Abstract
The antifungal flucytosine (5-fluorocytosine [5FC]) is a prodrug metabolized to its toxic form, 5-fluorouracil (5FU), only by organisms expressing cytosine deaminase. One such organism is Candida glabrata, which has emerged as the second most common agent of bloodstream and mucosal candidiasis. This emergence has been attributed to the high rate at which C. glabrata develops resistance to azole antifungals. As an oral agent, 5FC represents an attractive alternative or complement to azoles; however, the frequency of 5FC resistance mutations and the mechanisms by which these mutations confer resistance have been explored only minimally. On RPMI 1640 medium containing 1 μg/ml 5FC (32-fold above the MIC, but less than 1/10 of typical serum levels), resistant mutants occurred at a relatively low frequency (2 × 10−7). Three of six mutants characterized were 5FU cross-resistant, suggesting a mutation downstream of the Fcy1 gene (cytosine deaminase), which was confirmed by sequence analysis of the Fur1 gene (uracil phosphoribosyl transferase). The remaining three mutants had Fcy1 mutations. To ascertain the effects of 5FC resistance mutations on enzyme function, mutants were isolated in ura3 strains. Three of seven mutants harbored Fcy1 mutations and failed to grow in uridine-free, cytosine-supplemented medium, consistent with inactive Fcy1. The remainder grew in this medium and had wild-type Fcy1; further analysis revealed these to be mutated in the Fcy2L homolog of S. cerevisiae Fcy2 (purine-cytosine transporter). Based on this analysis, we characterized three 5FC-resistant clinical isolates, and mutations were identified in Fur1 and Fcy1. These data provide a framework for understanding 5FC resistance in C. glabrata and potentially in other fungal pathogens.
Candida glabrata has emerged in recent years as the second most common agent of mucosal and invasive candidiasis (16, 17, 20, 25). This emergence can be attributed largely to the intrinsically low susceptibility of C. glabrata to azole antifungals and its high capacity for acquired azole resistance. Azoles such as fluconazole, introduced in 1990, are used widely due to their low toxicity, availability in both oral and intravenous formulations, and excellent activity versus most other yeasts, including Candida albicans. With one exception, the remaining classes of antifungals are deficient in one or more of these properties; specifically, polyenes such as amphotericin B are toxic, echinocandins such as caspofungin can only be administered intravenously, and allylamines such as terbinafine lack anticandidal activity. The exception is the pyrimidine analog flucytosine (5-fluorocytosine [5FC]), which represents an attractive alternative or complement to azoles, with excellent activity against most C. glabrata isolates (MIC90 of ≤0.12 μg/ml) (21) and the capacity for both oral and intravenous administration (although the latter formulations are not currently available in the United States). 5FC is also well tolerated when moderately dosed (e.g., serum level of 25 μg/ml) (29); however, higher, potentially toxic doses are often used in attempts to counter resistance (see below) or to broaden the spectrum of activity to fungi with intrinsically low susceptibility, such as Cryptococcus spp. (MIC90 = 2 to 16 μg/ml) (22, 24).
5FC is unique among antifungals in being a prodrug and in targeting a nonessential salvage pathway (Fig. 1). Studies of susceptible fungi, primarily the genetic model Saccharomyces cerevisiae, have shown that 5FC is taken up by one or more cytosine permeases (the most relevant is encoded by FCY2) (19, 32) and modified to 5-fluorouracil (5FU) by cytosine deaminase (encoded by FCY1 [also known as FCA1 in C. albicans]) (3). Subsequent modifications to 5-fluoro-UMP by uracil phosphoribosyl transferase (UPRT; encoded by FUR1) (11, 12) and to 5-fluoro-dUMP ultimately result in the disruption of protein and DNA synthesis. The absence of cytosine deaminase in mammalian cells provides the basis for selective toxicity. 5FU, in contrast, is highly toxic, and indeed, the conversion of 5FC to 5FU by gut bacteria may be largely responsible for 5FC toxicity (29).
FIG. 1.
