Abstract
Bacterial protein synthesis is the target for numerous natural and synthetic antibacterial agents. We have developed a poly(U) mRNA-directed aminoacylation/translation protein synthesis system composed of phenyl-tRNA synthetases, ribosomes, and ribosomal factors from Escherichia coli. This system, utilizing purified components, has been used for high-throughput screening of a small-molecule chemical library. We have identified a series of compounds that inhibit protein synthesis with 50% inhibitory concentrations (IC50s) ranging from 3 to 14 μM. This series of compounds all contained the same central scaffold composed of tetrahydropyrido[4,3-d]pyrimidin-4-ol (e.g., 4H-pyridopyrimidine). All analogs contained an ortho pyridine ring attached to the central scaffold in the 2 position and either a five- or a six-member ring tethered to the 6-methylene nitrogen atom of the central scaffold. These compounds inhibited the growth of E. coli, Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis, with MICs ranging from 0.25 to 32 μg/ml. Macromolecular synthesis (MMS) assays with E. coli and S. aureus confirmed that antibacterial activity resulted from specific inhibition of protein synthesis. Assays were developed for the steps performed by each component of the system in order to ascertain the target of the compounds, and the ribosome was found to be the site of inhibition.
Bacterial infections continue to represent a major worldwide health problem. Infections range from the relatively innocuous, such as skin rashes and common ear infections in infants, to serious and potentially lethal infections in immune-compromised patients. Resistance to antibacterial agents has increased in many pathogenic bacteria and can occur through a variety of mechanisms, such as target mutation, induction of efflux pumps, or induction of metabolic pathways leading to the degradation of the compound. Resistance developed in one cell can be transferred to other bacteria by horizontal gene transfer. The need for new antibiotics to address the increase in resistance has become critical.
Antibacterial agents interfere with cellular processes that are essential for the survival of the cell (for a complete list, see reference 4). For both naturally occurring and synthetic antibiotics, protein synthesis is a major target of antibiotic action. Bacterial protein synthesis inhibitors include the macrolides (e.g., erythromycin, clarithromycin, and azithromycin), clindamycin, chloramphenicol, the aminoglycosides (e.g., streptomycin, gentamicin, and amikacin), and the tetracyclines (2, 18, 49). The newest class of antibacterials, the synthetic oxazolidinones (exemplified by linezolid, the only novel and approved ribosomal inhibitor), also inhibit protein synthesis (21, 45). Protein synthesis is the cellular process most frequently targeted by naturally occurring antibacterials, providing compelling evolutionary evidence for the susceptibility of this process to antibiotic intervention (21).
There has been much well-deserved ado recently concerning access to the crystal structures of ribosomes, either alone or bound to a variety of antibiotics (2, 17, 35, 44). This work has led to great progress in refining the effectiveness of these classes of inhibitors via structure-based drug design (19). However, extant resistance mechanisms may also be circumvented by identifying new structural classes that bind in substantially different ways or at different sites on the ribosome. In addition, certain molecular inhibitors bind to and inhibit their targets in an induced-fit mode (12), and this has been seen with some ribosomal inhibitors (10). Since this type of interaction may not be immediately recognized in a structure-based design process, the discovery of inhibitors of function remains a useful method for novel drug discovery.
The ribosome is a well-established target for drug discovery, but other components that are essential for protein synthesis also offer attractive targets. Elongation factor Tu (EF-Tu) delivers the charged tRNA to the A-site of the ribosome in a ternary complex with GTP and an aminoacylated tRNA, hydrolyzing the GTP to GDP in the process (14, 39). Elongation factor Ts (EF-Ts) then interacts with EF-Tu to regenerate EF-Tu to an active form, facilitating the replacement of bound GDP with GTP (50). Elongation factor G (EF-G) plays a central role in the elongation phase of protein synthesis by catalyzing GTP-dependent translocation (1, 13, 40). EF-G is also one of the proteins involved in the termination of protein synthesis in a GTP-dependent fashion (47). Amino-acyl tRNA synthetases (aaRS) catalyze the attachment of amino acids to their cognate tRNAs. They are essential components in protein synthesis and individually provide attractive targets for the discovery of antibiotics (42).
Recently, attempts have been made to screen chemical-compound libraries by using cell extracts containing native transcription and translation systems from Escherichia coli (37), Streptococcus pneumoniae (9, 37), and Staphylococcus aureus (28). This approach has had only limited success. The use of cell extracts for screening can be problematic due to the presence of nucleases, degraded nucleic acids, soluble but denatured proteins, and turbidity (22). In addition, different preparations of S30 fractions can differ in activity and are therefore undependable (23). To avoid these problems, we have developed a poly(U)-directed aminoacylation/translation (A/T) protein synthesis system composed of phenyl-tRNA synthetases, ribosomes, and ribosomal factors from E. coli. Using this system as a platform for screening, we have discovered a compound series capable of inhibiting protein synthesis in vitro and in whole-cell assays. The development of the screening system and the characterization of the resulting inhibitors is described.
MATERIALS AND METHODS
Gel electrophoresis and protein analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using either 12% or 4 to 12% polyacrylamide precast gels (Novex NuPAGE; Invitrogen) with morpholinepropanesulfonic acid (MOPS) running buffer (Invitrogen). Benchmark unstained protein molecular weight markers were from Invitrogen. Gels were stained with SimplyBlue Safe stain (Invitrogen). Photography and densitometry were performed using a Kodak Image Station, model 440CF. Protein concentrations were determined by the method of Bradford (3) using bovine serum albumin as the standard.
Purification of ribosomes and proteins.
Early-phase ribosomes from E. coli strain MRE600 were prepared in the Hill laboratory at the University of Montana (Missoula) as previously described (46). Native E. coli EF-Tu was purified from cells grown to an optical density of 2.0. The cells were first lysed with a homogenizer (Niro) and then clarified by centrifugation (22,000 × g, 60 min, 4°C), and the protein was precipitated using ammonium sulfate. The protein precipitating between 35 and 73% saturated ammonium sulfate was collected by centrifugation (23,000 × g, 60 min, 4°C) and further purified using DEAE ion exchange (GE Healthcare) and Superdex-200 (GE Healthcare) size exclusion chromatography on an AKTA liquid chromatography system (GE Healthcare). The resulting protein was more than 98% homogeneous.
The gene encoding E. coli EF-G was PCR amplified from E. coli genomic DNA using forward primer 5′-CACCATGCATCATCATCATCATCATGGTGGCGCTCGTACAACACCCATCG and reverse primer 5′-CTATTATTTACCACGGGCTTCAA, which was designed to add six histidine amino acid residues to the N terminus. The PCR product was inserted into a pET101D/TOPO vector (Invitrogen), transformed into Rosetta (DE3) (Novagen) cells, and expressed as an N-terminally His tagged protein. EF-G was purified to more than 98% homogeneity using nickel-nitrilotriacetic acid (NTA) affinity chromatography (Qiagen).
The genes encoding the E. coli PheRS α and β subunits were PCR amplified from genomic DNA as a natural operon using 5′-CACCATGTCACATCTCGCAGAACTG as a forward primer and 5′-ACTAGTTCAATCCCTCAATGATGCC as a reverse primer, and the PCR product was inserted into pET101D/TOPO (Invitrogen). The resulting plasmid was transformed into BL21 Star (DE3) cells (Novagen), and the two subunits were expressed in their native forms. The cells were lysed; the lysate was clarified by centrifugation; and the protein was precipitated using ammonium sulfate (as described above). The proteins precipitating between 40 and 55% saturated ammonium sulfate were further purified using DEAE ion exchange and Superdex-200 size exclusion chromatography. PheRS was purified to more than 98% homogeneity.
