenem plus tigecycline combination was tested. The rest of the clones showed either indifference or additivity at one-fourth the MIC and synergy at one-half the MIC of tigecycline when tested with this combination. With the imipenem plus colestimethate combination, seven out of eight clones showed synergy. One clone (number 4) showed additivity at one-fourth the MIC and synergy at one-half the MIC of colestimethate. For the amikacin plus tigecycline combination, all clones demonstrated indifference (Table 1).
TABLE 1.
Summary of antimicrobial synergy testing of PDR A. baumanii clones by Etest 2, checkerboard, and time-kill methods
| Clone no. | Susceptibility results with indicated test and antibiotic combinationa |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Etest 2b |
Checkerboardc |
Time-killd |
||||||||||
| TGC+CST | IPM+TGC | IPM+CST | AMK+TGC | TGC+CST | IPM+TGC | IPM+CST | AMK+TGC | IPM+TGC+AMK | IPM+TGC | IPM+CST | IPM+TGC+AMK | |
| 1 | I/I/I | I/S | S/S | I/I | I/I | A/I | A/A | I/I | A/I | NT | SND | 6 he |
| 2 | I/I | I/S | S/S | I/I | I/I | A/A | S/A | I/I | I/I | 6 he | 6, 24 h | 6, 24 h |
| 3 | I/I | S/S | S/S | I/I | I/I | S/A | A/A | I/I | I/I | 6, 24 h | SND | NT |
| 4 | I/I | I/S | A/S | I/I | I/I | A/I | A/I | A/A | S/I | 6 he | SND | 6 he |
| 5 | I/I | A/S | S/S | I/I | I/I | I/I | A/A | I/I | A/I | NT | 6, 24 h | 6, 24 h |
| 6 | I/I | S/S | S/S | I/I | I/I | A/I | A/I | I/I | I/I | NT | SND | 6 he |
| 7 | I/I | S/S | S/S | I/I | A/A | A/A | A/I | A/A | S/I | SND | SND | 6 he |
| 8 | I/I | S/S | S/S | I/I | A/A | A/A | S/A | I/I | I/I | 6, 24 h | 6 he | 6, 24 h |
TGC, tigecycline; CST, colistimethate; IPM, imipenem; AMK, amikacin; S, synergy; I, indifference; A, additiveness; SND, synergy not detected at 6 or 24 h; NT, not tested.
Etest 2 synergy testing was performed for all eight clonal types with the combinations TGC+CST, IPM+TGC, IPM+CST, and AMK+TGC. For Etest method 2, TGC and CST were added at 0.25 and 0.5 times the MIC into the agar medium. Results represent the interpretation of the change in the MIC of the drug in the E strip when added to the agar medium containing TGC or CST at 0.25 or 0.5 times the MIC compared to the MIC of the drug in the E strip in the agar medium containing no antibiotic.
Checkerboard synergy testing was performed for all eight clonal types with the indicated combinations. Results represent the interpretations of the minimum and maximum FICI values.
Time-kill analysis was performed only for the clones that showed synergy or additivity with Etest 2 and in more than two wells with CB testing. Results represent the duration (in hours) for which synergy was detected.
Synergy was not detected at 24 h.
For CB, Table 1 represents the interpretations for the minimum and maximum FICI values. For the tigecycline plus colestimethate combination, two clones (numbers 7 and 8) exhibited additivity in ≥1 clear well, while all other clones showed indifference (Table 1). With the imipenem plus tigecycline combination, 7/8 clones showed additivity or synergism in ≥1 clear well. With the imipenem plus colestimethate combination, all clones showed additivity or synergy in ≥1 clear well (Table 1). With the amikacin plus tigecycline combination, all clones demonstrated indifference and two clones (number 4 and 7) showed additivity in ≥1 clear well (Table 1). With the triple-drug combination of imipenem plus tigecycline plus amikacin, four clones (numbers 1, 4, 5, and 7) showed either synergy or additivity in ≥1 clear well. The remaining clones showed indifference to this combination (Table 1).