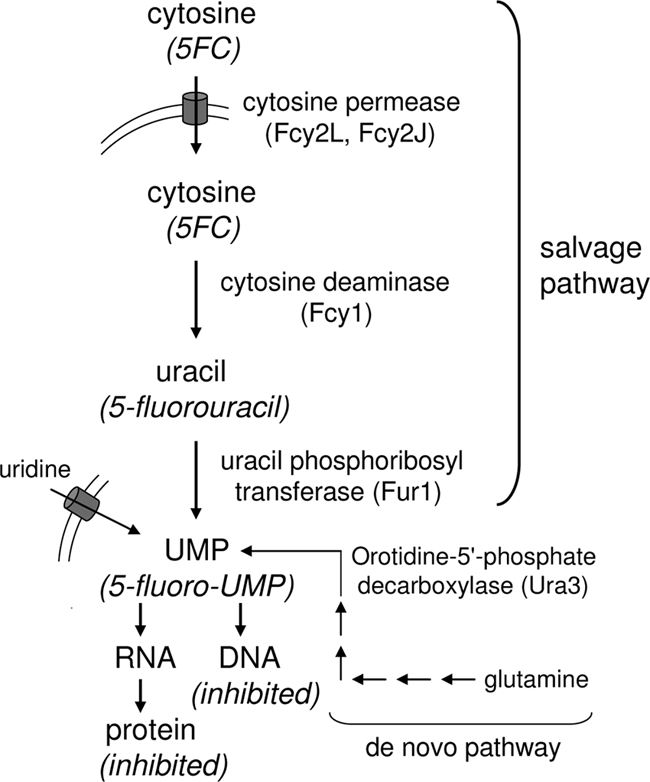
Salvage pathway for cytosine (or 5FC) uptake and conversion to UMP (or 5-fluoro-UMP) in yeast (and in the case of 5FC, the downstream consequences on RNA, DNA, and protein synthesis). Also shown (in abbreviated form) are the alternative pathways for UMP production via the de novo pathway or uridine uptake.
High rates of acquired resistance during monotherapy are considered a major limitation of 5FC therapy and are part of the rationale for the use of high, potentially toxic doses (29). However, these rates may differ significantly between the haploid species C. glabrata and diploid Candida species such as C. albicans and Candida tropicalis. In a large global survey, most isolates of these three species exhibited 5FC MICs of ≤0.25 μg/ml, and using MICs of ≥8 μg/ml as a breakpoint, only 1% of C. glabrata isolates were 5FC resistant (21). For C. albicans and C. tropicalis, the rate of resistant isolates increased, to 3 and 8%, respectively. Moreover, in a subset of C. albicans isolates (serotype B), the majority of isolates exhibit intermediate susceptibility or resistance (24). The predominant mechanism behind this was elucidated by Dodgson et al. (2) and Hope et al. (7). C. albicans isolates can be grouped genotypically into five major clades, and intermediate susceptibility and resistance strongly correlate with clade 1 (23). Dodgson et al. identified the mutation Arg101Cys (R101C) in Fur1, which is present in heterozygous form in intermediate isolates and in homozygous form in resistant isolates. The analogous mutation R99S (formerly labeled R134S based on an erroneous start site) was previously identified in an S. cerevisiae laboratory mutant (12). Thus, 5FC resistance results from a relatively high-frequency mitotic gene conversion event, as originally envisioned by Whelan (33), rather than requiring a much less frequent point mutation. Hope et al. confirmed these findings and, furthermore, identified specific mutations in Fca1 (G28D and S29L) responsible for 5FC resistance or intermediate susceptibility. The basis for 5FC resistance has also been explored in Candida dubliniensis (where it was associated with the Fca1 S29L mutation) (18), Candida lusitaniae (Fcy2 truncation or Fcy1 M9T mutation) (5), and most recently, C. glabrata (Fur1 G190D mutation, Fcy1 W148R mutation, and Fcy2 G246S mutation) (1, 28).
The mechanism by which these mutations confer resistance can be inferred from previous work with S. cerevisiae. Specifically, null mutants (including disruptants) of the genes encoding Fcy1, Fur1, and Fcy2 confer 5FC resistance (3, 9, 11, 19). This is feasible because the salvage pathway employing these enzymes is nonessential, i.e., UMP can also be synthesized from exogenously acquired uridine or de novo from l-glutamine (Fig. 1). An understanding of 5FC resistance mechanisms in pathogenic yeast would facilitate rapid, molecular technology-based detection and may also suggest novel ways to reduce or reverse resistance. Toward this goal, we present here an analysis of 5FC resistance mechanisms in C. glabrata. In laboratory mutants, resistance occurred at a moderately low frequency, and mutations were distributed between Fcy1, Fur1, and the Fcy2 paralog Fcy2L. Mutations in Fcy1, Fur1, and the Fcy2 paralog Fcy2J were subsequently identified in a small set of clinical isolates exhibiting reduced 5FC susceptibility.
MATERIALS AND METHODS
Strains, media, and drugs.