Bacteria overexpressing an N-terminally His tagged form of E. coli EF-Ts were a gift from the Spremulli laboratory (University of North Carolina, Chapel Hill). EF-Ts was purified to more than 98% homogeneity using Ni-NTA affinity chromatography (Qiagen).
A/T assays.
A scintillation proximity assay (SPA) was developed for the A/T assay. The complete assay mixture contained 50 mM Tris-HCl (pH 7.5), 40 mM KCl, 10 mM MgCl2, 0.1 mM spermine, 1.5 mM ATP, 0.5 mM GTP, 25 μM [3H]phenylalanine (100 cpm/pmol), and 0.25 mg/ml poly(U). To maintain constant levels of ATP and GTP, the assay mixture contained a nucleotide regeneration system composed of 4.75 mM phosphoenolpyruvate (PEP) and 0.026 U/μl pyruvate kinase (PK). The concentrations of ribosomes and proteins in the assay were as follows: ribosome, 0.11 μM; PheRS, 0.025 μM; EF-Tu, 0.9 μM; EF-Ts, 0.03 μM; EF-G, 0.16 μM. These concentrations were arrived at through sequential rounds of optimization: each concentration is just below the saturation point of the titration.
The screening reactions were carried out in 96-well microtiter plates (Costar). Test compounds were equilibrated by the addition of 39 μl of the protein-substrate mixture (without tRNA) to 1 μl of the chemical compound (3.2 mM) dissolved in 100% dimethyl sulfoxide (DMSO). This mixture was allowed to incubate at ambient temperature for 15 min, and then reactions were initiated by the addition of 10 μl of E. coli tRNA (150 μM), followed by a 2-h incubation at room temperature (comparable to 1 h at 37°C). Reactions were stopped by the addition of 5 μl of 0.5 M EDTA. Two hundred micrograms of SPA beads (RNA binding beads [YSi]; Perkin-Elmer) in 150 μl of 300 mM citrate buffer (pH 6.2) was added. The plates were analyzed using a Packard Topcount NXT scintillation counter. Assays to determine 50% inhibitory concentrations (IC50s) were carried out as described above with the test compounds serially diluted from 200 μM to 0.39 μM. The concentration ranges of antibiotics in control plates were as follows: spiramycin, 0.07 μM to 18.0 μM; tylosin, 0.05 μM to 13.0 μM.
PheRS assay.
SPAs to determine PheRS inhibition by chemical compounds were carried out as described previously (6), with the exception that the enzyme mixture was preincubated with 0.4 to 200 μM compound for 15 min prior to the addition of tRNA. The reactions were stopped by the addition of 5 μl of 0.5 M EDTA. Four hundred micrograms of SPA beads (polyethyleneimine [PEI]-polyvinyl toluene [PVT] beads; Perkin-Elmer) in 150 μl of 300 mM citrate buffer (pH 2.0) was added, and the plates were analyzed as described above.
EF-Tu GDP exchange assay.
Nitrocellulose binding assays were used to determine inhibition of GDP exchange by EF-Tu as previously described (5, 38), with the exception that the enzyme (12.0 μM) was preincubated with 0.4 to 200 μM compound for 15 min prior to the addition of [3H]GDP. EF-Ts stimulates the exchange of GDP bound by EF-Tu. The ability of compounds to inhibit EF-Ts stimulation of GDP exchange by EF-Tu was measured in assays as described for EF-Tu/GDP exchange, with the exceptions that EF-Ts was present (0.0075 μM), the concentration of EF-Tu was reduced to 0.75 μM, and the time for the reaction was decreased from 30 min to 30 s. The concentration of the compound ranged from 0.8 to 400 μM.
EF-Tu ternary-complex formation assay.
In a ternary complex, the acceptor stem and attached amino acid are protected by EF-Tu from hydrolysis by RNase A (24). Inhibition of EF-Tu in ternary-complex formation and in the protection of tRNA from hydrolysis was analyzed using filter binding assays as previously described (5). Enzyme mixtures contained 6 μM EF-Tu and 1.5 mM [3H]Phe-tRNAPhe and were preincubated with 0.8 to 400 μM compound for 15 min at 37°C prior to digestion with RNase A.
Ternary-complex-ribosome binding assay.
Aminoacylated tRNAs are delivered to the A-site on the ribosome in a ternary complex composed of EF-Tu, GTP, and Phe-tRNAPhe. To determine ternary-complex binding to the ribosome, a mixture similar to the A/T assay mixture was used except that phenylalanine, PheRS, and ATP were removed, and deacylated tRNA was replaced by [3H]Phe-tRNAPhe. GTP was also replaced with the nonhydrolyzable analog guanosine 5′-[β,γ-imido]triphosphate (GDPNP). All components except for the ribosomes were preincubated with 0.8 to 400 μM compound at 37°C for 15 min. Ribosomes were then added (0.5 μM), and incubation was continued for an additional 15 min. Assay products were analyzed using glass microfiber filter binding (Whatman) as previously described (7).
EF-G GTPase assay.
Assay mixtures for ribosome-dependent GTP hydrolysis by EF-G contained 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 70 mM NH4Cl, 1 mM dithiothreitol (DTT), 1.8 mM [35S]GTPγS, 0.04 μM ribosomes, and 2.2 μM EF-G. Mixtures were assembled on ice, and 48 μl of the mixture was added to 2 μl of the compound, transferred to 37°C, and incubated for 30 min. The final concentration of the compound ranged from 0.8 to 400 μM. The reaction was stopped by spotting 6.0 μl onto PEI-cellulose plates (Selecto Scientific). GTP and PPi were separated by thin-layer chromatography (TLC) using 4 M urea-0.75 M KPi (pH 3.5) as a mobile phase (26). GTP, PPi, and Pi were quantified by phosphorimaging with a Storm 840 phosphorimager (Molecular Dynamics).
Eukaryotic protein synthesis assay.
Reaction mixtures contained 60% wheat germ extract (Promega), 3.0 μg poly(U), 75.0 μM [3H]phenylalanine (100 cpm/pmol), 4.0 μM yeast tRNAPhe, and 6.0 mM magnesium acetate (MgOAc). All components were assembled on ice, mixed with the compound, and incubated at ambient temperature for 2 h. Reactions were stopped by the addition of 2 ml of 5% trichloroacetic acid (TCA), and reaction products were heated to 90°C for 15 min and filtered through glass fiber filters (Whatman). The concentrations of the compound ranged from 0.3 to 300 μM, and the concentration of the control, cycloheximide, ranged from 0.3 to 300 μM.
Microbiological assays.
Broth microdilution MIC testing was performed in 96-well microtiter plates according to National Committee for Clinical Laboratory Standards (NCCLS; now CLSI) document M7-A6 (30). MICs were determined for E. coli tolC mutants, E. coli ATCC 25922, polymyxin B nonapeptide (PMBN)-treated E. coli, S. aureus, and Streptococcus pneumoniae as previously described (31). Secondary tests were carried out against Haemophilus influenzae, Enterococcus faecalis, and Moraxella catarrhalis.