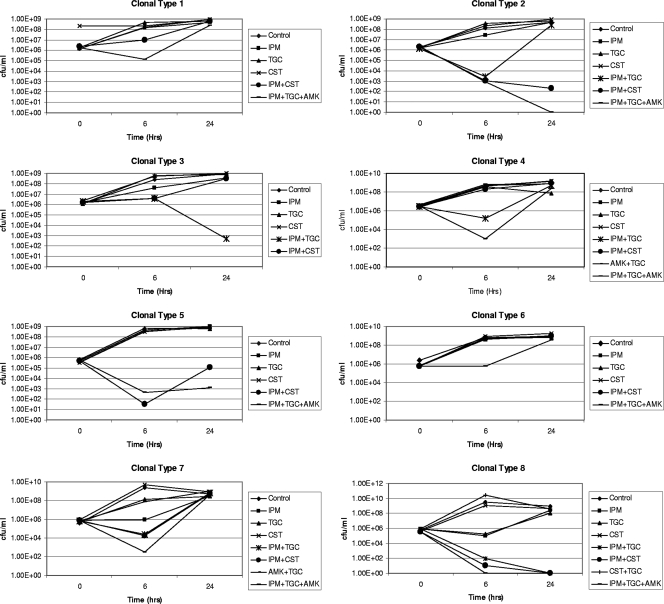
To validate the synergy detected by Etest 2 and CB, time-kill analysis was performed only for the clones that depicted synergy or additivity. Time-kill analysis showed synergy in four clones (numbers 2, 3, 4, and 8) with the imipenem plus tigecycline combination at 6 h. Synergy was detected at 6 h in three clones (numbers 2, 5, and 8) with the imipenem plus colestimethate combination. Synergy was also demonstrated with the three-drug combination of imipenem plus amikacin plus tigecycline at 6 h for seven clones tested (Table 1). Individual time-kill curves for all the clones are shown in Fig. 1.
FIG. 1.
Time-kill analyses of test isolates. Time-kill analysis was performed only for the clones that showed synergy or additiveness with Etest 2 and in more than two wells with CB testing. The log10 decreases in colony counts at 6 and 24 h with the antimicrobial combination compared to the most active single agent are represented for each clonal type. IP, imipenem; TG, tigecycline; CO, colistin; AK, amikacin.
In conclusion, the Etest 2 and CB seemed to correlate well with all the combinations tested. Among 32 opportunities for disagreement between the results of Etest 2 and CB, we only found 5 disagreements. Four clones that were indifferent by Etest 2 (tigecycline plus colestimethate [clones 7 and 8] and amikacin plus tigecycline [clones 4 and 7]) showed additivity by CB, and one clone (number 5) that showed additivity by Etest 2 to imipenem plus tigecycline was indifferent by CB (Table 1). The drug combinations imipenem plus tigecycline and imipenem plus colestimethate showed synergy or additivity with both of these tests and also demonstrated synergy by the time-kill method against some of the clones that were tested. The three-drug combination of imipenem plus tigecycline plus amikacin showed synergy at 6 h in all of the seven clones tested by time-kill analysis.
DISCUSSION
The goal of the in vitro study was to assess synergies among antibiotic combinations that are commonly used clinically for treatment of PDR A. baumannii infections and to validate synergy testing by Etest and CB with time-kill testing. We selected PDR A. baumannii isolates that caused clinical infections in our patient population to evaluate the potential of synergy testing. Eight distinct clones were selected on the basis of PFGE analysis to get a broad representation for this study. The phenotypic resistance patterns depicted the clones to be uniformly resistant to amikacin, imipenem, piperacillin-tazobactam, ampicillin-sulbactam, cefepime, and ciprofloxacin and susceptible or intermediate only to tigecycline and/or colistin. The selection of antibiotics for the synergy studies was based on our hospital antibiograms, formulary, and clinical practice in treating these infections. Antibiotic combinations of imipenem plus colestimethate, imipenem plus tigecycline, and tigecycline plus colestimethate were most commonly used for treating the PDR A. baumannii infections in our hospital (20). Antibiotics which differed in the mechanism and site of action were selected. The tigecycline plus amikacin combination was an exception, as both antibiotics have similar mechanisms of action. However, this combination was shown to be synergistic in another study (17). In addition, we also tested a three-drug combination, imipenem plus tigecycline plus amikacin, to assess whether there was an additional benefit of adding a third drug to a combination that has been shown to have in vitro synergy (17).