C. glabrata strains were obtained from the following sources: 66032, ATCC, Manassas, VA; BG14, B. Cormack, Johns Hopkins University; 20251.021 and 20408.055, M. Pfaller and D. Diekema, University of Iowa; and TE34-75 and TE34-78, P. Nyirjesy, Drexel University College of Medicine. Strain 66032u was previously described (31). Media included YPD (1% yeast extract, 2% peptone, 2% dextrose) and RPMI (RPMI 1640, 0.165 M morpholinepropanesulfonic acid [MOPS], pH 7.0, 2% dextrose). Where indicated, RPMI was supplemented with 10 μg/ml uridine or 30 μg/ml cytosine. Stocks of 5FC and 5FU were prepared in 50% dimethyl sulfoxide (DMSO) at 5 to 15 mg/ml and stored at −20°C; in all experiments, DMSO was diluted to ≤0.5%. Medium components and drugs were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma-Aldrich (St. Louis, MO).
Selection of 5FC-resistant mutants.
Approximately 1 × 107 cells from log-phase cultures were spread on RPMI agar containing 1 μg/ml 5FC and then incubated at 35°C for 3 to 4 days. For ura3 strains, the RPMI medium was supplemented with 10 μg/ml uridine. Colonies were streaked for isolation on drug-free YPD.
Susceptibility assays.
Broth microdilution assays with 5FC or 5FU were performed as previously described (10), substituting RPMI for YPD. Following incubation at 35°C for the indicated times, absorbance at 630 nm was read in a microplate reader. The MIC was defined as the lowest concentration inhibiting growth ≥80% relative to that of the drug-free control.
Cytosine assimilation assay.
Cultures were diluted to approximately 100 cells/μl in sterile water, and 3 μl of each was spotted onto an RPMI plate supplemented with 30 μg/ml cytosine. For negative and positive growth controls, plates were not supplemented or were supplemented with 10 μg/ml uridine. Growth was examined after 24 h at 35°C.
Sequence analysis.
A genomic DNA template was prepared and PCR amplified with gene-specific primer pairs (Table 1) as previously described (10). Following Exo-SAP-IT treatment (USB, Cleveland, OH), PCR products were sequenced (Genewiz, South Plainfield, NJ) with the indicated primers (Table 1). Sequences were compared by BLASTN and BLASTX searches of the Génolevures (www.genolevures.org) and GenBank (www.ncbi.nlm.nih.gov/GenBank) databases. All mutations were unambiguously confirmed by visual inspection of the chromatograms.
TABLE 1.
Primers used in this study
| Primer | Application | Sequence (5′-3′) |
|---|---|---|
| CgFCY1uF1 | PCR | ACTTAGTGCTGGTGGGCAGAAT |
| CgFCY1uF2 | Sequencing | TGGCACTGACTCCTGGTATTCA |
| CgFCY1dR | PCR | TCCTATCCGTATTTTCTTCGTCACT |
| CgFUR1uF1 | PCR | CCAGTCAGTATTCCACAGAAAGC |
| CgFUR1uF2 | Sequencing | ATGCACAGTTGATCCTGGAACAT |
| CgFUR1dR | PCR | ACAGTAAACGTTGGACTTAATGGAT |
| CgFCY2JuF | PCR/sequencing | GTAGAAAATAGATATAACATCCCC |
| CgFCY2JiF1 | Sequencing | CAAACTTCGCTGTCTTCCTAG |
| CgFCY2JiF2 | Sequencing | CCAAACATGTACACCATCGC |
| CgFCY2JdR | PCR/sequencing | GTATGAAATATTGATGCAAAGTGG |
| CgFCY2LuF | PCR/sequencing | GTTACGTTATCTTTATTTGTCGGGT |
| CgFCY2LiF | Sequencing | TGGGGTGTCGTTAACACTGT |
| CgFCY2LdR | PCR/sequencing | GTGTATGATTTCATTATGAAAGAGG |
RESULTS AND DISCUSSION
Laboratory mutants resistant to 5FC occur at a relatively low frequency.