Macromolecular synthesis (MMS) assays were performed in cultures of E. coli tolC mutants and in cultures of wild-type S. aureus as described previously (31). Briefly, E. coli ΔtolC5 CGSC5633 was obtained from the E. coli Genetic Stock Center (Yale University). This strain is an efflux mutant and was used to evaluate the global mode of action of the hit compounds. Assays were performed using the radiolabeled precursors [methyl-3H]thymidine, [5-3H]uridine, and l-[4,5-3H]leucine or l-[2,6-3H]phenylalanine (Amersham Biosciences Corp., Piscataway, NJ) to determine the effect of the representative compound REP323219 on the synthesis of DNA, RNA, and protein, respectively. For protein synthesis, the 10-min labeling reaction with l-[4,5-3H]leucine was followed by 5 min of chasing with 10 mM cold leucine to decrease the background due to tRNA-bound leucine. The effect of a selected compound (REP323370) on cell wall and lipid synthesis was also evaluated using radiolabeled precursors [3H]N-acetyl-d-glucosamine and [1,3-3H]glycerol, respectively.
Time-kill experiments were performed using three different bacteria according to the NCCLS guidelines (29). Bacterial strains H. influenzae ATCC 49766, S. pneumoniae ATCC 49619, and M. catarrhalis ATCC 25238 were from the American Type Culture Collection (Manassas, VA). Growth media were Haemophilus test medium (HTM) and Mueller-Hinton Broth (MHB) with or without 3% lysed horse blood from Remel (Lenexa, KS). For the experiments, 10 ml of broth medium was inoculated with 0.1 ml of a fresh overnight culture, and the mixture was grown at 35°C with shaking (200 rpm) for 2 to 3 h. Prewarmed flasks containing 10 ml of the medium alone or 10 ml of the medium with a compound at 4× MIC were then inoculated with 0.1 ml of the exponentially growing cultures. Samples were removed at 0, 2, 4, 6, and 24 h, and serial dilutions were plated on blood agar to allow for colony enumeration and the calculation of the live-cell density.
Assays for bacterial cell wall lysis were conducted with Live/Dead BacLight bacterial viability kits (Invitrogen) according to the manufacturer's instructions. Potential hemolytic activity was assessed by exposing equine erythrocytes (1% in Tris-buffered saline) to serially diluted test compounds for 10 min, followed by centrifugation and visual observation of hemolysis.
RESULTS
Development and optimization of the A/T assay for screening.
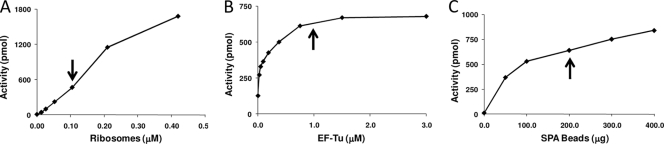
An aminoacylation/translation (A/T) system that contained the components required for the translation of poly(U) mRNA—ribosomes, EF-Tu, EF-Ts, EF-G, and PheRS—was developed. All the components of the A/T assay were purified to near-homogeneity as described in Materials and Methods. The coupled A/T reaction was adapted from separate aminoacylation and translation assays (5, 6, 11). The assay was optimized for inhibitor screening in a 96-well microtiter plate format. Ribosomes were initially titrated in the presence of saturating amounts of the other components (Fig. 1A), and 0.1 μM was chosen as the screening concentration that yielded sufficient signal over the background level. At this concentration of ribosomes, all other components of the system were then individually titrated into the system to determine the inflection point of saturation on a titration curve (see Fig. 1B for an example). Concentrations were set just below the saturation points to facilitate the maximum sensitivity to inhibition of each and every component of the system. In the initial screening assays, crude E. coli tRNA was used, but secondary-assay mixtures contained purified E. coli tRNAPhe. A scintillation proximity assay (SPA) was developed for the initial screening assay. The RNA portion of ribosomes was used to localize the ribosome to scintillation beads, enabling the detection of the nascent radiolabeled polyphenylalanine [poly(Phe)] peptide still attached to the ribosome. Yttrium silicate RNA binding beads (Perkin-Elmer) at 200 μg/well gave optimal results. Figure 1C shows the titration of SPA beads in assays to determine the amount of beads required for the capture of the maximum amounts of ribosomes allowing analysis of the activity of the system. The optimal pH for ribosome-bead binding was determined to be 6.2. In the absence of ribosomes, negligible amounts of tRNA charged with [3H]phenylalanine or free [3H]phenylalanine bound to the beads.
FIG. 1.
Optimization of the coupled aminoacylation/translation (A/T) assay. (A) Titration of E. coli ribosomes in the A/T protein synthesis system assay. Ribosomes (0.013 to 0.42 μM) were assayed in the presence of saturating amounts of the other components. The arrow indicates the concentration of ribosomes used in the screening assay. (B) Plot of poly(Phe) synthesis as a function of increasing concentrations of E. coli EF-Tu (0.023 to 3.0 μM) in the A/T system. The arrow indicates the concentration of EF-Tu used in the screening assay. (C) Determination of the amount of SPA beads needed to quantify the signal from poly(Phe) synthesis in the A/T reactions. The arrow indicates the amount of SPA beads used in the screening reactions.
Rationale and development of positive controls.
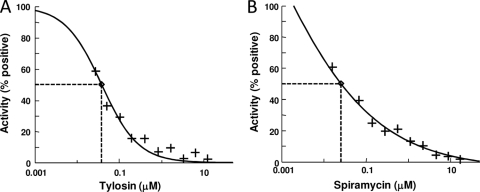
We selected the macrolides spiramycin and tylosin, each containing a 16-member lactone ring and a disaccharide at the C-5 position on the lactone ring, as control antibiotics. Both of these antibiotics inhibited the synthesis of poly(Phe), apparently by binding the 50S ribosomal subunit near the peptidyl transferase (PT) center, thereby stopping peptide synthesis (16). Spiramycin and tylosin were both effective at inhibiting poly(Phe) synthesis in the A/T assay, with IC50s of 0.022 μM and 0.038 μM, respectively (Fig. 2).
FIG. 2.
IC50 determination for positive controls in the A/T assay. Titrations of tylosin (A) and spiramycin (B) in the A/T assay are shown. The concentrations of tylosin and spiramycin ranged from 0.05 to 13.0 μM and 0.07 to 18 μM, respectively. The antibiotics were diluted into DMSO so that the concentration of DMSO was the same as that in the screening assays. Positive controls were assays in which there were no antibiotics and approximately 1,000 pmol of phenylalanine was synthesized. The curve fits and IC50s were determined by using XLfit (version 4.1; IDBS) as part of Microsoft Excel.
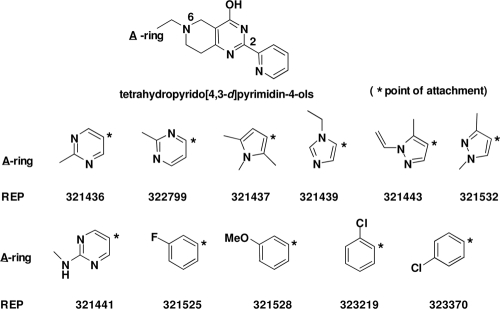
Screening of chemical compounds yields primary hits composed of compounds containing 4H-pyridopyrimidine central scaffolds.