Different mechanisms are usually involved in the development of A. baumannii resistance to cephalosporins and carbapenems, such as altered penicillin-binding proteins, presence of various β-lactamases, and loss of porins (2, 4, 5, 11, 15, 16, 19, 27, 28, 29). PCR amplification of the genetic determinants of resistance revealed class 1 integrons in all of our clones, along with those for oxacillin β-lactamases (but not the extended-spectrum β-lactamases [blaPER and blaTEM]), which can hydrolyze carbapenems along with the acetyltransferase genes aacA4, aac(6′)-Iad, and aacC6 and the phosphotransferase gene aphA1, which all impart resistance to aminoglycosides (data not shown).
Our study demonstrated synergy of the imipenem plus tigecycline and the imipenem plus colestimethate combinations by Etest 2 and showed additivity/synergy by CB. Synergism was not observed with the tigecycline plus colestimethate combination (Table 1). In contrast to the published study by Petersen et al. (17), the amikacin plus tigecycline combination in our study did not show in vitro synergy or additivity with the majority of the PDR A. baumannii clones by Etest 2 or CB. Addition of imipenem to tigecycline or colestimethate in two-drug combinations or to amikacin plus tigecycline in a three-drug combination enhanced the synergistic activity of the antibiotic combination as confirmed by time-kill analysis (Table 1). Based on these in vitro results, imipenem appears to be a potent drug when used in combination against PDR A. baumannii strains that are imipenem resistant. The combinations with antimicrobials that have different sites of action (imipenem plus colistin and imipenem plus tigecycline) have shown synergy in at least some of the tested isolates. However, synergy was not seen with the tigecycline plus colistin combination despite the different sites of action of these two antimicrobials.
With time-kill testing, decreases in viable cell counts were observed at 6 h, but regrowth was noted at 24 h (Table 1).
Another goal of this study was to find the method with the best correlation with time-kill analysis results and that could be easily performed in our clinical microbiology laboratory. Hence, we systematically analyzed the performance of the Etest for synergy testing in comparison to the CB dilution method, since these methodologies are widely used to assess synergy between antibiotics (7, 13, 14, 17, 18, 30). Etest 1 was the least discriminatory between the two Etest methods and showed the effects of the active drug rather than the combination, resulting in indifference in all the antibiotic combinations tested for all the clones (data not shown). We do not recommend the Etest 1 method for synergy testing if the organism is susceptible to at least one of the antibiotics being tested in combination. While both the Etest 2 and CB correlated well with time-kill analysis in demonstrating synergy for two-drug combinations, there was better correlation of the Etest 2 with time-kill results. All the antibiotic combinations that showed synergy in the time-kill analysis in all tested clones also showed synergy in Etest 2 (Table 1). On the other hand, most of these clones showed additivity (no synergy) in the CB test. Etest 2 was also easier to perform, less time-consuming, and less expensive.
One limitation of our study is that some combinations, such as tigecycline plus colestimethate and tigecycline plus amikacin, that showed indifference by both Etest 2 and CB were not tested by time-kill analysis. We do not know if these combinations would have been synergistic if tested by time-kill analysis. However, the major goal of our study was to find an agreement in synergy between Etest and CB results and then to confirm the agreed synergy or additivity via time-kill analysis. Among 32 opportunities for disagreement between the results of Etest 2 and CB, there were only 5 disagreements (Table 1). None of these involved a major categoric change, for example, from synergy to antagonism.