The 5FC MIC for C. glabrata strain 66032 was 0.03 μg/ml in RPMI medium, which is comparable to the MICs we obtained for 7 randomly selected clinical isolates (data not shown) and those reported in a large survey (21). This MIC increased 16-fold in YPD, consistent with the presence in this medium of cytosine, which competes with 5FC for uptake. Based on these data, we selected resistant mutants on RPMI medium containing 1 μg/ml 5FC, i.e., a concentration 32-fold above the MIC. On these plates, mutants occurred at a relatively low frequency of 2 × 10−7. This is about 50-fold lower than the frequency of spontaneous fluconazole resistance but about 10-fold higher than the frequency of spontaneous echinocandin resistance in the same strains (30; our unpublished data). Six mutants were streaked on drug-free medium for isolation and were tested for 5FC susceptibility. All were fully resistant, with 5FC MICs of ≥32 μg/ml (Table 2). They were further tested for 5FU susceptibility, which should distinguish between mutations upstream (5FU susceptible) and downstream (5FU resistant) of the Fur1 gene. The 6 mutants proved to be divided equally into 5FU-susceptible and -resistant strains (Table 2).
TABLE 2.
Characteristics of 5FC-resistant C. glabrata laboratory mutants and clinical isolates
| Strain | 5FC MIC (μg/ml) | 5FU susceptibilitya | Cytosine assimilationb | Mutated protein | Mutation |
|---|---|---|---|---|---|
| 66032-based strains | |||||
| Parent | 0.03 | S | |||
| 66-FC4 | ≥32 | S | Fcy1 | A15D | |
| 66-FC5 | ≥32 | S | Fcy1 | A15D | |
| 66-FC6 | ≥32 | S | Fcy1 | G11D | |
| 66-FC1 | ≥32 | R | Fur1 | I83K | |
| 66-FC2 | ≥32 | R | Fur1 | ΔG73-V81 | |
| 66-FC3 | ≥32 | R | Fur1 | G71V | |
| 66032u (ura3)-based strains | |||||
| Parent | 0.008 | S | + | ||
| 66u-FC3 | ≥32 | S | − | Fcy1 | H59D |
| 66u-FC6 | ≥32 | S | − | Fcy1 | T92Δstop |
| 66u-FC1 | 1 | S | + | Fcy2L | W291stop |
| 66u-FC5 | 1 | S | + | Fcy2L | E342stop |
| 66u-FC10 | 1 | S | + | Fcy2L | Q396stop |
| BG14 (ura3)-based strains | |||||
| Parent | 0.03 | S | + | ||
| BG-FC4 | ≥32 | S | − | Fcy1 | E17stop |
| BG-FC8 | 1 | S | + | Fcy2L | E238-V277dup |
| Clinical isolates | |||||
| 20251.021 | ≥32 | S | Fcy1 | T84L | |
| 20408.055 | ≥32 | R | Fur1 | G210D | |
| TE34-78 | ≥32 | R | Fur1 | L136R | |
| TE34-75 | 0.25 | S | Fcy2J | I384F |
S, susceptible to 5FU (MIC < 4 μg/ml); R, resistant to 5FU (MIC ≥ 4 μg/ml).
Cytosine assimilation by ura3 strains and mutants was tested on RPMI plates supplemented with 30 μg/ml cytosine. +, growth; −, no growth.
Fur1 and Fcy1 mutations correlate with 5FU resistance and susceptibility, respectively.
As predicted, all 3 mutants demonstrating 5FC/5FU cross-resistance harbored mutations in the C. glabrata homolog of Fur1 (Génolevures accession no. CAGL0H09064g; GenBank accession no. XP_447193). These Fur1 mutations were nonconservative in all cases: I83K and D193G mutations involved replacement of charged with uncharged residues, and the ΔG73-V81 mutation involved a 9-amino-acid deletion. The remaining 3 mutants, which were not 5FU cross-resistant, harbored 2 different mutations in the homolog of Fcy1 (Génolevures accession no. CAGL0D01562g; GenBank accession no. XP_445483). Both the A15D and G11D mutations were nonconservative substitutions of charged for uncharged residues.
Fcy1 mutants of ura3 strains fail to assimilate cytosine, confirming null mutation.
The most likely effect of mutations conferring 5FC resistance is inactivation of enzyme activity, i.e., the fcy1 and fur1 mutants are null. This was genetically tested by isolating mutants in ura3 strains and then testing their ability to assimilate cytosine. The ura3 parent strains 66032u and BG14 can assimilate cytosine, that is, grow on RPMI medium lacking uridine but supplemented with cytosine, since they have a functional Fcy2-Fcy1-Fur1 pathway for conversion of cytosine to UMP. Seven 5FC-resistant mutants of these ura3 strains were isolated and characterized: all were 5FU susceptible, consistent with studies of S. cerevisiae demonstrating synthetic lethality of a fur1-ura3 mutant combination (14). Three mutants were fully 5FC resistant (MICs of ≥32 μg/ml) and failed to assimilate cytosine (Table 2). Consistent with these data, sequencing identified Fcy1 mutations in all three strains that are likely null mutations, including a premature stop codon (E17stop), a nonconservative substitution (H59D) involving a catalytically critical residue (see below), and a frameshift at codon 92 followed shortly by a stop codon (T92Δstop).