A chemical-compound library containing 2,100 compounds from Asinex (Moscow, Russia) was tested in a high-throughput format. The screening concentration was 64 μM, and the reactions were carried out as described in Materials and Methods. Nine compounds were observed to inhibit more than 50% of poly(Phe) synthesis. Inhibition of more than 50% of poly(Phe) synthesis defines a hit compound. The ability to inhibit poly(Phe) synthesis was confirmed in triplicate assays for all nine compounds. Structural inspection of the nine compounds revealed that all had the same central scaffold, a tetrahydropyrido[4,3-d]pyrimidin-4-ol (e.g., 4H-pyridopyrimidine) (Fig. 3). In addition, all the structures contained an ortho-pyridine in position 2 of the central scaffold and held all structural variations in a heteroaromatic ring (A-ring) that connected to the N-6 atom via a methylene group. The 2,100-individual-sample library contained numerous chemotypes, of which approximately 113, or 5.4%, were of the 4H-pyridopyrimidine class. This small set of 113 4H-pyridopyrimidines contained an approximately equal distribution of regioisomers (e.g., ortho, meta, para) of the pyridyl ring connected at position 2 of the central scaffold. Other than the nine hit compounds, a subset exhibited activity inhibiting less than 50% of poly(Phe) synthesis, and another subset exhibited no inhibitory activity.
FIG. 3.
Chemical structures of screening hits. The first nine compounds are hits identified in the initial A/T screening assays. These inhibitors were identified in the A/T screening system from a library of 2,100 compounds. REP323219 and REP323370 are compounds modified from the original nine compounds and were used in macromolecular synthesis (MMS) assays.
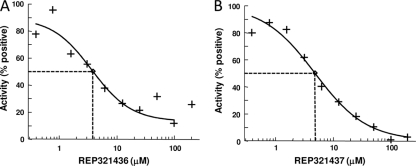
The nine compounds were serially diluted from 200 μM to 0.4 μM, and IC50s were determined. The most potent inhibitors of the A/T reaction were REP321436 and REP321437, with IC50s of 3.7 and 4.9 μM, respectively (Fig. 4). The IC50s of all the test compounds ranged from 3.7 to 14.0 μM (Table 1).
FIG. 4.
IC50 determination for protein synthesis inhibitors. The titration of representative 4H-pyridopyrimidines is shown. REP321436 (A) and REP321437 (B) exhibited IC50s of 3.7 and 4.9 μM, respectively, in A/T reactions. The reactions were carried out as described in Materials and Methods, with the concentrations of the compounds ranging from 0.4 to 400 μM. Positive controls were assays in which there were no antibiotics and approximately 1,000 pmol of phenylalanine was synthesized. The curve fits and IC50s were determined using XLfit (version 4.1; IDBS) as part of Microsoft Excel.
TABLE 1.
Inhibitory potencies of 4H-pyridopyrimidines against bacterial protein synthesis
| Compound | IC50 (μM) |
|---|---|
| REP321436 | 3.70 |
| REP321437 | 4.90 |
| REP321439 | 4.60 |
| REP321141 | 9.12 |
| REP321443 | 9.48 |
| REP321525 | 6.28 |
| REP321528 | 6.30 |
| REP321532 | 13.4 |
| REP321799 | 14.0 |
Determination of the target of 4H-pyridopyrimidines.
In our system, poly(Phe) synthesis is inhibited by the 4H-pyridopyrimidines. Other than the ribosome, four accessory proteins are required for poly(Phe) synthesis. We used specialty assays to rule out the possibility that the function (or functions) of one of the accessory proteins was inhibited (see below). The 4H-pyridopyrimidines were not observed to inhibit any of the four accessory proteins.
First, the compounds were tested to determine if they inhibited the activity of PheRS. The assay tested the ability of PheRS to attach phenylalanine to its cognate tRNAPhe. Purified E. coli tRNAPhe was used in these assays as described in Materials and Methods. No inhibition of PheRS activity by any of the compounds was observed at compound concentrations as high as 200 μM (data not shown).
Next, the abilities of the 4H-pyridopyrimidines to inhibit the function of EF-Tu were tested. In the absence of EF-Ts and GTP, EF-Tu binds GDP, and the exchange of the bound GDP for free GDP can be monitored. This binding has historically been used to characterize the activities of EF-Tu molecules from various species (33). In these assays, 3H-labeled GDP was used to track the amount of GDP exchanged by EF-Tu and the abilities of the test compounds to inhibit this exchange. We observed no reduction in the exchange of bound and free GDP by EF-Tu in the presence of any of the test compounds at concentrations as high as 200 μM (data not shown). In the presence of EF-Ts, the turnover of GDP binding by EF-Tu is stimulated 5-fold (51). Thus, when EF-Ts is added to the EF-Tu/GDP exchange assay mixture in the presence of the test compounds, inhibition of the exchange stimulated by EF-Ts may be monitored. The concentration of EF-Tu was reduced from 12 μM to 0.75 μM, and the time for completion of the assay was reduced from 30 min to 30 s. EF-Ts was added to a concentration equal to 1% of the concentration of EF-Tu (0.0075 μM). The stimulatory activity of EF-Ts was not affected by the presence of any of the test compounds at concentrations as high as 400 μM (data not shown).
When EF-Tu, GTP, and aminoacylated tRNA are associated in a ternary complex, EF-Tu protects the acceptor stem of the tRNA and the attached amino acid from nuclease digestion (RNase A). The RNase A assay, as described in Materials and Methods, was used to monitor the ability of EF-Tu to form a ternary complex and protect aminoacylated tRNA (5). E. coli tRNAPhe was aminoacylated using [3H]Phe, yielding [3H]Phe-tRNAPhe, and the radiolabeled aminoacylated tRNA was subsequently used to test the effects of the test compounds on ternary-complex formation. The compounds were assayed to concentrations as high as 400 μM, and no effect on the ability of EF-Tu to form a ternary complex and protect the bound tRNA was observed (data not shown).
EF-Tu delivers the aminoacylated tRNA to the A-site of the ribosome in the form of a ternary complex. The binding of the cognate tRNA to the A-site stimulates the hydrolysis of GTP by EF-Tu, and the tRNA is subsequently released for sole binding at the A-site. When GTP is replaced with a nonhydrolyzable analog (GDPNP), the ternary complex binds the A-site of the ribosome, but since GTP is not hydrolyzed, the ternary complex remains bound to the ribosome, and this interaction can be monitored. In these reactions, there is a certain amount of residual binding of the ternary complex to the filters in the absence of the ribosome. Using glass microfiber filters, we were able to achieve a 7-fold discrimination between the binding of the ternary complex and that of ternary-complex-ribosome complexes. The reaction was carried out in serial dilutions of the test compounds to 400 μM. We observed no effect on ribosome binding by the ternary complex in the presence of any of the test compounds.
Assays to determine the direct interaction of EF-G with its substrate, GTP, proved to be quite challenging, since the dissociation constant for the binary complex is on the order of 10 μM (8). In filter binding assays, the complex was unstable during the washing step, resulting in high variability. Therefore, we assayed the GTPase activity of EF-G in the presence of ribosomes in order to determine the effects of the compounds on the function of EF-G. In these experiments, the release of 35S-labeled inorganic phosphate from [35S]GTPγS was monitored using thin-layer chromatography (TLC) as described in Materials and Methods. We were unable to detect any inhibition of the GTPase activity of EF-G by any of the test compounds to a concentration up to 400 μM (data not shown).