There are challenges associated with performing synergy testing in a typical microbiology laboratory, especially since there is a lack of accepted standards for synergy testing. The testing process is laborious, time-consuming, and requires expertise in the specific procedures. The ability of in vitro combination testing to predict clinical synergy is unknown. Further clinical studies determining the relevance of these data are warranted. The clinical benefits of these antibiotic combinations in vivo can only be determined by assessing synergies through carefully designed pharmacokinetic studies and through multicenter randomized clinical trials.
Acknowledgments
We thank bioMérieux, Marcy l'Etoile, France, for providing the E-strips and Wyeth Pharmaceuticals for providing an educational grant to conduct this research.
We thank our microbiology staff, especially Michell Raczkowski for the supportive assistance provided for this project and John Gunn and Vijay Pancholi for their expert advice in the conduct of the time-kill studies.
Footnotes
Published ahead of print on 16 August 2010.
REFERENCES
- 1.Afzal-Shah, M., and D. M. Livermore. 1998. Worldwide emergence of carbapenem-resistant Acinetobacter. J. Antimicrob. Chemother. 41:576-577. [DOI] [PubMed] [Google Scholar]
- 2.Amyes, S. G. B., and H.-K. Young. 1996. Mechanism of antibiotic resistance in Acinetobacter spp.: genetics of resistance, p. 185-223. In E. Bergogne-Berezin, M. L. Joly-Guillou, and K. J. Towner (ed.), Acinetobacter: microbiology, epidemiology, infections, management. CRC Press, New York, NY.
- 3.Bergogne-Berezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bou, G., and J. Martïnez-Beltran. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, S., C. Bantar, H. K. Young, and S. G. B. Amyes. 1998. Limitation of Acinetobacter baumannii treatment by plasmid-mediated carbapenemase ARI-2. Lancet 351:186-187. [DOI] [PubMed] [Google Scholar]
- 6.CLSI. 2009. M100-S19: performance standards for antimicrobial susceptibility testing; nineteenth informational supplement, vol. 29, no. 3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Eliopoulos, G. M., and R. C. Mollering, Jr. 1996. Antimicrobial combinations, p. 330-396. In V. Lorain (ed.), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, MD.
- 8.Fournier, P. E., and H. Richet. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42:692-699. [DOI] [PubMed] [Google Scholar]
- 9.Giacometti, A., O. Cirioni, M. S. Del Prete, F. Barchiesi, A. Mataloni Paggi, E. Petrelli, and G. Scalise. 2000. Comparative activities of polycationic peptides and clinically used antimicrobial agents against multidrug-resistant nosocomial isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 46:807-810. [DOI] [PubMed] [Google Scholar]
- 10.Go, E., C. Urban, J. Burns, B. Kreiswirth, W. Eisner, N. Mariano, K. Mosinka-Snipas, and J. J. Rahal. 1994. Clinical and molecular epidemiology of Acinetobacter sensitive only to polymyxin B and sulbactam. Lancet 344:1329-1332. [DOI] [PubMed] [Google Scholar]
- 11.Hornstein, M., C. Sautjeau-Rostoker, J. Peduzzi, A. Vessieres, L. T. Hong, M. Barthelemy, M. Scavizzi, and R. Labia. 1997. Oxacillin-hydrolyzing beta-lactamase involved in resistance to imipenem in Acinetobacter baumannii. FEMS Microbiol. Lett. 153:333-339. [DOI] [PubMed] [Google Scholar]
- 12.Manikal, V. M., D. Landman, G. Saurina, E. Oydna, H. Lai, and J. Quale. 2000. Endemic carbapenem resistant Acinetobacter species in Brooklyn, New York: citywide prevalence, inter-institutional spread, and relation to antibiotic usage. Clin. Infect. Dis. 31:101-106. [DOI] [PubMed] [Google Scholar]
- 13.Moody, J. A. 1992. Synergy testing: broth microdilution checkerboard and broth macrodilution methods, p. 5.18.1-5.18.23. In H. D. Eisenberg (ed.), Clinical microbiology procedures handbook. American Society for Microbiology, Washington, DC.
