Abstract
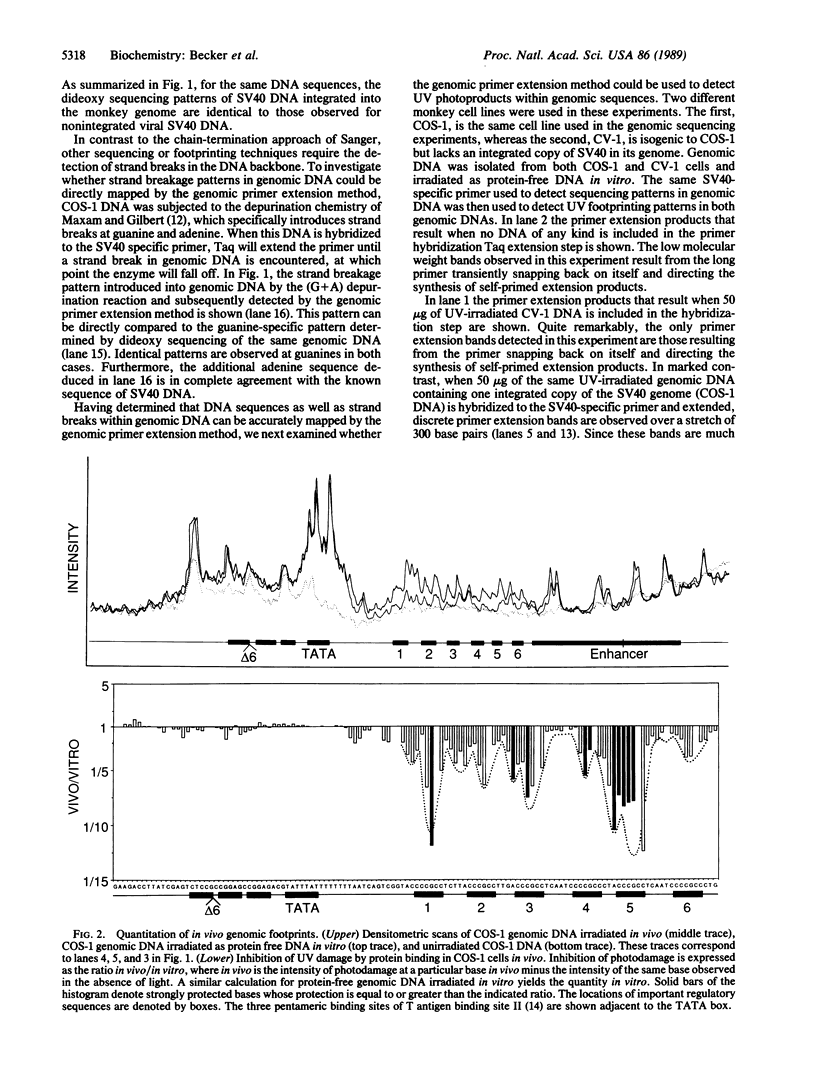
A simple and accurate genomic primer extension method has been developed to detect ultraviolet footprinting patterns of regulatory protein-DNA interactions in mammalian genomic DNA. The technique can also detect footprinting or sequencing patterns introduced into genomic DNA by other methods. Purified genomic DNA, containing either damaged bases or strand breaks introduced by footprinting or sequencing reactions, is first cut with a convenient restriction enzyme to reduce its molecular weight. A highly radioactive single-stranded DNA primer that is complementary to a region of genomic DNA whose sequence or footprint one wishes to examine is then mixed with 50 micrograms of restriction enzyme-cut genomic DNA. The primer is approximately 100 bases long and contains 85 radioactive phosphates, each of specific activity 3000 Ci/mmol (1 Ci = 37 GBq). A simple and fast method for preparing such primers is described. Following brief heat denaturation at 100 degrees C, the solution of genomic DNA and primer is cooled to 74 degrees C and a second solution containing Taq polymerase (Thermus aquaticus DNA polymerase) and the four deoxynucleotide triphosphates is added to initiate primer extension of genomic DNA. Taq polymerase extends genomic hybridized primer until its polymerization reaction is terminated either by a damaged base or strand break in genomic DNA or by the addition of dideoxynucleotide triphosphates in the polymerization reaction. The concurrent primer hybridization-extension reaction is terminated after 5 hr and unhybridized primer is digested away by mung bean nuclease. Primer-extended genomic DNA is then denatured and electrophoresed on a polyacrylamide sequencing gel, and radioactive primer extension products are revealed by autoradiography. By using this method we demonstrate that it is possible to footprint with ultraviolet light, in intact monkey cells, regulatory protein--DNA interactions along a single copy of a simian virus 40 viral genome integrated into the monkey genome.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axelrod J. D., Majors J. An improved method for photofootprinting yeast genes in vivo using Taq polymerase. Nucleic Acids Res. 1989 Jan 11;17(1):171–183. doi: 10.1093/nar/17.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Saldana H., Takahashi K., Vigneron M., Wildeman A., Davidson I., Chambon P. All six GC-motifs of the SV40 early upstream element contribute to promoter activity in vivo and in vitro. EMBO J. 1985 Dec 30;4(13B):3839–3849. doi: 10.1002/j.1460-2075.1985.tb04156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M. M., Lesser D., Kurpiewski M., Baranger A., Jen-Jacobson L. "Ultraviolet footprinting" accurately maps sequence-specific contacts and DNA kinking in the EcoRI endonuclease-DNA complex. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6247–6251. doi: 10.1073/pnas.85.17.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M. M., Wang J. C. Use of light for footprinting DNA in vivo. Nature. 1984 Jun 21;309(5970):682–687. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- Becker M. M., Wang Z. B----A transitions within a 5 S ribosomal RNA gene are highly sequence-specific. J Biol Chem. 1989 Mar 5;264(7):4163–4167. [PubMed] [Google Scholar]
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Chien A., Edgar D. B., Trela J. M. Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. J Bacteriol. 1976 Sep;127(3):1550–1557. doi: 10.1128/jb.127.3.1550-1557.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucia A. L., Lewton B. A., Tjian R., Tegtmeyer P. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J Virol. 1983 Apr;46(1):143–150. doi: 10.1128/jvi.46.1.143-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Gidoni D., Kadonaga J. T., Barrera-Saldaña H., Takahashi K., Chambon P., Tjian R. Bidirectional SV40 transcription mediated by tandem Sp1 binding interactions. Science. 1985 Nov 1;230(4725):511–517. doi: 10.1126/science.2996137. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. In vivo "photofootprint" changes at sequences between the yeast GAL1 upstream activating sequence and "TATA" element require activated GAL4 protein but not a functional TATA element. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5399–5403. doi: 10.1073/pnas.85.15.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. Photofootprinting in vivo detects transcription-dependent changes in yeast TATA boxes. Nature. 1987 Jan 8;325(7000):173–177. doi: 10.1038/325173a0. [DOI] [PubMed] [Google Scholar]
- Wang Z., Becker M. M. Selective visualization of gene structure with ultraviolet light. Proc Natl Acad Sci U S A. 1988 Feb;85(3):654–658. doi: 10.1073/pnas.85.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Matthes H., Wintzerith M., Chambon P. Transcription from the SV40 early-early and late-early overlapping promoters in the absence of DNA replication. EMBO J. 1983;2(9):1605–1611. doi: 10.1002/j.1460-2075.1983.tb01631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Zinn K., Maniatis T. Detection of factors that interact with the human beta-interferon regulatory region in vivo by DNAase I footprinting. Cell. 1986 May 23;45(4):611–618. doi: 10.1016/0092-8674(86)90293-x. [DOI] [PubMed] [Google Scholar]