The 4H-pyridopyrimidines did not inhibit any of the accessory protein activities represented in the coupled A/T screening assay. The inhibition of poly(Phe) synthesis in the biochemical assay therefore leads us to conclude that the likely mechanism of action of this compound class is direct inhibition of the ribosome itself.
4H-pyridopyrimidines specifically inhibit bacterial protein synthesis.
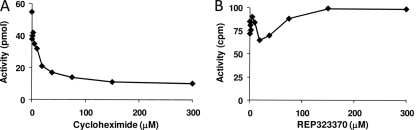
An ideal antibacterial compound would show potent inhibition of its bacterial target but little or no inhibition of the corresponding eukaryotic system. Wheat germ extract assays were used to determine whether the 4H-pyridopyrimidines inhibited protein synthesis in a eukaryotic system. Poly(U) mRNA, yeast tRNAPhe, [3H]phenylalanine, and Mg2+ concentrations were optimized for poly(Phe) synthesis in wheat germ extract assays. The 4H-pyridopyrimidines were tested along with a known inhibitor of protein synthesis, cycloheximide (41). In these assays, cycloheximide inhibited 80% of the protein synthesis at 30 μM. In contrast, none of the test compounds inhibited protein synthesis at concentrations up to 300 μM (Fig. 5). The level of poly(Phe) synthesis in the wheat germ assays is only approximately 10% of that seen in the A/T assays; this is due to the limited number of ribosomes present in the wheat germ lysate.
FIG. 5.
Inhibition of protein synthesis in wheat germ extract. Poly(U) mRNA, yeast tRNAPhe, [3H]phenylalanine, and Mg2+ concentrations were optimized for activity as described in Materials and Methods. Titrations of cycloheximide (A) and Rep323370 (B) are shown.
The compounds were negative in assays carried out to determine if they nonspecifically bind nucleic acid and thereby inhibit mRNA interactions with the ribosome. None of the compounds were shown to interact with nucleic acid nonspecifically (data not shown).
Microbiological testing of the 4H-pyridopyrimidines.
The 4H-pyridopyrimidines were tested in broth microdilution assays to determine their MICs. Despite the similarity in the biochemical potencies of the compounds, their abilities to inhibit bacterial growth differed widely. None of the compounds inhibited wild-type E. coli at compound concentrations below 128 μg/ml. The compounds showing the highest levels of activity were REP321525 and REP321528. REP321525 inhibited the growth of an E. coli tolC efflux pump mutant, E. coli treated with the permeabilizing agent PMBN, S. aureus, and S. pneumoniae at 8, 32, 32, and 8 μg/ml, respectively. REP321528 was slightly less active; it inhibited the four bacteria at 32, 128, >128, and 32 μg/ml, respectively (Table 2 ). The other 4H-pyridopyrimidines inhibited S. pneumoniae to a lesser extent.
TABLE 2.
Antibacterial activities of 4H-pyridopyrimidines against selected pathogens
| Pathogen | MIC (μg/ml)a of: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| REP321436 | REP321437 | REP321439 | REP321441 | REP321443 | REP321525 | REP321528 | REP321532 | REP322799 | |
| E. coli tolC mutant | >128 | >128 | >128 | >128 | >128 | 8 | 32 | >128 | >128 |
| E. coli ATCC 25922 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| PMBN-treated E. coli ATCC 25922 | >128 | >128 | >128 | >128 | >128 | 32 | 128 | >128 | >128 |
| S. aureus ATCC 29213 | >128 | >128 | >128 | >128 | >128 | 32 | >128 | >128 | >128 |
| S. pneumoniae ATCC 49619 | 128 | 128 | 128 | 128 | 128 | 8 | 32 | 128 | 128 |
| H. influenzae ATCC 49766 | ND | ND | ND | ND | ND | 4 | 8 | ND | ND |
| E. faecalis ATCC 29212 | ND | ND | ND | ND | ND | 32 | 128 | ND | ND |
| M. catarrhalis ATCC 25238 | ND | ND | ND | ND | ND | 0.25 | 2 | ND | ND |
ND, not determined.
REP321528 and REP321525 showed activity against three additional pathogens: H. influenzae, E. faecalis, and M. catarrhalis. The MICs of REP321525 against these three bacteria were 4, 32, and 0.25 μg/ml, respectively, while the MICs for REP321528 were 8, 128, and 2 μg/ml, respectively. REP321528 and REP321525 demonstrated broad-spectrum activity and had good to moderate activity against the major respiratory pathogens S. aureus, S. pneumoniae, H. influenzae, and M. catarrhalis. However, the growth of the Gram-negative bacterium E. coli was affected only if the bacteria were additionally compromised, either by the presence of a permeabilizing agent or by an efflux mutation, to allow the compounds access to the interiors of the cells.
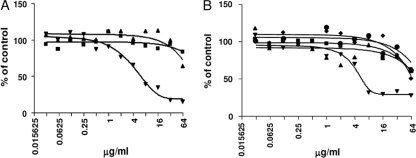
Macromolecular synthesis (MMS) assays can be used to determine the global mode of action of an inhibitor on the growth of bacteria (15, 31). MMS assays were carried out with the test compounds to determine if RNA, DNA, cell wall, lipid, or protein synthesis was inhibited in bacterial cultures. Assays were carried out in cultures containing the E. coli tolC mutant and also in cultures of S. aureus. The MMS data for two representative compounds, REP323219 and REP323370, show that the 4H-pyridopyrimidines are specific inhibitors of protein synthesis in the cell (Fig. 6). These molecules are similar to REP321525 except that the fluorine in the A-ring was replaced with chlorine and moved from the meta to the para or ortho position, respectively, relative to the attachment carbon (see Fig. 3). These compounds were tested as we had begun producing modifications of the original hits and focused on testing only the more potent compounds. REP323219 preferentially inhibited protein synthesis in E. coli tolC with an IC50 of 5.6 μg/ml. REP323370 demonstrated a similar MMS profile; measurements from the inhibition plots of the precursor incorporation assays resulted in IC50s of >64 μg/ml for the synthesis of RNA, DNA, the cell wall, and lipids, and an IC50 of 5.8 μg/ml for protein synthesis. The results did not change significantly if the cultures contained E. coli tolC mutants or S. aureus bacteria (data nor shown). This shows that the 4H-pyridopyrimidines are broad-spectrum inhibitors and preferentially inhibit protein synthesis in both Gram-negative and Gram-positive bacteria. The decreases in the synthesis of RNA, DNA, the cell wall, and lipids at high compound concentrations seen in Fig. 6 represent indirect effects of the inhibition of protein synthesis. Assays with the known protein synthesis inhibitor tylosin gave similar results (data not shown).
FIG. 6.
Macromolecular synthesis (MMS) in E. coli. Compounds REP323219 (A) and REP323370 (B) were tested in MMS assays to determine the effect of each compound on protein (▾), RNA (▪), DNA (▴), cell wall (⧫), and lipid (•) synthesis.
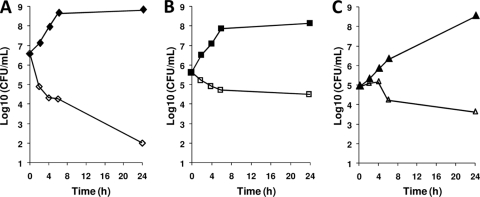
Minimum bactericidal concentration (MBC) testing of the 4H-pyridopyrimidines initially indicated that the compounds were bactericidal against H. influenzae but only bacteriostatic against S. pneumoniae. Time-kill assays with REP321525 at 4× MIC confirmed these results (Fig. 7). A >1,000-fold decrease in viable cell counts within 24 h, which is indicative of bactericidal effects, was achieved upon exposure of H. influenzae to REP321525. However, in the time-kill assays, the compound exhibited bacteriostatic effects against both S. pneumoniae and M. catarrhalis.