- 14.National Committee for Clinical Laboratory Standards. 1999. Methods for determining bactericidal activity of antimicrobial agents. Approved guideline M26-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 15.Paton, R., R. S. Miles, J. Hood, and S. G. B. Amyes. 1993. ARI-1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 2:81-88. [DOI] [PubMed] [Google Scholar]
- 16.Perilli, M., A. Felici, A. Oratore, G. Cornaglia, G. Bonfiglio, G. M. Rossolini, and G. Amicosante. 1996. Characterization of the chromosomal cephalosporinases produced by Acinetobacter lwoffii and Acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 40:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen, P. J., P. Labthavikul, C. H. Jones, and P. A. Bradford. 2006. In vitro antibacterial activities of tigecycline in combination with other antimicrobial agents determined by chequerboard and time-kill kinetic analysis. J. Antimicrob. Chemother. 57:573-576. [DOI] [PubMed] [Google Scholar]
- 18.Rand, K. H., H. J. Houck, P. Brown, et al. 1993. Reproducibility of the microdilution checkerboard method for antibiotic synergy. Antimicrob. Agents Chemother. 37:613-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato, K., and T. Nakae. 1991. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J. Antimicrob. Chemother. 28:33-45. [DOI] [PubMed] [Google Scholar]
- 20.Schafer, J. J., D. A. Goff, K. B. Stevenson, and J. E. Mangino. 2007. Early experience with tigecycline for ventilator associated pneumonia and bacteremia caused by multidrug-resistant Acinetobacter baumannii. Pharmacotherapy 27:980-987. [DOI] [PubMed] [Google Scholar]
- 21.Slover, C. M., K. A. Rodvold, and L. H. Danziger. 2007. Tigecycline: a novel broad-spectrum antimicrobial. Ann. Pharmacother. 41:965-972. [DOI] [PubMed] [Google Scholar]
- 22.Sopirala, M. M., A. Pope-Harman, D. R. Nunley, S. Moffatt-Bruce, P. Ross, and S. I. Martin. 2008. Multidrug-resistant Acinetobacter baumannii pneumonia in lung transplant recipients. J. Heart Lung Transplant. 27:804-807. [DOI] [PubMed] [Google Scholar]
- 23.Stevenson, K. B., P. Kulich, S. York, T. Bannerman, and J. E. Mangino. 2006. Interfacility clonal transmission of multidrug-resistant Acinetobacter baumanni, presentation 309. SHEA Annu. Sci. Meet. (18 to 21 March 2006). Society for Helathcare Epidemiology of America, Arlington, VA.
- 24.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urban, C., N. Mariano, J. J. Rahal, E. Tay, C. Ponio, T. Koprivnjak, and J. Weiss. 2001. Polymyxin B-resistant Acinetobacter baumannii clinical isolates susceptible to recombinant BPI21 and cecropin P1. Antimicrob. Agents Chemother. 45:994-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urban, C., S. Segal-Maurer, and J. J. Rahal. 2003. Considerations in control and treatment of nosocomial infection due to multi-drug-resistant Acinetobacter baumannii. Clin. Infect. Dis. 36:1268-1274. [DOI] [PubMed] [Google Scholar]
- 27.Vahaboglu, H., R. Öztürk, G. Aygün, F. Coşkunkan, A. Yaman, A. Kaygusuz, H. Leblebicioglu, I. Balik, K. Aydin, and M. Otkun. 1997. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob. Agents Chemother. 41:2265-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vila, J., A. Marcos, F. Marco, S. Abdalla, Y. Vergara, R. Reig, R. Gomez-Lus, and T. Jiménez De Anta. 1993. In vitro antimicrobial production of β-lactamases, aminoglycoside-modifying enzymes, and chloramphenicol acetyltransferase by and susceptibility of clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 37:138-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vila, J., M. Navia, J. Ruiz, and C. Casals. 1997. Cloning and nucleotide sequence analysis of a gene encoding an OXA-derived β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 41:2757-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, R., D. Burgess, M. Manduru, et al. 1996. Comparison of three different in vitro methods of detecting synergy: time kill, checkerboard, and E test. Antimicrob. Agents Chemother. 40:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]