Fcy2L mutants of ura3 strains are similarly null but retain ability to assimilate cytosine.
The remaining four mutants of the ura3 strains demonstrated low-level 5FC resistance (MIC = 1 μg/ml, i.e., 32- to 128-fold above the parent MICs) and retained the ability to assimilate cytosine (Table 2). Their Fcy1 genes were sequenced: no mutations were identified, consistent with the cytosine assimilation data. This suggested a mutation in 5FC uptake. In S. cerevisiae, the purine-cytosine permease Fcy2 has been implicated in 5FC transport, although two other paralogs, Fcy21 and Fcy22, are present; the genes for all three are closely linked on a single chromosome (19, 32). C. glabrata similarly encodes three Fcy2 homologs, although on separate chromosomes (L [Génolevures accession no. CAGL0L00671g], J [Génolevures accession no. CAGL0J02948g], and C [Génolevures accession no. CAGL0C00231g]); we refer to them as Fcy2L, Fcy2J, and Fcy2C, respectively. There is no unambiguous synteny between the S. cerevisiae and C. glabrata Fcy2 genes, although BLASTP analysis suggests that the order of relatedness to S. cerevisiae Fcy2 is C. glabrata Fcy2L (76% identity), then Fcy2J (71%), then Fcy2C (61%). We therefore sequenced the Fcy2L genes of the 4 mutants (and their 2 parents) and indeed observed mutations, all of which are very likely to be null; specifically, there were three premature stop codons and a 40-residue internal duplication in the middle third of the protein (Table 2). These data suggest that 5FC uptake is partially restricted in these Fcy2L mutants, conferring low-level resistance, but that cytosine uptake through the alternative Fcy2J or Fcy2C permease remains sufficient to confer growth. This is consistent with studies indicating multiple cytosine/5FC transporters in C. glabrata (4) and S. cerevisiae (19).
5FC-resistant clinical isolates demonstrate Fcy1, Fur1, and Fcy2J mutations.
Using the laboratory mutant data described above as a framework, we characterized four independent 5FC-resistant clinical isolates. Two were obtained from a large collection of yeast isolates characterized in terms of antifungal MICs but otherwise undefined (21), and two were isolated from vaginal swabs obtained from patients who had received and failed topical 5FC treatment for C. glabrata vaginitis (P. Nyirjesy, personal communication). Three of these isolates were fully 5FC resistant, while the fourth was only marginally so (i.e., the MIC was elevated 8-fold compared to those for 66032 and other wild-type strains). Two of the fully resistant isolates were 5FU cross-resistant and, as predicted, harbored Fur1 mutations, specifically G210D and L136R mutations (Table 2). One of the two 5FU-susceptible isolates harbored the Fcy1 T84L mutation, while the second was wild type for both Fcy1 and the Fcy2L permease. We therefore sequenced its Fcy2J gene and identified the mutation I384F (Table 2). In support of its role in 5FC resistance, a second Fcy2J mutation, G246S, was recently reported for a laboratory mutant (28).
Since 5FC-susceptible parents were not available for any of these clinical isolates, it was important to consider the possibility that these mutations were simply strain-specific polymorphisms unassociated with 5FC resistance. To do this, we compared the mutant sequences to panels of sequences for at least five susceptible C. glabrata isolates; in all cases, the mutations were limited to the resistant isolates (data not shown). Furthermore, in all four cases, the mutations were nonconservative, and the mutated residue was otherwise conserved among diverse fungal species. Nevertheless, direct evidence for the role of these specific mutations in 5FC resistance is currently lacking. This could be accomplished by their introduction into susceptible strains or by reversion of the clinical isolate mutation to the wild-type sequence.
Correlation of 5FC resistance-conferring mutations with Fcy1, Fur1, and Fcy2 structures.