FIG. 7.
Time-kill kinetics of 4H-pyridopyrimidines. REP321525 was added to bacterial cultures at four times the MIC. Samples were analyzed by plating for live-cell counts at 0, 2, 4, 6, and 24 h. Bacterial cultures contained H. influenzae (A), S. pneumoniae (B), or M. catarrhalis (C). Filled symbols represent control cultures grown in the absence of inhibitor, and open symbols represent cultures containing the test compound at 4× MIC.
The 4H-pyridopyrimidine compounds were also evaluated for their potential to cause nonspecific membrane damage and/or cell lysis. Live/Dead BacLight bacterial viability kits were used to determine whether the test compounds affected the integrity of the bacterial membrane. In E. coli tolC mutants, the test compounds did not cause cell lysis at concentrations as high as 128 μg/ml. Similarly, the compounds had no adverse effects on eukaryotic cell membranes, since hemolysis was not observed upon exposure of equine erythrocytes to this class of compounds at 128 μg/ml.
DISCUSSION
Biochemical screening and target identification of bacterial protein synthesis inhibitors.
The results presented here show that a coupled aminoacylation/translation (A/T) system constructed using purified components is functional in poly(Phe) synthesis and can be used to screen for compounds that inhibit protein synthesis in bacteria in a high-throughput format. We have used this system to screen a small-molecule compound library containing 2,100 compounds, and we identified a series of 4H-pyridopyrimidine analogs that are bacterial protein synthesis inhibitors. All of the compounds in the library had molecular masses of approximately 500 Da. Through the use of a set of assays to determine the effects of the 4H-pyridopyrimidines on the activities of the ribosomal ligands, we have determined that the likely target of the inhibitors is the ribosome. The activities of EF-Tu, EF-Ts, EF-G, and PheRS showed no sign of a reduction when assayed in the presence of test compounds, whereas in A/T assay mixtures containing ribosomes, protein synthesis was reduced in a dose-dependent manner.
Using macromolecular synthesis assays, the mode of action was confirmed to be inhibition of protein synthesis. In contrast, the 4H-pyridopyrimidines had little or no effect on DNA, RNA, cell wall, or lipid production in bacteria, and the synthesis of these macromolecules was affected only indirectly due to blocking of the production of proteins essential for their synthesis. In addition, the 4H-pyridopyrimidines did not block protein synthesis in eukaryotic cells. Inhibition of protein synthesis in bacteria as a mode of action with the ribosome as the target and specificity for bacteria makes the 4H-pyridoprymidines candidates for drug development against pathogenic bacterial infections.
4H-pyridopyrimidines are active in microbiological assays.
The compounds identified in the A/T screen were subjected to microbiological assays in which they inhibited bacterial growth. Initial assays indicated that the more potent compounds had activity against S. aureus, S. pneumoniae, and permeabilized or efflux-compromised E. coli. Secondary assays confirmed the broad-spectrum activity of this compound series against both Gram-positive and Gram-negative bacteria (Table 2). In particular, the 4H-pyridopyrimidines had good activity against all the respiratory pathogens tested, including S. aureus, S. pneumoniae, H. influenzae, and M. catarrhalis. In time-kill assays, the compounds were shown to have bacteriostatic activity against S. pneumoniae and M. catarrhalis but were bactericidal against H. influenzae. In general, inhibitors of protein biosynthesis tend to exhibit a static effect on the growth of bacteria; however, some protein synthesis inhibitors, such as certain aminoglycosides, do exhibit bactericidal effects (20). The bacteriostatic/bactericidal effects of 4H-pyridopyrimidines appear to be bacterial strain specific.
Antibiotics targeting the ribosome and mechanisms of action.
Previous work indicates that chloramphenicol and certain macrolides fail to inhibit poly(Phe) synthesis in cell extracts (25, 32, 34). We initially tested azithromycin and chloramphenicol in the A/T system, and a <5% decrease in poly(Phe) synthesis was detected. More-recent evidence suggested that macrolides containing a 14-member lactone ring structure inefficiently inhibited the peptidyl transferase reaction, and certain macrolides containing a 16-member lactone ring but only a single sugar moiety at C-5 also failed to inhibit peptidyl transfer completely (36). These macrolides apparently bind in the peptide exit tunnel adjacent to the PT center, and in normal protein synthesis, they allow 6 to 8 amino acids to be synthesized before completely arresting synthesis (4, 17, 43, 48). Other macrolides, such as tylosin and spiramycin, contain 16-member lactone rings. Besides a different number of carbon constituents, the 16-member macrolides have a disaccharide at C-5, whereas the 14- and 15-member macrolides have a single sugar moiety at that position. There are subtle differences in the 16-member macrolides, but the disaccharide at C-6 appears to be constant. In X-ray crystal structures of the large ribosomal subunit bound to these 16-member macrolides, tylosin or spiramycin, the disaccharide protrudes back up the exit tunnel toward the PT center (16). The 15-member macrolide azithromycin does not protrude as far into the PT ring structure as do the 16-member macrolides (16, 43).
Although the 15-member macrolide azithromycin was assayed for inhibition of the A/T system, less than a 5% decrease in the level of poly(Phe) synthesis was observed. In contrast, both tylosin and spiramycin showed potent inhibition in the A/T assay, although a level of residual poly(Phe) synthesis was detectable. The residual activity may have been the result of allowing 2 to 4 amino acids to be polymerized, as has been previously observed for both tylosin and spiramycin (4). This may be a characteristic of the synthesis of pure poly(Phe), which is atypically hydrophobic compared to a natural protein sequence (32, 34). We have not yet determined the exact mode of action of the 4H-pyridopyrimidines on the ribosome, but the levels of residual activity observed in poly(Phe) synthesis were similar to the levels observed with tylosin and spiramycin. One possibility is that the 4H-pyridopyrimidine may be binding in or near the exit tunnel for the polypeptide or near the PT center, allowing several amino acids to be polymerized before synthesis is completely blocked. Additional structure/function studies will be needed to elucidate the mechanism of action.
Other compounds in this class have a different mode of action.
Concurrent with the work described here, in an attempt to elucidate inhibitors of bacterial growth, Miller et al. at Pfizer (Ann Arbor, MI) screened a 1.6 million-compound library against E. coli (tolC imp) in whole-cell assays (27). A series of compounds with a pyridopyrimidine scaffold was identified and was shown to inhibit the activity of bacterial biotin carboxylase. By use of macromolecular synthesis assays, the mode of action was confirmed to be inhibition of fatty acid synthesis. In contrast, the 4H-pyridopyrimidines described here had no effect on the synthesis of lipids (Fig. 6B). Conversely, the Pfizer compounds had no effect on protein synthesis. There are subtle differences in the electronic configuration of the pyridine ring structure within the central scaffold between the two series of compounds. The Pfizer compounds also lack the pyridine ring attached to the central scaffold that is present in the 4H-pyridopyrimidine series, and even though the central scaffolds of the two compound series are structurally similar, the site of action is completely different.