The crystal structure of S. cerevisiae Fcy1 has been described, and catalytically critical residues that directly interact with the cytosine substrate or zinc cofactor have been identified (8, 13). Figure 2A presents an alignment of the S. cerevisiae and C. glabrata Fcy1 sequences with critical residues labeled, along with 5FC resistance-conferring mutations reported here and previously for other yeast species. The 10 mutations are not randomly distributed; rather, 6 involve (H59D and W152R) or are adjacent to (G28D, S29L, T84L, and T92Δstop) catalytically important residues. The four remaining mutations are clustered near the amino terminus (G11 to E17). While not directly involved in catalysis, in the crystal structure they reside within the α1 helix, in close proximity to the β1 strand, containing the substrate-binding residue I27 and resistance-conferring mutations G28D and S29L. Furthermore, the α1 helix forms a “flap” at the entrance to the active site (8), and thus mutations in the G11 to E17 region may block substrate entry.
FIG. 2.
(A) Alignment of Fcy1 sequences from C. glabrata (CgFcy1) and S. cerevisiae (ScFcy1). Residues involved in substrate binding (▵) or zinc binding and catalysis (▴), based on the ScFcy1 crystal structure (8, 13), are labeled. Residues that are mutated in 5FC-resistant C. glabrata (Table 2) are underlined, and the mutations are indicated (*, premature stop codon). Also shown are previously reported 5FC resistance-conferring mutations in Fcy1 orthologs of C. albicans (7), C. dubliniensis (18), C. lusitaniae (5), and C. glabrata (28), with the mutation preceded by Ca, Cd, Cl, and Cg, respectively. (B) Alignment of C. glabrata Fur1 (CgFur1) and T. gondii UPRT (TgUPRT) sequences (for space reasons, the 26 N-terminal residues of TgUPRT are not shown). Residues within the crystal structure (26) which bind the substrate phosphoribosyl pyrophosphate (▵) or the GTP regulator (⋄) and those which function as an active site general base (▴) or uracil hood (•) are labeled. Residues that are mutated in 5FC-resistant C. glabrata (Table 2) are underlined, and the mutations are indicated. Also shown are previously reported 5FC/5FU resistance-conferring Fur1 mutations in S. cerevisiae (11, 12), C. albicans (2, 7), and C. glabrata (1), with the mutation preceded by Sc, Ca, and Cg, respectively.
With respect to Fur1, a crystal structure has been determined for protozoan Toxoplasma gondii UPRT (26), which is closely related (58% identity) to C. glabrata Fur1, despite the evolutionary distance between these organisms. An alignment of these two sequences is presented in Fig. 2B, with T. gondii residues implicated in catalysis and substrate (uracil and phosphoribosyl pyrophosphate) binding indicated, along with the 5FC/5FU resistance-conferring mutations reported here and previously for other yeast species. Of the four mutations reported here, one (I83K) resides within the longest stretch of conserved residues and is adjacent to phosphoribosyl pyrophosphate-binding residue R85. The G71V and ΔG73-V81 mutations involve or are adjacent to residues that bind GTP, a regulator of UPRT tetramerization (26). Finally, the G210D mutation is adjacent to the “hood” residues capping the uracil binding site (26).
The Fcy2L and Fcy2J permeases are integral membrane proteins. Crystal structures are not available, but for both permeases, TMHMM analysis (15) predicts a 75- to 104-residue cytoplasmic N terminus followed by 12 closely spaced transmembrane helices. All four of the Fcy2L mutations conferring 5FC resistance are likely to be null, as they represent either premature stop codons or a 40-amino-acid insertion within the middle third of the protein. In contrast, the Fcy2J mutations reported here (I384F) and elsewhere (G246S) (28) represent nonconservative but relatively subtle changes within transmembrane helices 6 and 9, respectively. Further studies are required to determine the effects of these mutations on Fcy2J function.
Conclusions.
Although its clinical use has declined in recent years, 5FC has several features that make it an attractive alternative or complement to azoles, echinocandins, or amphotericin B for C. glabrata infections. These include oral administration, lack of cross-resistance with azoles, and low toxicity (when appropriately dosed) as a consequence of its fungus-specific mechanism of activity. A remaining concern is the perception that 5FC resistance readily occurs during monotherapy. The in vitro data presented here partly allay these concerns, as the resistance frequency (2 × 10−7) was well below that observed for fluconazole, though higher than that for echinocandins. However, in contrast to echinocandin resistance, which requires specific Fks1 hot spot mutations that minimally affect enzymatic activity (6), the data presented here and previously suggest that high-level 5FC resistance is conferred by a wide array of mutations conferring null phenotypes for Fcy1 or Fur1. Furthermore, low-level resistance may be conferred by a similarly wide range of mutations in one or more Fcy2 permeases. On the other hand, it is possible that null mutations in this cytosine assimilation pathway would have deleterious effects on C. glabrata viability or virulence in vivo, effectively reducing the resistance frequency. An additional, commonly used approach to reducing 5FC resistance frequency is to combine it with a second antifungal (29). While 5FC-azole combinations would seem particularly advantageous for treating C. glabrata infection, this combination may in fact be antagonistic (27); the basis for this is currently being explored. In any case, the data presented here provide a framework for mechanistic and epidemiological studies of 5FC resistance in C. glabrata. It is hoped that this framework will address some of the uncertainties that currently limit the clinical use of this much needed antifungal.
Acknowledgments
We thank B. Cormack, D. Diekema, P. Nyirjesy, and M. Pfaller for generously providing strains.
This work was supported by NIH/NIAID grant AI073794.
Footnotes
Published ahead of print on 7 September 2010.
REFERENCES
- 1.Chapeland-Leclerc, F., C. Hennequin, N. Papon, T. Noël, A. Girard, G. Socié, P. Ribaud, and C. Lacroix. 2010. Acquisition of flucytosine, azole, and caspofungin resistance in Candida glabrata bloodstream isolates serially obtained from a hematopoietic stem cell transplant recipient. Antimicrob. Agents Chemother. 54:1360-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodgson, A. R., K. J. Dodgson, C. Pujol, M. A. Pfaller, and D. R. Soll. 2004. Clade-specific flucytosine resistance is due to a single nucleotide change in the FUR1 gene of Candida albicans. Antimicrob. Agents Chemother. 48:2223-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erbs, P., F. Exinger, and R. Jund. 1997. Characterization of the Saccharomyces cerevisiae FCY1 gene encoding cytosine deaminase and its homologue FCA1 of Candida albicans. Curr. Genet. 31:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Fasoli, M. O., and D. Kerridge. 1990. Uptake of pyrimidines and their derivatives into Candida glabrata and Candida albicans. J. Gen. Microbiol. 136:1475-1481. [DOI] [PubMed] [Google Scholar]
- 5.Florent, M., T. Noel, G. Ruprich-Robert, B. Da Silva, V. Fitton-Ouhabi, C. Chastin, N. Papon, and F. Chapeland-Leclerc. 2009. Nonsense and missense mutations in FCY2 and FCY1 genes are responsible for flucytosine resistance and flucytosine-fluconazole cross-resistance in clinical isolates of Candida lusitaniae. Antimicrob. Agents Chemother. 53:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Effron, G., S. Lee, S. Park, J. D. Cleary, and D. S. Perlin. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 53:3690-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hope, W. W., L. Tabernero, D. W. Denning, and M. J. Anderson. 2004. Molecular mechanisms of primary resistance to flucytosine in Candida albicans. Antimicrob. Agents Chemother. 48:4377-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ireton, G. C., M. E. Black, and B. L. Stoddard. 2003. The 1.14 Å crystal structure of yeast cytosine deaminase: evolution of nucleotide salvage enzymes and implications for genetic chemotherapy. Structure 11:961-972. [DOI] [PubMed] [Google Scholar]
- 9.Jund, R., and F. Lacroute. 1970. Genetic and physiological aspects of resistance to 5-fluoropyrimidines in Saccharomyces cerevisiae. J. Bacteriol. 102:607-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katiyar, S. K., and T. D. Edlind. 2009. Role for Fks1 in the intrinsic echinocandin resistance of Fusarium solani as evidenced by hybrid expression in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 53:1772-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kern, L., J. de Montigny, R. Jund, and F. Lacroute. 1990. The FUR1 gene of Saccharomyces cerevisiae: cloning, structure and expression of wild-type and mutant alleles. Gene 88:149-157. [DOI] [PubMed] [Google Scholar]
- 12.Kern, L., J. de Montigny, F. Lacroute, and R. Jund. 1991. Regulation of the pyrimidine salvage pathway by the FUR1 gene product of Saccharomyces cerevisiae. Curr. Genet. 19:333-337. [DOI] [PubMed] [Google Scholar]
- 13.Ko, T. P., J. J. Lin, C. Y. Hu, Y. H. Hsu, A. H. Wang, and S. H. Liaw. 2003. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J. Biol. Chem. 278:19111-19117. [DOI] [PubMed] [Google Scholar]
- 14.Koren, A., S. Ben-Aroya, R. Steinlauf, and M. Kupiec. 2003. Pitfalls of the synthetic lethality screen in Saccharomyces cerevisiae: an improved design. Curr. Genet. 43:62-69. [DOI] [PubMed] [Google Scholar]
- 15.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 16.Lewis, R. E. 2009. Overview of the changing epidemiology of candidemia. Curr. Med. Res. Opin. 25:1732-1740. [DOI] [PubMed] [Google Scholar]