The A/T system as a screening platform.
Initially, nine compounds from a selected low-molecular-mass chemical-compound library were identified as inhibitory in the A/T system. The results shown here demonstrate that a reconstituted protein synthesis system composed entirely of purified components can be an effective screening system. The resulting hits can be characterized through a series of assays to identify the target component. This system can be applied to the screening of larger chemical-compound libraries and offers significant advantages over reactions carried out using crude extracts. The A/T system has the potential to be adapted to a 384-well microtiter plate format with further optimization of enzymatic and detection components, allowing higher-throughput screening. In addition, this system can be used to aid in the identification of targets of lead compounds identified in whole-cell screens with a mode of action of inhibiting protein synthesis.
The A/T system has obvious limitations in that only inhibitors of poly(Phe) synthesis will be detected. In the translation of a natural mRNA, 19 additional aminoacyl-tRNA synthetases and several additional initiation and termination factors are required. To detect inhibitors of all components of protein synthesis, a more complete system will be required.
To proceed in the development of the 4H-pyridopyrimidines as drug candidates requires in-depth development. During follow-up studies, the 4H-pyridopyrimidines were subjected to further medicinal chemistry optimization to maximize antibacterial activity, and during the course of this work, more than 100 additional analogs have been synthesized. The IC50s for these optimized compounds have been reduced in the A/T protein synthesis system to the low nanomolar range, and the MICs for H. influenzae, S. pneumoniae, M. catarrhalis, and S. aureus have been reduced to 1, 2, <0.12, and 4 μg/ml, respectively. These studies will be discussed elsewhere.
Acknowledgments
We thank Linda Spremulli of the University of North Carolina and Frank Dean of the University of Texas—Pan American for critical reading of the manuscript.
Footnotes
Published ahead of print on 9 August 2010.
REFERENCES
- 1.Arai, N., and Y. Kaziro. 1975. Mechanism of the ribosome-dependent uncoupled GTPase reaction catalyzed by polypeptide chain elongation factor G. J. Biochem. 77:439-447. [DOI] [PubMed] [Google Scholar]
- 2.Auerbach, T., A. Bashan, J. Harms, F. Schluenzen, R. Zarivach, H. Bartels, I. Agmon, M. Kessler, M. Pioletti, F. Franceschi, and A. Yonath. 2002. Antibiotics targeting ribosomes: crystallographic studies. Curr. Drug Targets Infect. Disord. 2:169-186. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bryskier, Andrè. 2005. Antimicrobial agents: antibacterials and antifungals. ASM Press, Washington, DC.
- 5.Bullard, J. M., Y. C. Cai, Y. Zhang, and L. L. Spremulli. 1999. Effects of domain exchanges between Escherichia coli and mammalian mitochondrial EF-Tu on interactions with guanine nucleotides, aminoacyl-tRNA and ribosomes. Biochim. Biophys. Acta 1446:102-114. [DOI] [PubMed] [Google Scholar]
- 6.Bullard, J. M., Y. C. Cai, B. Demeler, and L. L. Spremulli. 1999. Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J. Mol. Biol. 288:567-577. [DOI] [PubMed] [Google Scholar]
- 7.Bullard, J. M., J. C. Williams, W. K. Acker, C. Jacobi, N. Janjic, and C. S. McHenry. 2002. DNA polymerase III holoenzyme from Thermus thermophilus identification, expression, purification of components, and use to reconstitute a processive replicase. J. Biol. Chem. 277:13401-13408. [DOI] [PubMed] [Google Scholar]
- 8.Chládek, S., K. Quiggle, G. Chinali, J. Kohut III, and J. Ofengand. 1977. Synthesis and properties of nucleoside 5′-phosphoazidates derived from guanosine and adenosine nucleotides: effective on elongation factors G and Tu dependent reactions. Biochemistry 16:4312-4319. [DOI] [PubMed] [Google Scholar]
- 9.Dandliker, P. J., S. D. Pratt, A. M. Nilius, C. Black-Schaefer, X. Ruan, D. L. Towne, R. F. Clark, E. E. Englund, R. Wagner, M. Weitzberg, L. E. Chovan, R. K. Hickman, M. M. Daly, S. Kakavas, P. Zhong, Z. Cao, C. A. David, X. Xuei, C. G. Lerner, N. B. Soni, M. Bui, L. L. Shen, Y. Cai, P. J. Merta, A. Y. Saiki, and B. A. Beutel. 2003. Novel antibacterial class. Antimicrob. Agents Chemother. 47:3831-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidovich, C., A. Bashan, T. Auerbach-Nevo, R. D. Yaggie, R. R. Gontarek, and A. Yonath. 2007. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc. Natl. Acad. Sci. U. S. A. 104:4291-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberly, S. L., V. Locklear, and L. L. Spremulli. 1985. Bovine mitochondrial ribosomes. Elongation factor specificity. J. Biol. Chem. 260:8721-8725. [PubMed] [Google Scholar]
- 12.Evans, R., L. Green, X. Sun, J. Guiles, D. Lorimer, A. Burgin, N. Janjic, T. Jarvis, and D. Davies. 2007. Co-crystal structure of REP3123 bound to Clostridium difficile methionyl tRNA synthetase, poster F1-2114. Abstr. 47th Int. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 13.Gavrilova, L. P., V. E. Kotelianskii, and A. S. Spirin. 1975. Studies on the mechanism of translocation in ribosomes. IV. The role of ribosomal proteins S12 in translocation. Mol. Biol. (Mosk.) 9:609-621. [PubMed] [Google Scholar]
- 14.Gordon, J. 1968. A stepwise reaction yielding a complex between a supernatant fraction from E. coli, guanosine 5′-triphosphate, and aminoacyl-sRNA. Proc. Natl. Acad. Sci. U. S. A. 59:179-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenwood, R. C., and D. R. Gentry. 2002. Confirmation of the antibacterial mode of action of SB-219383, a novel tyrosyl tRNA synthetase inhibitor from a Micromonospora sp. J. Antibiot. (Tokyo) 55:423-426. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, J. L., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, J. L., P. B. Moore, and T. A. Steitz. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061-1075. [DOI] [PubMed] [Google Scholar]
- 18.Hermann, T. 2005. Drugs targeting the ribosome. Curr. Opin. Struct. Biol. 15:355-366. [DOI] [PubMed] [Google Scholar]
- 19.Ippolito, J. A., Z. F. Kanyo, D. Wang, F. J. Franceschi, P. B. Moore, T. A. Steitz, and E. M. Duffy. 2008. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 51:3353-3356. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, G. G., V. T. Lolans, and G. L. Daikos. 1990. The inductive role of ionic binding in the bactericidal and postexposure effects of aminoglycoside antibiotics with implications for dosing. J. Infect. Dis. 162:408-413. [DOI] [PubMed] [Google Scholar]
- 21.Knowles, D. J., N. Foloppe, N. B. Matassova, and A. I. Murchie. 2002. The bacterial ribosome, a promising focus for structure-based drug design. Curr. Opin. Pharmacol. 2:501-506. [DOI] [PubMed] [Google Scholar]
- 22.Kudlicki, W., G. Kramer, and B. Hardesty. 1992. High efficiency cell-free synthesis of proteins: refinement of the coupled transcription/translation system. Anal. Biochem. 206:389-393. [DOI] [PubMed] [Google Scholar]
- 23.Kuriki, Y. 1986. Stimulation in vitro of expression of the amp gene of pBR322 by soluble protein fractions isolated from E. coli. Biochem. Int. 12:593-602. [PubMed] [Google Scholar]
- 24.Louie, A., and F. Jurnak. 1985. Kinetic studies of Escherichia coli elongation factor Tu-guanosine 5′-triphosphate-aminoacyl-tRNA complexes. Biochemistry 24:6433-6439. [DOI] [PubMed] [Google Scholar]
- 25.Mao, J. C., and E. E. Robishaw. 1971. Effects of macrolides on peptide-bond formation and translocation. Biochemistry 10:2054-2061. [DOI] [PubMed] [Google Scholar]
- 26.Martinis, S. A., and G. E. Fox. 1997. Non-standard amino acid recognition by Escherichia coli leucyl-tRNA synthetase. Nucleic Acids Symp. Ser. 36:125-128. [PubMed] [Google Scholar]
- 27.Miller, J. R., S. Dunham, I. Mochalkin, C. Banotai, M. Bowman, S. Buist, B. Dunkle, D. Hanna, H. J. Harwood, M. D. Huband, A. Karnovsky, M. Kuhn, C. Limberakis, J. Y. Liu, S. Mehrens, W. T. Mueller, L. Narasimhan, A. Ogden, J. Ohren, J. V. Prasad, J. A. Shelly, L. Skerlos, M. Sulavik, V. H. Thomas, S. VanderRoest, L. Wang, Z. Wang, A. Whitton, T. Zhu, and C. K. Stover. 2009. A class of selective antibacterials derived from a protein kinase inhibitor pharmacophore. Proc. Natl. Acad. Sci. U. S. A. 106:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray, R. W., E. P. Melchior, J. C. Hagadorn, and K. R. Marotti. 2001. Staphylococcus aureus cell extract transcription-translation assay: firefly luciferase reporter system for evaluating protein translation inhibitors. Antimicrob. Agents Chemother. 45:1900-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 2002. Methods for determining bactericidal activity of antimicrobial agents: approved guideline M26-A. NCCLS, Wayne, PA.
- 30.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically: approved standard M7-A6. NCCLS, Wayne, PA.
- 31.Ochsner, U. A., C. L. Young, K. C. Stone, F. B. Dean, N. Janjic, and I. A. Critchley. 2005. Mode of action and biochemical characterization of REP8839, a novel inhibitor of methionyl-tRNA synthetase. Antimicrob. Agents Chemother. 49:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odom, O. W., W. D. Picking, T. Tsalkova, and B. Hardesty. 1991. The synthesis of polyphenylalanine on ribosomes to which erythromycin is bound. Eur. J. Biochem. 198:713-722. [DOI] [PubMed] [Google Scholar]
- 33.Ohta, S., M. Nakanishi, M. Tsuboi, K. Arai, and Y. Kaziro. 1977. Structural fluctuation of the polypeptide-chain elongation factor Tu. A comparison of factors from Escherichia coli and Thermus thermophilus HB8. Eur. J. Biochem. 78:599-608. [DOI] [PubMed] [Google Scholar]
- 34.Picking, W., W. D. Picking, and B. Hardesty. 1991. The use of synthetic tRNAs as probes for examining nascent peptides on Escherichia coli ribosomes. Biochimie 73:1101-1107. [DOI] [PubMed] [Google Scholar]
- 35.Poehlsgaard, J., and S. Douthwaite. 2005. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 3:870-881. [DOI] [PubMed] [Google Scholar]
- 36.Poulsen, S. M., C. Kofoed, and B. Vester. 2000. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 304:471-481. [DOI] [PubMed] [Google Scholar]
- 37.Pratt, S. D., C. A. David, C. Black-Schaefer, P. J. Dandliker, X. Xuei, U. Warrior, D. J. Burns, P. Zhong, Z. Cao, A. Y. Saiki, C. G. Lerner, L. E. Chovan, N. B. Soni, A. M. Nilius, F. L. Wagenaar, P. J. Merta, L. M. Traphagen, and B. A. Beutel. 2004. A strategy for discovery of novel broad-spectrum antibacterials using a high-throughput Streptococcus pneumoniae transcription/translation screen. J. Biomol. Screen. 9:3-11. [DOI] [PubMed] [Google Scholar]
- 38.Ravel, J. M., R. L. Shorey, S. Froehner, and W. Shive. 1968. A study of the enzymic transfer of aminoacyl-RNA to Escherichia coli ribosomes. Arch. Biochem. Biophys. 125:514-526. [DOI] [PubMed] [Google Scholar]
- 39.Ravel, J. M., R. L. Shorey, C. W. Garner, R. C. Dawkins, and W. Shive. 1969. The role of an aminoacyl-tRNA-GTP-protein complex in polypeptide synthesis. Cold Spring Harbor Symp. Quant. Biol. 34:321-330. [DOI] [PubMed] [Google Scholar]
- 40.Sander, G., R. C. Marsh, J. Voigt, and A. Parmeggiani. 1975. A comparative study of the 50S ribosomal subunit and several 50S subparticles in EF-T-and EF-G-dependent activities. Biochemistry 14:1805-1814. [DOI] [PubMed] [Google Scholar]
- 41.Satav, J. G., S. S. Katyare, P. Fatterparker, and A. Sreenivasan. 1977. Study of protein synthesis in rat liver mitochondria: use of cycloheximide. Eur. J. Biochem. 73:287-296. [DOI] [PubMed] [Google Scholar]
- 42.Schimmel, P., J. Tao, and J. Hill. 1998. Aminoacyl tRNA synthetases as targets for new anti-infectives. FASEB J. 12:1599-1609. [PubMed] [Google Scholar]
- 43.Schlünzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 44.Schmeing, T. M., and V. Ramakrishnan. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461:1234-1242. [DOI] [PubMed] [Google Scholar]
- 45.Sutcliffe, J. A. 2005. Improving on nature: antibiotics that target the ribosome. Curr. Opin. Microbiol. 8:534-542. [DOI] [PubMed] [Google Scholar]
- 46.Tam, M. F., J. A. Dodd, and W. E. Hill. 1981. Physical characteristics of 16S rRNA under reconstitution conditions. J. Biol. Chem. 256:6430-6434. [PubMed] [Google Scholar]
- 47.Tate, W. P., A. L. Beaudet, and C. T. Caskey. 1973. Influence of guanine nucleotides and elongation factors on interaction of release factors with the ribosome. Proc. Natl. Acad. Sci. U. S. A. 70:2350-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu, D., G. Blaha, P. B. Moore, and T. A. Steitz. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257-270. [DOI] [PubMed] [Google Scholar]
- 49.Yonath, A. 2005. Ribosomal crystallography: peptide bond formation, chaperone assistance and antibiotics activity. Mol. Cells. 20:1-16. [PubMed] [Google Scholar]
- 50.Zhang, Y., N. J. Yu, and L. L. Spremulli. 1998. Mutational analysis of the roles of residues in Escherichia coli elongation factor Ts in the interaction with elongation factor Tu. J. Biol. Chem. 273:4556-4562. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, Y., V. Sun, and L. L. Spremulli. 1997. Role of domains in Escherichia coli and mammalian mitochondrial elongation factor Ts in the interaction with elongation factor Tu. J. Biol. Chem. 272:21956-21963. [DOI] [PubMed] [Google Scholar]