- 17.Li, L., S. Redding, and A. Dongari-Bagtzoglou. 2007. Candida glabrata: an emerging oral opportunistic pathogen. J. Dent. Res. 86:204-215. [DOI] [PubMed] [Google Scholar]
- 18.McManus, B. A., G. P. Moran, J. A. Higgins, D. J. Sullivan, and D. C. Coleman. 2009. A Ser29Leu substitution in the cytosine deaminase Fca1p is responsible for clade-specific flucytosine resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 53:4678-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paluszynski, J. P., R. Klassen, M. Rohe, and F. Meinhardt. 2006. Various cytosine/adenine permease homologues are involved in the toxicity of 5-fluorocytosine in Saccharomyces cerevisiae. Yeast 23:707-715. [DOI] [PubMed] [Google Scholar]
- 20.Pfaller, M. A., and D. J. Diekema. 2007. The epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaller, M. A., S. A. Messer, L. Boyken, H. Huynh, R. J. Hollis, and D. J. Diekema. 2002. In vitro activities of 5-fluorocytosine against 8,803 clinical isolates of Candida spp.: global assessment of primary resistance using National Committee for Clinical Laboratory Standards susceptibility testing methods. Antimicrob. Agents Chemother. 46:3518-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, G. V. Doern, and D. J. Diekema. 2005. Global trends in the antifungal susceptibility of Cryptococcus neoformans (1990 to 2004). J. Clin. Microbiol. 43:2163-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pujol, C., M. A. Pfaller, and D. R. Soll. 2004. Flucytosine resistance is restricted to a single genetic clade of Candida albicans. Antimicrob. Agents Chemother. 48:262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quindós, G., M. T. Ruesga, E. Martín-Mazuelos, R. Salesa, R. Alonso-Vargas, A. J. Carrillo-Muñoz, S. Brena, R. San Millán, and J. Pontón. 2004. In-vitro activity of 5-fluorocytosine against 1,021 Spanish clinical isolates of Candida and other medically important yeasts. Rev. Iberoam. Micol. 21:63-69. [PubMed] [Google Scholar]
- 25.Richter, S. S., R. P. Galask, S. A. Messer, R. J. Hollis, D. J. Diekema, and M. A. Pfaller. 2005. Antifungal susceptibilities of Candida species causing vulvovaginitis and epidemiology of recurrent cases. J. Clin. Microbiol. 30:2155-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher, M. A., C. J. Bashor, M. H. Song, K. Otsu, S. Zhu, R. J. Parry, B. Ullman, and R. G. Brennan. 2002. The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase. Proc. Natl. Acad. Sci. U. S. A. 99:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siau, H., and D. Kerridge. 1999. 5-Fluorocytosine antagonizes the action of sterol biosynthesis inhibitors in Candida glabrata. J. Antimicrob. Chemother. 43:767-775. [DOI] [PubMed] [Google Scholar]
- 28.Vandeputte, P., L. Pineau, G. Larcher, T. Noel, D. Brethes, D. Chabasse, and J.-P. Bouchara. 9 July 2010, posting date. Molecular mechanisms of resistance to 5-fluorocytosine in laboratory mutants of Candida glabrata. Mycopathologia. [Epub ahead of print.] doi: 10.1007/s11046-010-9342-1. [DOI] [PubMed]
- 29.Vermes, A., H. J. Guchelaar, and J. Dankert. 2000. Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 46:171-179. [DOI] [PubMed] [Google Scholar]
- 30.Vermitsky, J.-P., and T. D. Edlind. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 48:3773-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vermitsky, J.-P., K. D. Earhart, W. L. Smith, R. Homayouni, T. D. Edlind, and P. D. Rogers. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol. Microbiol. 61:704-722. [DOI] [PubMed] [Google Scholar]
- 32.Weber, E., C. Rodriguez, M. R. Chevallier, and R. Jund. 1990. The purine-cytosine permease gene of Saccharomyces cerevisiae: primary structure and deduced protein sequence of the FCY2 gene product. Mol. Microbiol. 4:585-596. [DOI] [PubMed] [Google Scholar]
- 33.Whelan, W. L. 1987. The genetic basis of resistance to 5-fluorocytosine in Candida species and Cryptococcus neoformans. Crit. Rev. Microbiol. 15:45-56. [DOI] [PubMed] [Google Scholar]



